
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PTK2-MKLN1 (FusionGDB2 ID:70042) |
Fusion Gene Summary for PTK2-MKLN1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PTK2-MKLN1 | Fusion gene ID: 70042 | Hgene | Tgene | Gene symbol | PTK2 | MKLN1 | Gene ID | 5747 | 4289 |
| Gene name | protein tyrosine kinase 2 | muskelin 1 | |
| Synonyms | FADK|FAK|FAK1|FRNK|PPP1R71|p125FAK|pp125FAK | TWA2 | |
| Cytomap | 8q24.3 | 7q32.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | focal adhesion kinase 1FADK 1FAK-related non-kinase polypeptidePTK2 protein tyrosine kinase 2focal adhesion kinase isoform FAK-Del33focal adhesion kinase-related nonkinaseprotein phosphatase 1 regulatory subunit 71 | muskelinmuskelin 1, intracellular mediator containing kelch motifs | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | . | Q9UL63 | |
| Ensembl transtripts involved in fusion gene | ENST00000340930, ENST00000395218, ENST00000517887, ENST00000519419, ENST00000519465, ENST00000521059, ENST00000522684, ENST00000535192, ENST00000538769, ENST00000430260, ENST00000517712, ENST00000519635, ENST00000519881, ENST00000520151, ENST00000520892, ENST00000522950, | ENST00000352689, ENST00000429546, ENST00000498778, ENST00000421797, | |
| Fusion gene scores | * DoF score | 39 X 32 X 15=18720 | 17 X 15 X 7=1785 |
| # samples | 57 | 18 | |
| ** MAII score | log2(57/18720*10)=-5.03747470541866 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(18/1785*10)=-3.30985526258679 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PTK2 [Title/Abstract] AND MKLN1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PTK2(141745350)-MKLN1(131147976), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | PTK2-MKLN1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PTK2-MKLN1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. PTK2-MKLN1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PTK2 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 24036928 |
| Hgene | PTK2 | GO:0007229 | integrin-mediated signaling pathway | 24036928 |
| Hgene | PTK2 | GO:0010763 | positive regulation of fibroblast migration | 26763945 |
| Hgene | PTK2 | GO:0018108 | peptidyl-tyrosine phosphorylation | 10655584|11331870 |
| Hgene | PTK2 | GO:0022408 | negative regulation of cell-cell adhesion | 21703394 |
| Hgene | PTK2 | GO:0030335 | positive regulation of cell migration | 11331870|21703394 |
| Hgene | PTK2 | GO:0033628 | regulation of cell adhesion mediated by integrin | 10655584 |
| Hgene | PTK2 | GO:0046777 | protein autophosphorylation | 10655584|11331870 |
| Hgene | PTK2 | GO:0048013 | ephrin receptor signaling pathway | 10655584 |
| Hgene | PTK2 | GO:0060396 | growth hormone receptor signaling pathway | 10925297 |
| Hgene | PTK2 | GO:0090303 | positive regulation of wound healing | 26763945 |
 Fusion gene breakpoints across PTK2 (5'-gene) Fusion gene breakpoints across PTK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
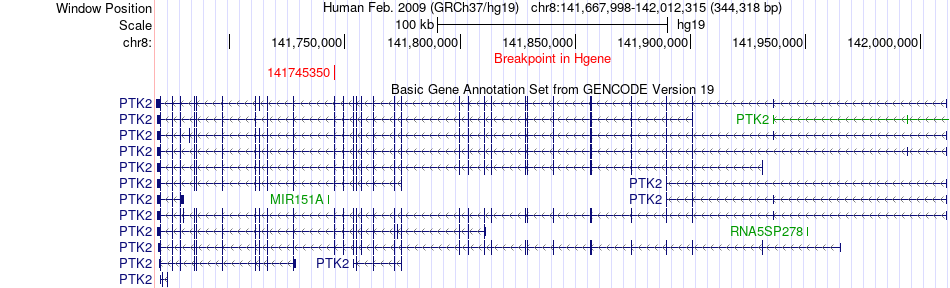 |
 Fusion gene breakpoints across MKLN1 (3'-gene) Fusion gene breakpoints across MKLN1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
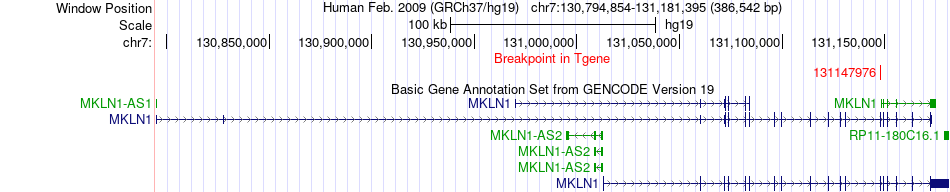 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-AX-A2H4-01A | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
Top |
Fusion Gene ORF analysis for PTK2-MKLN1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000340930 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000340930 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000340930 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000395218 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000395218 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000395218 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000517887 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000517887 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000517887 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000519419 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000519419 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000519419 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000519465 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000519465 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000519465 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000521059 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000521059 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000521059 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000522684 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000522684 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000522684 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000535192 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000535192 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000535192 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000538769 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000538769 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| 5CDS-intron | ENST00000538769 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| Frame-shift | ENST00000517887 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| Frame-shift | ENST00000519465 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| Frame-shift | ENST00000521059 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| Frame-shift | ENST00000535192 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| Frame-shift | ENST00000538769 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| In-frame | ENST00000340930 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| In-frame | ENST00000395218 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| In-frame | ENST00000519419 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| In-frame | ENST00000522684 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000430260 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000517712 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000519635 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000519881 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000520151 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000520892 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-3CDS | ENST00000522950 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000430260 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000430260 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000430260 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000517712 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000517712 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000517712 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000519635 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000519635 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000519635 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000519881 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000519881 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000519881 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000520151 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000520151 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000520151 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000520892 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000520892 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000520892 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000522950 | ENST00000352689 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000522950 | ENST00000429546 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
| intron-intron | ENST00000522950 | ENST00000498778 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000522684 | PTK2 | chr8 | 141745350 | - | ENST00000421797 | MKLN1 | chr7 | 131147976 | + | 3059 | 2260 | 230 | 2296 | 688 |
| ENST00000395218 | PTK2 | chr8 | 141745350 | - | ENST00000421797 | MKLN1 | chr7 | 131147976 | + | 3059 | 2260 | 230 | 2296 | 688 |
| ENST00000340930 | PTK2 | chr8 | 141745350 | - | ENST00000421797 | MKLN1 | chr7 | 131147976 | + | 2984 | 2185 | 155 | 2221 | 688 |
| ENST00000519419 | PTK2 | chr8 | 141745350 | - | ENST00000421797 | MKLN1 | chr7 | 131147976 | + | 3273 | 2474 | 270 | 2510 | 746 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000522684 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + | 0.001403 | 0.99859697 |
| ENST00000395218 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + | 0.001403 | 0.99859697 |
| ENST00000340930 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + | 0.00139768 | 0.99860233 |
| ENST00000519419 | ENST00000421797 | PTK2 | chr8 | 141745350 | - | MKLN1 | chr7 | 131147976 | + | 0.001184833 | 0.9988152 |
Top |
Fusion Genomic Features for PTK2-MKLN1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PTK2 | chr8 | 141745349 | - | MKLN1 | chr7 | 131147975 | + | 0.00010015 | 0.99989986 |
| PTK2 | chr8 | 141745349 | - | MKLN1 | chr7 | 131147975 | + | 0.00010015 | 0.99989986 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
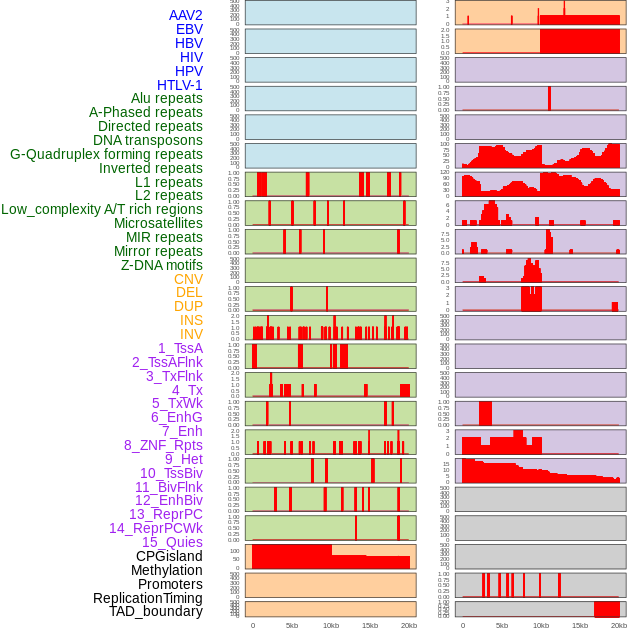 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
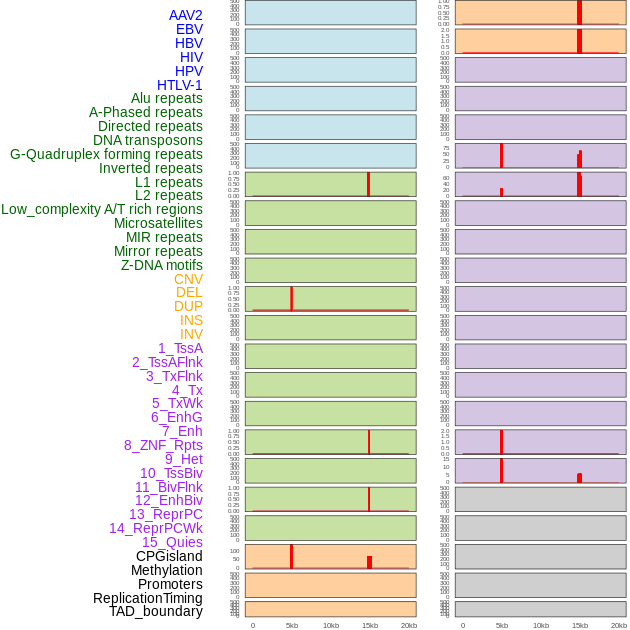 |
Top |
Fusion Protein Features for PTK2-MKLN1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:141745350/chr7:131147976) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MKLN1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Component of the CTLH E3 ubiquitin-protein ligase complex that selectively accepts ubiquitin from UBE2H and mediates ubiquitination and subsequent proteasomal degradation of the transcription factor HBP1 (PubMed:29911972). Required for internalization of the GABA receptor GABRA1 from the cell membrane via endosomes and subsequent GABRA1 degradation (By similarity). Acts as a mediator of cell spreading and cytoskeletal responses to the extracellular matrix component THBS1 (PubMed:18710924). {ECO:0000250|UniProtKB:O89050, ECO:0000269|PubMed:18710924, ECO:0000269|PubMed:29911972}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 35_355 | 676 | 1066.0 | Domain | FERM |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 35_355 | 676 | 1053.0 | Domain | FERM |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 35_355 | 676 | 1053.0 | Domain | FERM |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 428_434 | 676 | 1066.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 500_502 | 676 | 1066.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 428_434 | 676 | 1053.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 500_502 | 676 | 1053.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 428_434 | 676 | 1053.0 | Nucleotide binding | ATP |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 500_502 | 676 | 1053.0 | Nucleotide binding | ATP |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 526_578 | 508 | 736.0 | Repeat | Note=Kelch 5 | |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 597_651 | 508 | 736.0 | Repeat | Note=Kelch 6 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 712_733 | 676 | 1066.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 863_913 | 676 | 1066.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 712_733 | 676 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 863_913 | 676 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 712_733 | 676 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 863_913 | 676 | 1053.0 | Compositional bias | Note=Pro-rich |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 422_680 | 676 | 1066.0 | Domain | Protein kinase |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 422_680 | 676 | 1053.0 | Domain | Protein kinase |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 422_680 | 676 | 1053.0 | Domain | Protein kinase |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 172_204 | 508 | 736.0 | Domain | LisH | |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 206_258 | 508 | 736.0 | Domain | CTLH | |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 284_330 | 508 | 736.0 | Repeat | Note=Kelch 1 | |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 339_391 | 508 | 736.0 | Repeat | Note=Kelch 2 | |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 408_458 | 508 | 736.0 | Repeat | Note=Kelch 3 | |
| Tgene | MKLN1 | chr8:141745350 | chr7:131147976 | ENST00000352689 | 11 | 18 | 469_515 | 508 | 736.0 | Repeat | Note=Kelch 4 |
Top |
Fusion Gene Sequence for PTK2-MKLN1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >70042_70042_1_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000340930_MKLN1_chr7_131147976_ENST00000421797_length(transcript)=2984nt_BP=2185nt TGCCTTCGCTTCACGGCGCCGAGCCGCGGTCCGAGCAGAACTGGGGCTCCCTTGCATCTTCCAGTTACAAATTCAGTGCCTTCTGCAGTT TCCCCAGAGCTCCTCAAGAATAACGGAAGGGAGAATATGACAGATACCTAGCATCTAGCAAAATAATGGCAGCTGCTTACCTTGACCCCA ACTTGAATCACACACCAAATTCGAGTACTAAGACTCACCTGGGTACTGGTATGGAACGTTCTCCTGGTGCAATGGAGCGAGTATTAAAGG TCTTTCATTATTTTGAAAGCAATAGTGAGCCAACCACCTGGGCCAGTATTATCAGGCATGGAGATGCTACTGATGTCAGGGGCATCATTC AGAAGATAGTGGACAGTCACAAAGTAAAGCATGTGGCCTGCTATGGATTCCGCCTCAGTCACCTGCGGTCAGAGGAGGTTCACTGGCTTC ACGTGGATATGGGCGTCTCCAGTGTGAGGGAGAAGTATGAGCTTGCTCACCCACCAGAGGAGTGGAAATATGAATTGAGAATTCGTTATT TGCCAAAAGGATTTCTAAACCAGTTTACTGAAGATAAGCCAACTTTGAATTTCTTCTATCAACAGGTGAAGAGCGATTATATGTTAGAGA TAGCTGATCAAGTGGACCAGGAAATTGCTTTGAAGTTGGGTTGTCTAGAAATACGGCGATCATACTGGGAGATGCGGGGCAATGCACTAG AAAAGAAGTCTAACTATGAAGTATTAGAAAAAGATGTTGGTTTAAAGCGATTTTTTCCTAAGAGTTTACTGGATTCTGTCAAGGCCAAAA CACTAAGAAAACTGATCCAACAAACATTTAGACAATTTGCCAACCTTAATAGAGAAGAAAGTATTCTGAAATTCTTTGAGATCCTGTCTC CAGTCTACAGATTTGATAAGGAATGCTTCAAGTGTGCTCTTGGTTCAAGCTGGATTATTTCAGTGGAACTGGCAATCGGCCCAGAAGAAG GAATCAGTTACCTAACGGACAAGGGCTGCAATCCCACACATCTTGCTGACTTCACTCAAGTGCAAACCATTCAGTATTCAAACAGTGAAG ACAAGGACAGAAAAGGAATGCTACAACTAAAAATAGCAGGTGCACCCGAGCCTCTGACAGTGACGGCACCATCCCTAACCATTGCGGAGA ATATGGCTGACCTAATAGATGGGTACTGCCGGCTGGTGAATGGAACCTCGCAGTCATTTATCATCAGACCTCAGAAAGAAGGTGAACGGG CTTTGCCATCAATACCAAAGTTGGCCAACAGCGAAAAGCAAGGCATGCGGACACACGCCGTCTCTGTGTCAGAAACAGATGATTATGCTG AGATTATAGATGAAGAAGATACTTACACCATGCCCTCAACCAGGGATTATGAGATTCAAAGAGAAAGAATAGAACTTGGACGATGTATTG GAGAAGGCCAATTTGGAGATGTACATCAAGGCATTTATATGAGTCCAGAGAATCCAGCTTTGGCGGTTGCAATTAAAACATGTAAAAACT GTACTTCGGACAGCGTGAGAGAGAAATTTCTTCAAGAAGCCTTAACAATGCGTCAGTTTGACCATCCTCATATTGTGAAGCTGATTGGAG TCATCACAGAGAATCCTGTCTGGATAATCATGGAGCTGTGCACACTTGGAGAGCTGAGGTCATTTTTGCAAGTAAGGAAATACAGTTTGG ATCTAGCATCTTTGATCCTGTATGCCTATCAGCTTAGTACAGCTCTTGCATATCTAGAGAGCAAAAGATTTGTACACAGGGACATTGCTG CTCGGAATGTTCTGGTGTCCTCAAATGATTGTGTAAAATTAGGAGACTTTGGATTATCCCGATATATGGAAGATAGTACTTACTACAAAG CTTCCAAAGGAAAATTGCCTATTAAATGGATGGCTCCAGAGTCAATCAATTTTCGACGTTTTACCTCAGCTAGTGACGTATGGATGTTTG GTGTGTGTATGTGGGAGATACTGATGCATGGTGTGAAGCCTTTTCAAGGAGTGAAGAACAATGATGTAATCGGTCGAATTGAAAATGGGG AAAGATTACCAATGCCTCCAAATTGTCCTCCTACCCTCTACAGCCTTATGACGAAATGCTGGGCCTATGACCCCAGCAGGCGGCCCAGGT TTACTGAACTTAAAGCTCAGCTCAGTTCCAATGACAGGATTTACACAGAGAGCAACTATTGATCCAGAACTGAATGAAATACACGTCTTA TCTGGACTCAGCAAAGATAAGGAAAAGAGGGAAGAAAATGTTAGAAATTCATTCTGGATTTATGACATTGTGAGGAATAGTTGGTCTTGT GTCTATAAGAATGATCAAGCTGCAAAGGATAATCCAACTAAAAGTCTTCAGGAAGAAGAACCATGTCCAAGGTTTGCCCATCAGCTTGTA TACGATGAGCTACACAAGGTTCATTACTTATTTGGTGGGAATCCAGGAAAATCTTGCTCTCCAAAGATGAGATTAGATGACTTCTGGTCA CTGAAGTTGTGTAGACCTTCAAAAGATTATTTACTGAGGCATTGCAAGTACCTCATAAGAAAACACAGGTTTGAAGAAAAGGCCCAAGTG GATCCCCTTAGTGCTCTGAAATATTTACAAAATGATCTTTATATAACTGTGGATCATTCAGACCCAGAAGAGACAAAAGAGTTTCAGCTC CTGGCATCAGCTCTATTCAAATCTGGTTCAGATTTTACAGCTCTGGGCTTTTCTGATGTGGATCACACCTATGCTCAAAGAACTCAGCTC TTTGACACCTTAGTAAATTTCTTTCCTGACAGCATGACTCCTCCTAAAGGCAACCTGGTAGACCTCATCACACTGTAACTGAAGAGTCAC TGGACACAGAAATGGAAAACAGGAGTCGATTTTCCGTCTTTTGGATTGCAGCTCCACTGACTGACAGTAAAGCTGCAGTGATTGAGGACT >70042_70042_1_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000340930_MKLN1_chr7_131147976_ENST00000421797_length(amino acids)=688AA_BP= MAAAYLDPNLNHTPNSSTKTHLGTGMERSPGAMERVLKVFHYFESNSEPTTWASIIRHGDATDVRGIIQKIVDSHKVKHVACYGFRLSHL RSEEVHWLHVDMGVSSVREKYELAHPPEEWKYELRIRYLPKGFLNQFTEDKPTLNFFYQQVKSDYMLEIADQVDQEIALKLGCLEIRRSY WEMRGNALEKKSNYEVLEKDVGLKRFFPKSLLDSVKAKTLRKLIQQTFRQFANLNREESILKFFEILSPVYRFDKECFKCALGSSWIISV ELAIGPEEGISYLTDKGCNPTHLADFTQVQTIQYSNSEDKDRKGMLQLKIAGAPEPLTVTAPSLTIAENMADLIDGYCRLVNGTSQSFII RPQKEGERALPSIPKLANSEKQGMRTHAVSVSETDDYAEIIDEEDTYTMPSTRDYEIQRERIELGRCIGEGQFGDVHQGIYMSPENPALA VAIKTCKNCTSDSVREKFLQEALTMRQFDHPHIVKLIGVITENPVWIIMELCTLGELRSFLQVRKYSLDLASLILYAYQLSTALAYLESK RFVHRDIAARNVLVSSNDCVKLGDFGLSRYMEDSTYYKASKGKLPIKWMAPESINFRRFTSASDVWMFGVCMWEILMHGVKPFQGVKNND -------------------------------------------------------------- >70042_70042_2_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000395218_MKLN1_chr7_131147976_ENST00000421797_length(transcript)=3059nt_BP=2260nt GACCACTGTGAGCCCGCGGCGTGAGGCGTGGGAGGAAGCGCGGCTGCTGTCGCCCAGCGCCGCCCCGTCGTCGTCTGCCTTCGCTTCACG GCGCCGAGCCGCGGTCCGAGCAGAACTGGGGCTCCCTTGCATCTTCCAGTTACAAATTCAGTGCCTTCTGCAGTTTCCCCAGAGCTCCTC AAGAATAACGGAAGGGAGAATATGACAGATACCTAGCATCTAGCAAAATAATGGCAGCTGCTTACCTTGACCCCAACTTGAATCACACAC CAAATTCGAGTACTAAGACTCACCTGGGTACTGGTATGGAACGTTCTCCTGGTGCAATGGAGCGAGTATTAAAGGTCTTTCATTATTTTG AAAGCAATAGTGAGCCAACCACCTGGGCCAGTATTATCAGGCATGGAGATGCTACTGATGTCAGGGGCATCATTCAGAAGATAGTGGACA GTCACAAAGTAAAGCATGTGGCCTGCTATGGATTCCGCCTCAGTCACCTGCGGTCAGAGGAGGTTCACTGGCTTCACGTGGATATGGGCG TCTCCAGTGTGAGGGAGAAGTATGAGCTTGCTCACCCACCAGAGGAGTGGAAATATGAATTGAGAATTCGTTATTTGCCAAAAGGATTTC TAAACCAGTTTACTGAAGATAAGCCAACTTTGAATTTCTTCTATCAACAGGTGAAGAGCGATTATATGTTAGAGATAGCTGATCAAGTGG ACCAGGAAATTGCTTTGAAGTTGGGTTGTCTAGAAATACGGCGATCATACTGGGAGATGCGGGGCAATGCACTAGAAAAGAAGTCTAACT ATGAAGTATTAGAAAAAGATGTTGGTTTAAAGCGATTTTTTCCTAAGAGTTTACTGGATTCTGTCAAGGCCAAAACACTAAGAAAACTGA TCCAACAAACATTTAGACAATTTGCCAACCTTAATAGAGAAGAAAGTATTCTGAAATTCTTTGAGATCCTGTCTCCAGTCTACAGATTTG ATAAGGAATGCTTCAAGTGTGCTCTTGGTTCAAGCTGGATTATTTCAGTGGAACTGGCAATCGGCCCAGAAGAAGGAATCAGTTACCTAA CGGACAAGGGCTGCAATCCCACACATCTTGCTGACTTCACTCAAGTGCAAACCATTCAGTATTCAAACAGTGAAGACAAGGACAGAAAAG GAATGCTACAACTAAAAATAGCAGGTGCACCCGAGCCTCTGACAGTGACGGCACCATCCCTAACCATTGCGGAGAATATGGCTGACCTAA TAGATGGGTACTGCCGGCTGGTGAATGGAACCTCGCAGTCATTTATCATCAGACCTCAGAAAGAAGGTGAACGGGCTTTGCCATCAATAC CAAAGTTGGCCAACAGCGAAAAGCAAGGCATGCGGACACACGCCGTCTCTGTGTCAGAAACAGATGATTATGCTGAGATTATAGATGAAG AAGATACTTACACCATGCCCTCAACCAGGGATTATGAGATTCAAAGAGAAAGAATAGAACTTGGACGATGTATTGGAGAAGGCCAATTTG GAGATGTACATCAAGGCATTTATATGAGTCCAGAGAATCCAGCTTTGGCGGTTGCAATTAAAACATGTAAAAACTGTACTTCGGACAGCG TGAGAGAGAAATTTCTTCAAGAAGCCTTAACAATGCGTCAGTTTGACCATCCTCATATTGTGAAGCTGATTGGAGTCATCACAGAGAATC CTGTCTGGATAATCATGGAGCTGTGCACACTTGGAGAGCTGAGGTCATTTTTGCAAGTAAGGAAATACAGTTTGGATCTAGCATCTTTGA TCCTGTATGCCTATCAGCTTAGTACAGCTCTTGCATATCTAGAGAGCAAAAGATTTGTACACAGGGACATTGCTGCTCGGAATGTTCTGG TGTCCTCAAATGATTGTGTAAAATTAGGAGACTTTGGATTATCCCGATATATGGAAGATAGTACTTACTACAAAGCTTCCAAAGGAAAAT TGCCTATTAAATGGATGGCTCCAGAGTCAATCAATTTTCGACGTTTTACCTCAGCTAGTGACGTATGGATGTTTGGTGTGTGTATGTGGG AGATACTGATGCATGGTGTGAAGCCTTTTCAAGGAGTGAAGAACAATGATGTAATCGGTCGAATTGAAAATGGGGAAAGATTACCAATGC CTCCAAATTGTCCTCCTACCCTCTACAGCCTTATGACGAAATGCTGGGCCTATGACCCCAGCAGGCGGCCCAGGTTTACTGAACTTAAAG CTCAGCTCAGTTCCAATGACAGGATTTACACAGAGAGCAACTATTGATCCAGAACTGAATGAAATACACGTCTTATCTGGACTCAGCAAA GATAAGGAAAAGAGGGAAGAAAATGTTAGAAATTCATTCTGGATTTATGACATTGTGAGGAATAGTTGGTCTTGTGTCTATAAGAATGAT CAAGCTGCAAAGGATAATCCAACTAAAAGTCTTCAGGAAGAAGAACCATGTCCAAGGTTTGCCCATCAGCTTGTATACGATGAGCTACAC AAGGTTCATTACTTATTTGGTGGGAATCCAGGAAAATCTTGCTCTCCAAAGATGAGATTAGATGACTTCTGGTCACTGAAGTTGTGTAGA CCTTCAAAAGATTATTTACTGAGGCATTGCAAGTACCTCATAAGAAAACACAGGTTTGAAGAAAAGGCCCAAGTGGATCCCCTTAGTGCT CTGAAATATTTACAAAATGATCTTTATATAACTGTGGATCATTCAGACCCAGAAGAGACAAAAGAGTTTCAGCTCCTGGCATCAGCTCTA TTCAAATCTGGTTCAGATTTTACAGCTCTGGGCTTTTCTGATGTGGATCACACCTATGCTCAAAGAACTCAGCTCTTTGACACCTTAGTA AATTTCTTTCCTGACAGCATGACTCCTCCTAAAGGCAACCTGGTAGACCTCATCACACTGTAACTGAAGAGTCACTGGACACAGAAATGG >70042_70042_2_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000395218_MKLN1_chr7_131147976_ENST00000421797_length(amino acids)=688AA_BP= MAAAYLDPNLNHTPNSSTKTHLGTGMERSPGAMERVLKVFHYFESNSEPTTWASIIRHGDATDVRGIIQKIVDSHKVKHVACYGFRLSHL RSEEVHWLHVDMGVSSVREKYELAHPPEEWKYELRIRYLPKGFLNQFTEDKPTLNFFYQQVKSDYMLEIADQVDQEIALKLGCLEIRRSY WEMRGNALEKKSNYEVLEKDVGLKRFFPKSLLDSVKAKTLRKLIQQTFRQFANLNREESILKFFEILSPVYRFDKECFKCALGSSWIISV ELAIGPEEGISYLTDKGCNPTHLADFTQVQTIQYSNSEDKDRKGMLQLKIAGAPEPLTVTAPSLTIAENMADLIDGYCRLVNGTSQSFII RPQKEGERALPSIPKLANSEKQGMRTHAVSVSETDDYAEIIDEEDTYTMPSTRDYEIQRERIELGRCIGEGQFGDVHQGIYMSPENPALA VAIKTCKNCTSDSVREKFLQEALTMRQFDHPHIVKLIGVITENPVWIIMELCTLGELRSFLQVRKYSLDLASLILYAYQLSTALAYLESK RFVHRDIAARNVLVSSNDCVKLGDFGLSRYMEDSTYYKASKGKLPIKWMAPESINFRRFTSASDVWMFGVCMWEILMHGVKPFQGVKNND -------------------------------------------------------------- >70042_70042_3_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000519419_MKLN1_chr7_131147976_ENST00000421797_length(transcript)=3273nt_BP=2474nt TCTCTGACTGCAATGGAAAGAGGAAAGAGAAATTAGAAGTATTTTTTTAAATTGTCATTTGATTTCTTCAGAGTTATTGTGGGCTTCTGA ACTATTGCGGAAGCATGAGAAGATTCTGTAACTGAGAGGCATTCTTCAAGCTGCTGGAAAATATTACAGAAGGAACACCTTCCAAATTTT GCTTCAAGCTCTGAAAAAAGGAGCACACAGGGGAACCTTAGTCTTGCTTGTGGATCGAAGCAAATACTATCACAGCGAGGAACTGGGACT TTGTGTGCAGACTCCTGGCTTGATCTTTATAGGATTGGGTGTATGTTGTTGGAACTGGCAGGCCAAGAGGCCCTGAAACCAGCAGGTGCC ATCTATATGGAGAAAAGCGGATGTAGTCCATTTCCAGTGTGTTGGGCTAAAGAATATGACAGATACCTAGCATCTAGCAAAATAATGGCA GCTGCTTACCTTGACCCCAACTTGAATCACACACCAAATTCGAGTACTAAGACTCACCTGGGTACTGGTATGGAACGTTCTCCTGGTGCA ATGGAGCGAGTATTAAAGGTCTTTCATTATTTTGAAAGCAATAGTGAGCCAACCACCTGGGCCAGTATTATCAGGCATGGAGATGCTACT GATGTCAGGGGCATCATTCAGAAGATAGTGGACAGTCACAAAGTAAAGCATGTGGCCTGCTATGGATTCCGCCTCAGTCACCTGCGGTCA GAGGAGGTTCACTGGCTTCACGTGGATATGGGCGTCTCCAGTGTGAGGGAGAAGTATGAGCTTGCTCACCCACCAGAGGAGTGGAAATAT GAATTGAGAATTCGTTATTTGCCAAAAGGATTTCTAAACCAGTTTACTGAAGATAAGCCAACTTTGAATTTCTTCTATCAACAGGTGAAG AGCGATTATATGTTAGAGATAGCTGATCAAGTGGACCAGGAAATTGCTTTGAAGTTGGGTTGTCTAGAAATACGGCGATCATACTGGGAG ATGCGGGGCAATGCACTAGAAAAGAAGTCTAACTATGAAGTATTAGAAAAAGATGTTGGTTTAAAGCGATTTTTTCCTAAGAGTTTACTG GATTCTGTCAAGGCCAAAACACTAAGAAAACTGATCCAACAAACATTTAGACAATTTGCCAACCTTAATAGAGAAGAAAGTATTCTGAAA TTCTTTGAGATCCTGTCTCCAGTCTACAGATTTGATAAGGAATGCTTCAAGTGTGCTCTTGGTTCAAGCTGGATTATTTCAGTGGAACTG GCAATCGGCCCAGAAGAAGGAATCAGTTACCTAACGGACAAGGGCTGCAATCCCACACATCTTGCTGACTTCACTCAAGTGCAAACCATT CAGTATTCAAACAGTGAAGACAAGGACAGAAAAGGAATGCTACAACTAAAAATAGCAGGTGCACCCGAGCCTCTGACAGTGACGGCACCA TCCCTAACCATTGCGGAGAATATGGCTGACCTAATAGATGGGTACTGCCGGCTGGTGAATGGAACCTCGCAGTCATTTATCATCAGACCT CAGAAAGAAGGTGAACGGGCTTTGCCATCAATACCAAAGTTGGCCAACAGCGAAAAGCAAGGCATGCGGACACACGCCGTCTCTGTGTCA GAAACAGATGATTATGCTGAGATTATAGATGAAGAAGATACTTACACCATGCCCTCAACCAGGGATTATGAGATTCAAAGAGAAAGAATA GAACTTGGACGATGTATTGGAGAAGGCCAATTTGGAGATGTACATCAAGGCATTTATATGAGTCCAGAGAATCCAGCTTTGGCGGTTGCA ATTAAAACATGTAAAAACTGTACTTCGGACAGCGTGAGAGAGAAATTTCTTCAAGAAGCCTTAACAATGCGTCAGTTTGACCATCCTCAT ATTGTGAAGCTGATTGGAGTCATCACAGAGAATCCTGTCTGGATAATCATGGAGCTGTGCACACTTGGAGAGCTGAGGTCATTTTTGCAA GTAAGGAAATACAGTTTGGATCTAGCATCTTTGATCCTGTATGCCTATCAGCTTAGTACAGCTCTTGCATATCTAGAGAGCAAAAGATTT GTACACAGGGACATTGCTGCTCGGAATGTTCTGGTGTCCTCAAATGATTGTGTAAAATTAGGAGACTTTGGATTATCCCGATATATGGAA GATAGTACTTACTACAAAGCTTCCAAAGGAAAATTGCCTATTAAATGGATGGCTCCAGAGTCAATCAATTTTCGACGTTTTACCTCAGCT AGTGACGTATGGATGTTTGGTGTGTGTATGTGGGAGATACTGATGCATGGTGTGAAGCCTTTTCAAGGAGTGAAGAACAATGATGTAATC GGTCGAATTGAAAATGGGGAAAGATTACCAATGCCTCCAAATTGTCCTCCTACCCTCTACAGCCTTATGACGAAATGCTGGGCCTATGAC CCCAGCAGGCGGCCCAGGTTTACTGAACTTAAAGCTCAGCTCAGTTCCAATGACAGGATTTACACAGAGAGCAACTATTGATCCAGAACT GAATGAAATACACGTCTTATCTGGACTCAGCAAAGATAAGGAAAAGAGGGAAGAAAATGTTAGAAATTCATTCTGGATTTATGACATTGT GAGGAATAGTTGGTCTTGTGTCTATAAGAATGATCAAGCTGCAAAGGATAATCCAACTAAAAGTCTTCAGGAAGAAGAACCATGTCCAAG GTTTGCCCATCAGCTTGTATACGATGAGCTACACAAGGTTCATTACTTATTTGGTGGGAATCCAGGAAAATCTTGCTCTCCAAAGATGAG ATTAGATGACTTCTGGTCACTGAAGTTGTGTAGACCTTCAAAAGATTATTTACTGAGGCATTGCAAGTACCTCATAAGAAAACACAGGTT TGAAGAAAAGGCCCAAGTGGATCCCCTTAGTGCTCTGAAATATTTACAAAATGATCTTTATATAACTGTGGATCATTCAGACCCAGAAGA GACAAAAGAGTTTCAGCTCCTGGCATCAGCTCTATTCAAATCTGGTTCAGATTTTACAGCTCTGGGCTTTTCTGATGTGGATCACACCTA TGCTCAAAGAACTCAGCTCTTTGACACCTTAGTAAATTTCTTTCCTGACAGCATGACTCCTCCTAAAGGCAACCTGGTAGACCTCATCAC ACTGTAACTGAAGAGTCACTGGACACAGAAATGGAAAACAGGAGTCGATTTTCCGTCTTTTGGATTGCAGCTCCACTGACTGACAGTAAA >70042_70042_3_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000519419_MKLN1_chr7_131147976_ENST00000421797_length(amino acids)=746AA_BP= MCADSWLDLYRIGCMLLELAGQEALKPAGAIYMEKSGCSPFPVCWAKEYDRYLASSKIMAAAYLDPNLNHTPNSSTKTHLGTGMERSPGA MERVLKVFHYFESNSEPTTWASIIRHGDATDVRGIIQKIVDSHKVKHVACYGFRLSHLRSEEVHWLHVDMGVSSVREKYELAHPPEEWKY ELRIRYLPKGFLNQFTEDKPTLNFFYQQVKSDYMLEIADQVDQEIALKLGCLEIRRSYWEMRGNALEKKSNYEVLEKDVGLKRFFPKSLL DSVKAKTLRKLIQQTFRQFANLNREESILKFFEILSPVYRFDKECFKCALGSSWIISVELAIGPEEGISYLTDKGCNPTHLADFTQVQTI QYSNSEDKDRKGMLQLKIAGAPEPLTVTAPSLTIAENMADLIDGYCRLVNGTSQSFIIRPQKEGERALPSIPKLANSEKQGMRTHAVSVS ETDDYAEIIDEEDTYTMPSTRDYEIQRERIELGRCIGEGQFGDVHQGIYMSPENPALAVAIKTCKNCTSDSVREKFLQEALTMRQFDHPH IVKLIGVITENPVWIIMELCTLGELRSFLQVRKYSLDLASLILYAYQLSTALAYLESKRFVHRDIAARNVLVSSNDCVKLGDFGLSRYME DSTYYKASKGKLPIKWMAPESINFRRFTSASDVWMFGVCMWEILMHGVKPFQGVKNNDVIGRIENGERLPMPPNCPPTLYSLMTKCWAYD -------------------------------------------------------------- >70042_70042_4_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000522684_MKLN1_chr7_131147976_ENST00000421797_length(transcript)=3059nt_BP=2260nt GACCACTGTGAGCCCGCGGCGTGAGGCGTGGGAGGAAGCGCGGCTGCTGTCGCCCAGCGCCGCCCCGTCGTCGTCTGCCTTCGCTTCACG GCGCCGAGCCGCGGTCCGAGCAGAACTGGGGCTCCCTTGCATCTTCCAGTTACAAATTCAGTGCCTTCTGCAGTTTCCCCAGAGCTCCTC AAGAATAACGGAAGGGAGAATATGACAGATACCTAGCATCTAGCAAAATAATGGCAGCTGCTTACCTTGACCCCAACTTGAATCACACAC CAAATTCGAGTACTAAGACTCACCTGGGTACTGGTATGGAACGTTCTCCTGGTGCAATGGAGCGAGTATTAAAGGTCTTTCATTATTTTG AAAGCAATAGTGAGCCAACCACCTGGGCCAGTATTATCAGGCATGGAGATGCTACTGATGTCAGGGGCATCATTCAGAAGATAGTGGACA GTCACAAAGTAAAGCATGTGGCCTGCTATGGATTCCGCCTCAGTCACCTGCGGTCAGAGGAGGTTCACTGGCTTCACGTGGATATGGGCG TCTCCAGTGTGAGGGAGAAGTATGAGCTTGCTCACCCACCAGAGGAGTGGAAATATGAATTGAGAATTCGTTATTTGCCAAAAGGATTTC TAAACCAGTTTACTGAAGATAAGCCAACTTTGAATTTCTTCTATCAACAGGTGAAGAGCGATTATATGTTAGAGATAGCTGATCAAGTGG ACCAGGAAATTGCTTTGAAGTTGGGTTGTCTAGAAATACGGCGATCATACTGGGAGATGCGGGGCAATGCACTAGAAAAGAAGTCTAACT ATGAAGTATTAGAAAAAGATGTTGGTTTAAAGCGATTTTTTCCTAAGAGTTTACTGGATTCTGTCAAGGCCAAAACACTAAGAAAACTGA TCCAACAAACATTTAGACAATTTGCCAACCTTAATAGAGAAGAAAGTATTCTGAAATTCTTTGAGATCCTGTCTCCAGTCTACAGATTTG ATAAGGAATGCTTCAAGTGTGCTCTTGGTTCAAGCTGGATTATTTCAGTGGAACTGGCAATCGGCCCAGAAGAAGGAATCAGTTACCTAA CGGACAAGGGCTGCAATCCCACACATCTTGCTGACTTCACTCAAGTGCAAACCATTCAGTATTCAAACAGTGAAGACAAGGACAGAAAAG GAATGCTACAACTAAAAATAGCAGGTGCACCCGAGCCTCTGACAGTGACGGCACCATCCCTAACCATTGCGGAGAATATGGCTGACCTAA TAGATGGGTACTGCCGGCTGGTGAATGGAACCTCGCAGTCATTTATCATCAGACCTCAGAAAGAAGGTGAACGGGCTTTGCCATCAATAC CAAAGTTGGCCAACAGCGAAAAGCAAGGCATGCGGACACACGCCGTCTCTGTGTCAGAAACAGATGATTATGCTGAGATTATAGATGAAG AAGATACTTACACCATGCCCTCAACCAGGGATTATGAGATTCAAAGAGAAAGAATAGAACTTGGACGATGTATTGGAGAAGGCCAATTTG GAGATGTACATCAAGGCATTTATATGAGTCCAGAGAATCCAGCTTTGGCGGTTGCAATTAAAACATGTAAAAACTGTACTTCGGACAGCG TGAGAGAGAAATTTCTTCAAGAAGCCTTAACAATGCGTCAGTTTGACCATCCTCATATTGTGAAGCTGATTGGAGTCATCACAGAGAATC CTGTCTGGATAATCATGGAGCTGTGCACACTTGGAGAGCTGAGGTCATTTTTGCAAGTAAGGAAATACAGTTTGGATCTAGCATCTTTGA TCCTGTATGCCTATCAGCTTAGTACAGCTCTTGCATATCTAGAGAGCAAAAGATTTGTACACAGGGACATTGCTGCTCGGAATGTTCTGG TGTCCTCAAATGATTGTGTAAAATTAGGAGACTTTGGATTATCCCGATATATGGAAGATAGTACTTACTACAAAGCTTCCAAAGGAAAAT TGCCTATTAAATGGATGGCTCCAGAGTCAATCAATTTTCGACGTTTTACCTCAGCTAGTGACGTATGGATGTTTGGTGTGTGTATGTGGG AGATACTGATGCATGGTGTGAAGCCTTTTCAAGGAGTGAAGAACAATGATGTAATCGGTCGAATTGAAAATGGGGAAAGATTACCAATGC CTCCAAATTGTCCTCCTACCCTCTACAGCCTTATGACGAAATGCTGGGCCTATGACCCCAGCAGGCGGCCCAGGTTTACTGAACTTAAAG CTCAGCTCAGTTCCAATGACAGGATTTACACAGAGAGCAACTATTGATCCAGAACTGAATGAAATACACGTCTTATCTGGACTCAGCAAA GATAAGGAAAAGAGGGAAGAAAATGTTAGAAATTCATTCTGGATTTATGACATTGTGAGGAATAGTTGGTCTTGTGTCTATAAGAATGAT CAAGCTGCAAAGGATAATCCAACTAAAAGTCTTCAGGAAGAAGAACCATGTCCAAGGTTTGCCCATCAGCTTGTATACGATGAGCTACAC AAGGTTCATTACTTATTTGGTGGGAATCCAGGAAAATCTTGCTCTCCAAAGATGAGATTAGATGACTTCTGGTCACTGAAGTTGTGTAGA CCTTCAAAAGATTATTTACTGAGGCATTGCAAGTACCTCATAAGAAAACACAGGTTTGAAGAAAAGGCCCAAGTGGATCCCCTTAGTGCT CTGAAATATTTACAAAATGATCTTTATATAACTGTGGATCATTCAGACCCAGAAGAGACAAAAGAGTTTCAGCTCCTGGCATCAGCTCTA TTCAAATCTGGTTCAGATTTTACAGCTCTGGGCTTTTCTGATGTGGATCACACCTATGCTCAAAGAACTCAGCTCTTTGACACCTTAGTA AATTTCTTTCCTGACAGCATGACTCCTCCTAAAGGCAACCTGGTAGACCTCATCACACTGTAACTGAAGAGTCACTGGACACAGAAATGG >70042_70042_4_PTK2-MKLN1_PTK2_chr8_141745350_ENST00000522684_MKLN1_chr7_131147976_ENST00000421797_length(amino acids)=688AA_BP= MAAAYLDPNLNHTPNSSTKTHLGTGMERSPGAMERVLKVFHYFESNSEPTTWASIIRHGDATDVRGIIQKIVDSHKVKHVACYGFRLSHL RSEEVHWLHVDMGVSSVREKYELAHPPEEWKYELRIRYLPKGFLNQFTEDKPTLNFFYQQVKSDYMLEIADQVDQEIALKLGCLEIRRSY WEMRGNALEKKSNYEVLEKDVGLKRFFPKSLLDSVKAKTLRKLIQQTFRQFANLNREESILKFFEILSPVYRFDKECFKCALGSSWIISV ELAIGPEEGISYLTDKGCNPTHLADFTQVQTIQYSNSEDKDRKGMLQLKIAGAPEPLTVTAPSLTIAENMADLIDGYCRLVNGTSQSFII RPQKEGERALPSIPKLANSEKQGMRTHAVSVSETDDYAEIIDEEDTYTMPSTRDYEIQRERIELGRCIGEGQFGDVHQGIYMSPENPALA VAIKTCKNCTSDSVREKFLQEALTMRQFDHPHIVKLIGVITENPVWIIMELCTLGELRSFLQVRKYSLDLASLILYAYQLSTALAYLESK RFVHRDIAARNVLVSSNDCVKLGDFGLSRYMEDSTYYKASKGKLPIKWMAPESINFRRFTSASDVWMFGVCMWEILMHGVKPFQGVKNND -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PTK2-MKLN1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000340930 | - | 22 | 33 | 912_1052 | 676.6666666666666 | 1066.0 | ARHGEF28 |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000521059 | - | 22 | 32 | 912_1052 | 676.6666666666666 | 1053.0 | ARHGEF28 |
| Hgene | PTK2 | chr8:141745350 | chr7:131147976 | ENST00000522684 | - | 22 | 32 | 912_1052 | 676.6666666666666 | 1053.0 | ARHGEF28 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PTK2-MKLN1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PTK2-MKLN1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
