
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PTOV1-TRIP10 (FusionGDB2 ID:70140) |
Fusion Gene Summary for PTOV1-TRIP10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PTOV1-TRIP10 | Fusion gene ID: 70140 | Hgene | Tgene | Gene symbol | PTOV1 | TRIP10 | Gene ID | 53635 | 9322 |
| Gene name | PTOV1 extended AT-hook containing adaptor protein | thyroid hormone receptor interactor 10 | |
| Synonyms | ACID2|PTOV-1 | CIP4|HSTP|STOT|STP|TRIP-10 | |
| Cytomap | 19q13.33 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | prostate tumor-overexpressed gene 1 proteinactivator interaction domain-containing protein 2prostate tumor overexpressed 1 | cdc42-interacting protein 4TR-interacting protein 10protein Felicsalt tolerant proteinsalt toleratorthyroid receptor-interacting protein 10 | |
| Modification date | 20200320 | 20200320 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000598325, ENST00000221557, ENST00000391842, ENST00000599732, ENST00000600603, ENST00000601638, ENST00000601675, | ENST00000600428, ENST00000313244, ENST00000313285, ENST00000596543, ENST00000596758, | |
| Fusion gene scores | * DoF score | 5 X 5 X 4=100 | 2 X 2 X 2=8 |
| # samples | 5 | 2 | |
| ** MAII score | log2(5/100*10)=-1 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/8*10)=1.32192809488736 | |
| Context | PubMed: PTOV1 [Title/Abstract] AND TRIP10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PTOV1(50361371)-TRIP10(6746462), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across PTOV1 (5'-gene) Fusion gene breakpoints across PTOV1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
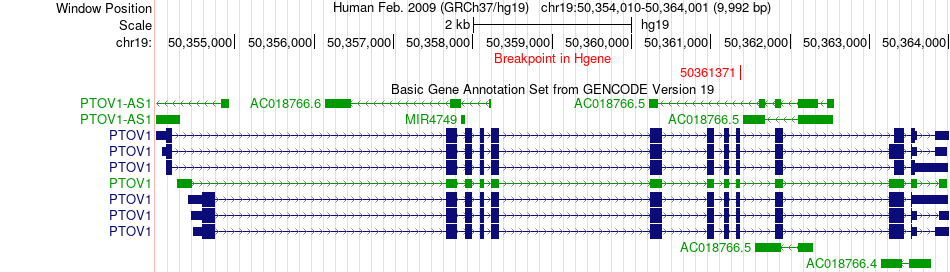 |
 Fusion gene breakpoints across TRIP10 (3'-gene) Fusion gene breakpoints across TRIP10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
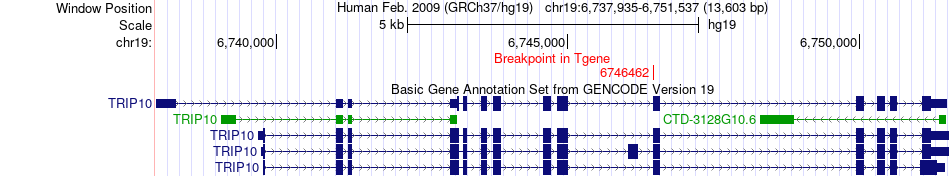 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 131Nd | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
Top |
Fusion Gene ORF analysis for PTOV1-TRIP10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000598325 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 3UTR-intron | ENST00000598325 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 3UTR-intron | ENST00000598325 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 3UTR-intron | ENST00000598325 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 3UTR-intron | ENST00000598325 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000221557 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000221557 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000221557 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000221557 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000391842 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000391842 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000391842 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000391842 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000599732 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000599732 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000599732 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000599732 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000600603 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000600603 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000600603 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000600603 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601638 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601638 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601638 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601638 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601675 | ENST00000313244 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601675 | ENST00000313285 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601675 | ENST00000596543 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| 5CDS-intron | ENST00000601675 | ENST00000596758 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| In-frame | ENST00000221557 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| In-frame | ENST00000391842 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| In-frame | ENST00000599732 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| In-frame | ENST00000600603 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| In-frame | ENST00000601638 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
| In-frame | ENST00000601675 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000600603 | PTOV1 | chr19 | 50361371 | + | ENST00000600428 | TRIP10 | chr19 | 6746462 | + | 1910 | 967 | 88 | 1620 | 510 |
| ENST00000601638 | PTOV1 | chr19 | 50361371 | + | ENST00000600428 | TRIP10 | chr19 | 6746462 | + | 1836 | 893 | 14 | 1546 | 510 |
| ENST00000221557 | PTOV1 | chr19 | 50361371 | + | ENST00000600428 | TRIP10 | chr19 | 6746462 | + | 1783 | 840 | 0 | 1493 | 497 |
| ENST00000391842 | PTOV1 | chr19 | 50361371 | + | ENST00000600428 | TRIP10 | chr19 | 6746462 | + | 2049 | 1106 | 47 | 1759 | 570 |
| ENST00000599732 | PTOV1 | chr19 | 50361371 | + | ENST00000600428 | TRIP10 | chr19 | 6746462 | + | 2017 | 1074 | 15 | 1727 | 570 |
| ENST00000601675 | PTOV1 | chr19 | 50361371 | + | ENST00000600428 | TRIP10 | chr19 | 6746462 | + | 1983 | 1040 | 104 | 1693 | 529 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000600603 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.026996773 | 0.9730032 |
| ENST00000601638 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.03423427 | 0.9657657 |
| ENST00000221557 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.036328748 | 0.96367127 |
| ENST00000391842 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.073270425 | 0.9267296 |
| ENST00000599732 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.07534205 | 0.92465794 |
| ENST00000601675 | ENST00000600428 | PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.07625515 | 0.9237448 |
Top |
Fusion Genomic Features for PTOV1-TRIP10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.10077409 | 0.89922595 |
| PTOV1 | chr19 | 50361371 | + | TRIP10 | chr19 | 6746462 | + | 0.10077409 | 0.89922595 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
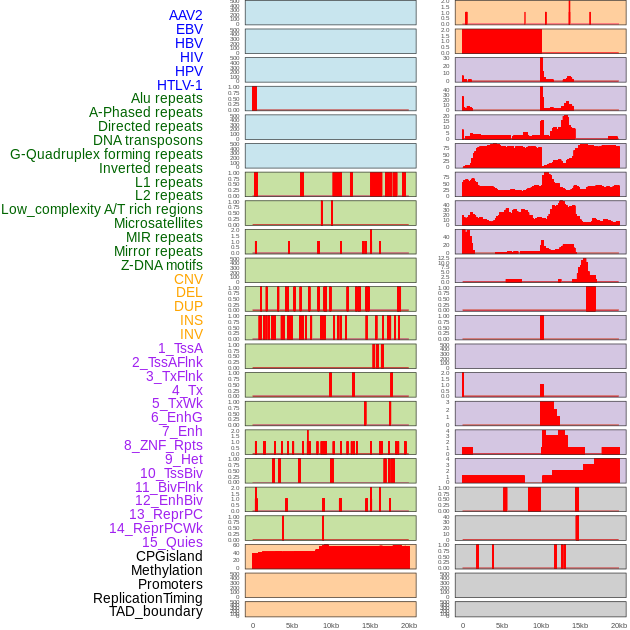 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
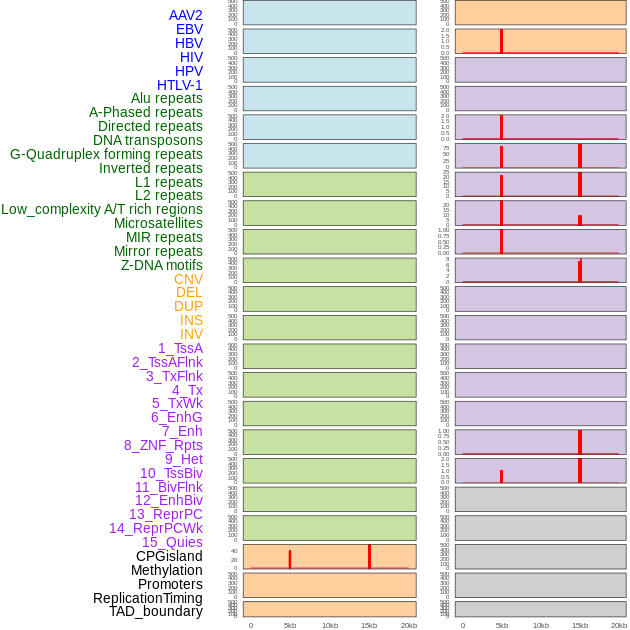 |
Top |
Fusion Protein Features for PTOV1-TRIP10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:50361371/chr19:6746462) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 388_481 | 384 | 602.0 | Coiled coil | . | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 388_481 | 328 | 546.0 | Coiled coil | . | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 393_470 | 384 | 602.0 | Domain | REM-1 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 540_601 | 384 | 602.0 | Domain | SH3 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 393_470 | 328 | 546.0 | Domain | REM-1 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 540_601 | 328 | 546.0 | Domain | SH3 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 538_601 | 384 | 602.0 | Region | Note=Required for podosome formation | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 538_601 | 328 | 546.0 | Region | Note=Required for podosome formation |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 67_259 | 384 | 602.0 | Coiled coil | . | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 67_259 | 328 | 546.0 | Coiled coil | . | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 1_264 | 384 | 602.0 | Domain | F-BAR | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 1_264 | 328 | 546.0 | Domain | F-BAR | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 1_117 | 384 | 602.0 | Region | Required for translocation to the plasma membrane in response to insulin | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 1_117 | 328 | 546.0 | Region | Required for translocation to the plasma membrane in response to insulin |
Top |
Fusion Gene Sequence for PTOV1-TRIP10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >70140_70140_1_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000221557_TRIP10_chr19_6746462_ENST00000600428_length(transcript)=1783nt_BP=840nt ATGAGGTCTCCCGCCGTCCCGACCCCCGCAAGGGGCCAGCTTGGTGTCGCCTTCGTTCTTCTGCCACCCCATTCGGAAGGTGCTCGGGTC TTCGGGGCACTGGGTCCCATCGGTCCCTCCTCACCTGGGCTCACCCTCGGGGGTCTGGCCGTGAGCGAGCACCGGCTCAGCAACAAGCTG CTGGCTTGGAGCGGCGTCCTCGAGTGGCAGGAGAAGCGCAGACCCTACTCTGACTCCACTGCAAAGCTGAAGCGGACCCTGCCCTGCCAA GCCTACGTGAACCAAGGCGAGAACCTGGAGACCGACCAGTGGCCGCAGAAGCTGATCATGCAGCTGATCCCTCAGCAGCTGCTGACCACC CTGGGCCCCCTGTTCCGGAACTCCCAGTTGGCACAGTTCCACTTCACCAACAGAGACTGCGACTCGCTCAAGGGGCTCTGCCGCATCATG GGCAACGGCTTCGCGGGCTGCATGCTGTTCCCCCACATCTCCCCCTGTGAGGTGCGCGTGCTCATGCTCCTGTACTCGTCCAAGAAGAAG ATCTTCATGGGCCTCATCCCCTACGACCAGAGCGGCTTCGTCAGTGCCATCCGGCAGGTCATCACCACCCGCAAGCAGGCAGTGGGACCT GGTGGTGTCAACTCAGGCCCAGTCCAGATCGTCAACAACAAGTTTCTGGCATGGAGTGGTGTCATGGAGTGGCAGGAGCCCAGGCCTGAG CCCAACAGTCGGTCCAAGAGGTGGCTGCCATCCCACGTCTACGTGAACCAGGGGGAGATCCTGAGGACCGAGCAGTGGCCAAGGAAGCTG TACATGCAGCTCATCCCGCAGCAGCTGCTGACAGTGGTGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAA CAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAGGAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACA CCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAGATCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTAT GAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTAGCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCT AGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCACAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACC CCCATTTACACGGAGTTTGATGAGGATTTCGAGGAGGAACCCACATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGGGTCC AGCGAGGGCACTATCTCTATGGCCGAGGGTGAAGACCTCAGTCTTATGGAAGAAGACAAAGGGGACGGCTGGACCCGGGTCAGGCGGAAA GAGGGAGGCGAGGGCTACGTGCCCACCTCCTACCTCCGAGTCACGCTCAATTGAACCCTGCCAGAGACGGGAAGAGGGGGGCTGTCGGCT GCTGCTTCTGGGCCACGGGGAGCCCCAGGACCTATGCACTTTATTTCTGACCCCGTGGCTTCGGCTGAGACCTGTGTAACCTGCTGCCCC CTCCACCCCCAACCCAGTCCTACCTGTCACACCGGACGGACCCGCTGTGCCTTCTACCATCGTTCCACCATTGATGTACATACTCATGTT >70140_70140_1_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000221557_TRIP10_chr19_6746462_ENST00000600428_length(amino acids)=497AA_BP=278 MRSPAVPTPARGQLGVAFVLLPPHSEGARVFGALGPIGPSSPGLTLGGLAVSEHRLSNKLLAWSGVLEWQEKRRPYSDSTAKLKRTLPCQ AYVNQGENLETDQWPQKLIMQLIPQQLLTTLGPLFRNSQLAQFHFTNRDCDSLKGLCRIMGNGFAGCMLFPHISPCEVRVLMLLYSSKKK IFMGLIPYDQSGFVSAIRQVITTRKQAVGPGGVNSGPVQIVNNKFLAWSGVMEWQEPRPEPNSRSKRWLPSHVYVNQGEILRTEQWPRKL YMQLIPQQLLTVVTEDFSHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQMGDPASLEPQIAETLSNIERLKLEVQKY EAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSPIGHCVAIYHFEGS -------------------------------------------------------------- >70140_70140_2_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000391842_TRIP10_chr19_6746462_ENST00000600428_length(transcript)=2049nt_BP=1106nt AGTGGTTCGGCCGCGGCGACCCCACTCCGGCGGCGCGTCCCCCGAGCTTGGTACGGCTCAGCCCGTCTCCCCCGAAGCCGCGCGCCCGCG CCCGCGCCCCTCAGTCGGTGGAGCCCGCAGCCCCCCTTGTGGCCCGCGGCAGCTCCCCGCCCGCTCGGCCCGCGCCCGCCATGGTCCGTC CGCGCCGTGCCCCGTACCGCTCCGGCGCCGGGGGCCCCCTCGGGGGTCGCGGCCGCCCTCCGCGGCCCCTCGTGGTGCGCGCCGTCCGCT CGCGCTCCTGGCCTGCCAGCCCCCGAGGCCCGCAGCCTCCGCGGATCCGGGCCCGCTCGGCCCCTCCCATGGAAGGTGCTCGGGTCTTCG GGGCACTGGGTCCCATCGGTCCCTCCTCACCTGGGCTCACCCTCGGGGGTCTGGCCGTGAGCGAGCACCGGCTCAGCAACAAGCTGCTGG CTTGGAGCGGCGTCCTCGAGTGGCAGGAGAAGCGCAGACCCTACTCTGACTCCACTGCAAAGCTGAAGCGGACCCTGCCCTGCCAAGCCT ACGTGAACCAAGGCGAGAACCTGGAGACCGACCAGTGGCCGCAGAAGCTGATCATGCAGCTGATCCCTCAGCAGCTGCTGACCACCCTGG GCCCCCTGTTCCGGAACTCCCAGTTGGCACAGTTCCACTTCACCAACAGAGACTGCGACTCGCTCAAGGGGCTCTGCCGCATCATGGGCA ACGGCTTCGCGGGCTGCATGCTGTTCCCCCACATCTCCCCCTGTGAGGTGCGCGTGCTCATGCTCCTGTACTCGTCCAAGAAGAAGATCT TCATGGGCCTCATCCCCTACGACCAGAGCGGCTTCGTCAGTGCCATCCGGCAGGTCATCACCACCCGCAAGCAGGCAGTGGGACCTGGTG GTGTCAACTCAGGCCCAGTCCAGATCGTCAACAACAAGTTTCTGGCATGGAGTGGTGTCATGGAGTGGCAGGAGCCCAGGCCTGAGCCCA ACAGTCGGTCCAAGAGGTGGCTGCCATCCCACGTCTACGTGAACCAGGGGGAGATCCTGAGGACCGAGCAGTGGCCAAGGAAGCTGTACA TGCAGCTCATCCCGCAGCAGCTGCTGACAGTGGTGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGC AGTTGGAAGAACGCAGTCGTGAACTTCAGAAGGAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTC AGATGGGGGACCCCGCCAGCTTGGAGCCCCAGATCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGG CGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTAGCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCG CCCCGCCAGACAGCAGCAGCAACAGCGCATCACAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCA TTTACACGGAGTTTGATGAGGATTTCGAGGAGGAACCCACATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGGGTCCAGCG AGGGCACTATCTCTATGGCCGAGGGTGAAGACCTCAGTCTTATGGAAGAAGACAAAGGGGACGGCTGGACCCGGGTCAGGCGGAAAGAGG GAGGCGAGGGCTACGTGCCCACCTCCTACCTCCGAGTCACGCTCAATTGAACCCTGCCAGAGACGGGAAGAGGGGGGCTGTCGGCTGCTG CTTCTGGGCCACGGGGAGCCCCAGGACCTATGCACTTTATTTCTGACCCCGTGGCTTCGGCTGAGACCTGTGTAACCTGCTGCCCCCTCC ACCCCCAACCCAGTCCTACCTGTCACACCGGACGGACCCGCTGTGCCTTCTACCATCGTTCCACCATTGATGTACATACTCATGTTTTAC >70140_70140_2_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000391842_TRIP10_chr19_6746462_ENST00000600428_length(amino acids)=570AA_BP=351 MVRLSPSPPKPRARARAPQSVEPAAPLVARGSSPPARPAPAMVRPRRAPYRSGAGGPLGGRGRPPRPLVVRAVRSRSWPASPRGPQPPRI RARSAPPMEGARVFGALGPIGPSSPGLTLGGLAVSEHRLSNKLLAWSGVLEWQEKRRPYSDSTAKLKRTLPCQAYVNQGENLETDQWPQK LIMQLIPQQLLTTLGPLFRNSQLAQFHFTNRDCDSLKGLCRIMGNGFAGCMLFPHISPCEVRVLMLLYSSKKKIFMGLIPYDQSGFVSAI RQVITTRKQAVGPGGVNSGPVQIVNNKFLAWSGVMEWQEPRPEPNSRSKRWLPSHVYVNQGEILRTEQWPRKLYMQLIPQQLLTVVTEDF SHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQMGDPASLEPQIAETLSNIERLKLEVQKYEAWLAEAESRVLSNRGD SLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSPIGHCVAIYHFEGSSEGTISMAEGEDLSLME -------------------------------------------------------------- >70140_70140_3_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000599732_TRIP10_chr19_6746462_ENST00000600428_length(transcript)=2017nt_BP=1074nt GCGCGTCCCCCGAGCTTGGTACGGCTCAGCCCGTCTCCCCCGAAGCCGCGCGCCCGCGCCCGCGCCCCTCAGTCGGTGGAGCCCGCAGCC CCCCTTGTGGCCCGCGGCAGCTCCCCGCCCGCTCGGCCCGCGCCCGCCATGGTCCGTCCGCGCCGTGCCCCGTACCGCTCCGGCGCCGGG GGCCCCCTCGGGGGTCGCGGCCGCCCTCCGCGGCCCCTCGTGGTGCGCGCCGTCCGCTCGCGCTCCTGGCCTGCCAGCCCCCGAGGCCCG CAGCCTCCGCGGATCCGGGCCCGCTCGGCCCCTCCCATGGAAGGTGCTCGGGTCTTCGGGGCACTGGGTCCCATCGGTCCCTCCTCACCT GGGCTCACCCTCGGGGGTCTGGCCGTGAGCGAGCACCGGCTCAGCAACAAGCTGCTGGCTTGGAGCGGCGTCCTCGAGTGGCAGGAGAAG CGCAGACCCTACTCTGACTCCACTGCAAAGCTGAAGCGGACCCTGCCCTGCCAAGCCTACGTGAACCAAGGCGAGAACCTGGAGACCGAC CAGTGGCCGCAGAAGCTGATCATGCAGCTGATCCCTCAGCAGCTGCTGACCACCCTGGGCCCCCTGTTCCGGAACTCCCAGTTGGCACAG TTCCACTTCACCAACAGAGACTGCGACTCGCTCAAGGGGCTCTGCCGCATCATGGGCAACGGCTTCGCGGGCTGCATGCTGTTCCCCCAC ATCTCCCCCTGTGAGGTGCGCGTGCTCATGCTCCTGTACTCGTCCAAGAAGAAGATCTTCATGGGCCTCATCCCCTACGACCAGAGCGGC TTCGTCAGTGCCATCCGGCAGGTCATCACCACCCGCAAGCAGGCAGTGGGACCTGGTGGTGTCAACTCAGGCCCAGTCCAGATCGTCAAC AACAAGTTTCTGGCATGGAGTGGTGTCATGGAGTGGCAGGAGCCCAGGCCTGAGCCCAACAGTCGGTCCAAGAGGTGGCTGCCATCCCAC GTCTACGTGAACCAGGGGGAGATCCTGAGGACCGAGCAGTGGCCAAGGAAGCTGTACATGCAGCTCATCCCGCAGCAGCTGCTGACAGTG GTGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAG GAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAG ATCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTT AGCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCA CAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAG GAACCCACATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGGGTCCAGCGAGGGCACTATCTCTATGGCCGAGGGTGAAGAC CTCAGTCTTATGGAAGAAGACAAAGGGGACGGCTGGACCCGGGTCAGGCGGAAAGAGGGAGGCGAGGGCTACGTGCCCACCTCCTACCTC CGAGTCACGCTCAATTGAACCCTGCCAGAGACGGGAAGAGGGGGGCTGTCGGCTGCTGCTTCTGGGCCACGGGGAGCCCCAGGACCTATG CACTTTATTTCTGACCCCGTGGCTTCGGCTGAGACCTGTGTAACCTGCTGCCCCCTCCACCCCCAACCCAGTCCTACCTGTCACACCGGA CGGACCCGCTGTGCCTTCTACCATCGTTCCACCATTGATGTACATACTCATGTTTTACATCTTTTCTTTCTGCCGCTCGGCTCCGGCCAT >70140_70140_3_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000599732_TRIP10_chr19_6746462_ENST00000600428_length(amino acids)=570AA_BP=351 MVRLSPSPPKPRARARAPQSVEPAAPLVARGSSPPARPAPAMVRPRRAPYRSGAGGPLGGRGRPPRPLVVRAVRSRSWPASPRGPQPPRI RARSAPPMEGARVFGALGPIGPSSPGLTLGGLAVSEHRLSNKLLAWSGVLEWQEKRRPYSDSTAKLKRTLPCQAYVNQGENLETDQWPQK LIMQLIPQQLLTTLGPLFRNSQLAQFHFTNRDCDSLKGLCRIMGNGFAGCMLFPHISPCEVRVLMLLYSSKKKIFMGLIPYDQSGFVSAI RQVITTRKQAVGPGGVNSGPVQIVNNKFLAWSGVMEWQEPRPEPNSRSKRWLPSHVYVNQGEILRTEQWPRKLYMQLIPQQLLTVVTEDF SHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQMGDPASLEPQIAETLSNIERLKLEVQKYEAWLAEAESRVLSNRGD SLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSPIGHCVAIYHFEGSSEGTISMAEGEDLSLME -------------------------------------------------------------- >70140_70140_4_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000600603_TRIP10_chr19_6746462_ENST00000600428_length(transcript)=1910nt_BP=967nt GCAATCCGTACCCTCAGTGGGTTCCCTTTCAGTGGGTTCCTTTGTCCCCAGGCCCATTATTCCGTCCTCCCCTCTTCCCTGATGTATTTT GGCGCGGTCTCCTGGCTCTGCGGGCCCAGGGCTCCGGATGAGGTCTCCCGCCGTCCCGACCCCCGCAAGGGGCCAGCTTGGTGTCGCCTT CGTTCTTCTGCCACCCCATTCGGAAGGTGCTCGGGTCTTCGGGGCACTGGGTCCCATCGGTCCCTCCTCACCTGGGCTCACCCTCGGGGG TCTGGCCGTGAGCGAGCACCGGCTCAGCAACAAGCTGCTGGCTTGGAGCGGCGTCCTCGAGTGGCAGGAGAAGCGCAGACCCTACTCTGA CTCCACTGCAAAGCTGAAGCGGACCCTGCCCTGCCAAGCCTACGTGAACCAAGGCGAGAACCTGGAGACCGACCAGTGGCCGCAGAAGCT GATCATGCAGCTGATCCCTCAGCAGCTGCTGACCACCCTGGGCCCCCTGTTCCGGAACTCCCAGTTGGCACAGTTCCACTTCACCAACAG AGACTGCGACTCGCTCAAGGGGCTCTGCCGCATCATGGGCAACGGCTTCGCGGGCTGCATGCTGTTCCCCCACATCTCCCCCTGTGAGGT GCGCGTGCTCATGCTCCTGTACTCGTCCAAGAAGAAGATCTTCATGGGCCTCATCCCCTACGACCAGAGCGGCTTCGTCAGTGCCATCCG GCAGGTCATCACCACCCGCAAGCAGGCAGTGGGACCTGGTGGTGTCAACTCAGGCCCAGTCCAGATCGTCAACAACAAGTTTCTGGCATG GAGTGGTGTCATGGAGTGGCAGGAGCCCAGGCCTGAGCCCAACAGTCGGTCCAAGAGGTGGCTGCCATCCCACGTCTACGTGAACCAGGG GGAGATCCTGAGGACCGAGCAGTGGCCAAGGAAGCTGTACATGCAGCTCATCCCGCAGCAGCTGCTGACAGTGGTGACCGAGGATTTTAG CCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAGGAGGTTGACCAGAGGGA AGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAGATCGCTGAAACCCTGAG CAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTAGCAACCGGGGAGACAG CCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCACAGGACACCAAGGAGAG CTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAGGAACCCACATCCCCCAT AGGTCACTGTGTGGCCATCTACCACTTTGAAGGGTCCAGCGAGGGCACTATCTCTATGGCCGAGGGTGAAGACCTCAGTCTTATGGAAGA AGACAAAGGGGACGGCTGGACCCGGGTCAGGCGGAAAGAGGGAGGCGAGGGCTACGTGCCCACCTCCTACCTCCGAGTCACGCTCAATTG AACCCTGCCAGAGACGGGAAGAGGGGGGCTGTCGGCTGCTGCTTCTGGGCCACGGGGAGCCCCAGGACCTATGCACTTTATTTCTGACCC CGTGGCTTCGGCTGAGACCTGTGTAACCTGCTGCCCCCTCCACCCCCAACCCAGTCCTACCTGTCACACCGGACGGACCCGCTGTGCCTT CTACCATCGTTCCACCATTGATGTACATACTCATGTTTTACATCTTTTCTTTCTGCCGCTCGGCTCCGGCCATTTTGTTTTATACAAAAA >70140_70140_4_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000600603_TRIP10_chr19_6746462_ENST00000600428_length(amino acids)=510AA_BP=291 MARSPGSAGPGLRMRSPAVPTPARGQLGVAFVLLPPHSEGARVFGALGPIGPSSPGLTLGGLAVSEHRLSNKLLAWSGVLEWQEKRRPYS DSTAKLKRTLPCQAYVNQGENLETDQWPQKLIMQLIPQQLLTTLGPLFRNSQLAQFHFTNRDCDSLKGLCRIMGNGFAGCMLFPHISPCE VRVLMLLYSSKKKIFMGLIPYDQSGFVSAIRQVITTRKQAVGPGGVNSGPVQIVNNKFLAWSGVMEWQEPRPEPNSRSKRWLPSHVYVNQ GEILRTEQWPRKLYMQLIPQQLLTVVTEDFSHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQMGDPASLEPQIAETL SNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSP -------------------------------------------------------------- >70140_70140_5_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000601638_TRIP10_chr19_6746462_ENST00000600428_length(transcript)=1836nt_BP=893nt TTCCCTGATGTATTTTGGCGCGGTCTCCTGGCTCTGCGGGCCCAGGGCTCCGGATGAGGTCTCCCGCCGTCCCGACCCCCGCAAGGGGCC AGCTTGGTGTCGCCTTCGTTCTTCTGCCACCCCATTCGGAAGGTGCTCGGGTCTTCGGGGCACTGGGTCCCATCGGTCCCTCCTCACCTG GGCTCACCCTCGGGGGTCTGGCCGTGAGCGAGCACCGGCTCAGCAACAAGCTGCTGGCTTGGAGCGGCGTCCTCGAGTGGCAGGAGAAGC GCAGACCCTACTCTGACTCCACTGCAAAGCTGAAGCGGACCCTGCCCTGCCAAGCCTACGTGAACCAAGGCGAGAACCTGGAGACCGACC AGTGGCCGCAGAAGCTGATCATGCAGCTGATCCCTCAGCAGCTGCTGACCACCCTGGGCCCCCTGTTCCGGAACTCCCAGTTGGCACAGT TCCACTTCACCAACAGAGACTGCGACTCGCTCAAGGGGCTCTGCCGCATCATGGGCAACGGCTTCGCGGGCTGCATGCTGTTCCCCCACA TCTCCCCCTGTGAGGTGCGCGTGCTCATGCTCCTGTACTCGTCCAAGAAGAAGATCTTCATGGGCCTCATCCCCTACGACCAGAGCGGCT TCGTCAGTGCCATCCGGCAGGTCATCACCACCCGCAAGCAGGCAGTGGGACCTGGTGGTGTCAACTCAGGCCCAGTCCAGATCGTCAACA ACAAGTTTCTGGCATGGAGTGGTGTCATGGAGTGGCAGGAGCCCAGGCCTGAGCCCAACAGTCGGTCCAAGAGGTGGCTGCCATCCCACG TCTACGTGAACCAGGGGGAGATCCTGAGGACCGAGCAGTGGCCAAGGAAGCTGTACATGCAGCTCATCCCGCAGCAGCTGCTGACAGTGG TGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAGG AGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAGA TCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTA GCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCAC AGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAGG AACCCACATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGGGTCCAGCGAGGGCACTATCTCTATGGCCGAGGGTGAAGACC TCAGTCTTATGGAAGAAGACAAAGGGGACGGCTGGACCCGGGTCAGGCGGAAAGAGGGAGGCGAGGGCTACGTGCCCACCTCCTACCTCC GAGTCACGCTCAATTGAACCCTGCCAGAGACGGGAAGAGGGGGGCTGTCGGCTGCTGCTTCTGGGCCACGGGGAGCCCCAGGACCTATGC ACTTTATTTCTGACCCCGTGGCTTCGGCTGAGACCTGTGTAACCTGCTGCCCCCTCCACCCCCAACCCAGTCCTACCTGTCACACCGGAC GGACCCGCTGTGCCTTCTACCATCGTTCCACCATTGATGTACATACTCATGTTTTACATCTTTTCTTTCTGCCGCTCGGCTCCGGCCATT >70140_70140_5_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000601638_TRIP10_chr19_6746462_ENST00000600428_length(amino acids)=510AA_BP=291 MARSPGSAGPGLRMRSPAVPTPARGQLGVAFVLLPPHSEGARVFGALGPIGPSSPGLTLGGLAVSEHRLSNKLLAWSGVLEWQEKRRPYS DSTAKLKRTLPCQAYVNQGENLETDQWPQKLIMQLIPQQLLTTLGPLFRNSQLAQFHFTNRDCDSLKGLCRIMGNGFAGCMLFPHISPCE VRVLMLLYSSKKKIFMGLIPYDQSGFVSAIRQVITTRKQAVGPGGVNSGPVQIVNNKFLAWSGVMEWQEPRPEPNSRSKRWLPSHVYVNQ GEILRTEQWPRKLYMQLIPQQLLTVVTEDFSHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQMGDPASLEPQIAETL SNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSP -------------------------------------------------------------- >70140_70140_6_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000601675_TRIP10_chr19_6746462_ENST00000600428_length(transcript)=1983nt_BP=1040nt CTCCCCCGAAGCCGCGCGCCCGCGCCCGCGCCCCTCAGTCGGTGGAGCCCGCAGCCCCCCTTGTGGCCCGCGGCAGCTCCCCGCCCGCTC GGCCCGCGCCCGCCATGGTCCGTCCGCGCCGTGCCCCGTACCGCTCCGGCGCCGGGGGCCCCCTCGGGGGTCGCGGCCGCCCTCCGCGGC CCCTCGTGGTGCGCGCCGTCCGCTCGCGCTCCTGGCCTGCCAGCCCCCGAGGCCCGCAGCCTCCGCGGATCCGGGCCCGCTCGGCCCCTC CCATGGAAGGTGCTCGGGTCTTCGGGGCACTGGGTCCCATCGGTCCCTCCTCACCTGGGCTCACCCTCGGGGGTCTGGCCGTGAGCGAGC ACCGGCTCAGCAACAAGCTGCTGGCTTGGAGCGGCGTCCTCGAGTGGCAGGAGAAGCGCAGACCCTACTCTGACTCCACTGCAAAGCTGA AGCGGACCCTGCCCTGCCAAGCCTACGTGAACCAAGGCGAGAACCTGGAGACCGACCAGTGGCCGCAGAAGCTGATCATGCAGCTGATCC CTCAGCAGCTGCTGACCACCCTGGGCCCCCTGTTCCGGAACTCCCAGTTGGCACAGTTCCACTTCACCAACAGAGACTGCGACTCGCTCA AGGGGCTCTGCCGCATCATGGGCAACGGCTTCGCGGGCTGCATGCTGTTCCCCCACATCTCCCCCTGTGAGGTGCGCGTGCTCATGCTCC TGTACTCGTCCAAGAAGAAGATCTTCATGGGCCTCATCCCCTACGACCAGAGCGGCTTCGTCAGTGCCATCCGGCAGGTCATCACCACCC GCAAGCAGGCAGTGGGACCTGGTGGTGTCAACTCAGGCCCAGTCCAGATCGTCAACAACAAGTTTCTGGCATGGAGTGGTGTCATGGAGT GGCAGGAGCCCAGGCCTGAGCCCAACAGTCGGTCCAAGAGGTGGCTGCCATCCCACGTCTACGTGAACCAGGGGGAGATCCTGAGGACCG AGCAGTGGCCAAGGAAGCTGTACATGCAGCTCATCCCGCAGCAGCTGCTGACAGTGGTGACCGAGGATTTTAGCCACTTGCCCCCAGAGC AGCAGCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAGGAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGA AGGATGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAGATCGCTGAAACCCTGAGCAACATTGAACGGCTGA AATTGGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTAGCAACCGGGGAGACAGCCTGAGCCGGCACGCCC GGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCACAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCT CAGAAGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAGGAACCCACATCCCCCATAGGTCACTGTGTGGCCA TCTACCACTTTGAAGGGTCCAGCGAGGGCACTATCTCTATGGCCGAGGGTGAAGACCTCAGTCTTATGGAAGAAGACAAAGGGGACGGCT GGACCCGGGTCAGGCGGAAAGAGGGAGGCGAGGGCTACGTGCCCACCTCCTACCTCCGAGTCACGCTCAATTGAACCCTGCCAGAGACGG GAAGAGGGGGGCTGTCGGCTGCTGCTTCTGGGCCACGGGGAGCCCCAGGACCTATGCACTTTATTTCTGACCCCGTGGCTTCGGCTGAGA CCTGTGTAACCTGCTGCCCCCTCCACCCCCAACCCAGTCCTACCTGTCACACCGGACGGACCCGCTGTGCCTTCTACCATCGTTCCACCA TTGATGTACATACTCATGTTTTACATCTTTTCTTTCTGCCGCTCGGCTCCGGCCATTTTGTTTTATACAAAAATGGGAAAAAAAAAAAAG >70140_70140_6_PTOV1-TRIP10_PTOV1_chr19_50361371_ENST00000601675_TRIP10_chr19_6746462_ENST00000600428_length(amino acids)=529AA_BP=310 MVRPRRAPYRSGAGGPLGGRGRPPRPLVVRAVRSRSWPASPRGPQPPRIRARSAPPMEGARVFGALGPIGPSSPGLTLGGLAVSEHRLSN KLLAWSGVLEWQEKRRPYSDSTAKLKRTLPCQAYVNQGENLETDQWPQKLIMQLIPQQLLTTLGPLFRNSQLAQFHFTNRDCDSLKGLCR IMGNGFAGCMLFPHISPCEVRVLMLLYSSKKKIFMGLIPYDQSGFVSAIRQVITTRKQAVGPGGVNSGPVQIVNNKFLAWSGVMEWQEPR PEPNSRSKRWLPSHVYVNQGEILRTEQWPRKLYMQLIPQQLLTVVTEDFSHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYE KTPQMGDPASLEPQIAETLSNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PTOV1-TRIP10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 546_601 | 384.0 | 602.0 | ARHGAP17%2C DAAM1%2C DIAPH1 and DIAPH2 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 546_601 | 328.0 | 546.0 | ARHGAP17%2C DAAM1%2C DIAPH1 and DIAPH2 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 529_601 | 384.0 | 602.0 | DNM1 and WASL | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 529_601 | 328.0 | 546.0 | DNM1 and WASL | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 487_541 | 384.0 | 602.0 | DNM2 and WASL | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 487_541 | 328.0 | 546.0 | DNM2 and WASL | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 544_601 | 384.0 | 602.0 | WAS | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 544_601 | 328.0 | 546.0 | WAS |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | PTOV1 | chr19:50361371 | chr19:6746462 | ENST00000221557 | + | 9 | 12 | 184_416 | 280.0 | 375.0 | FLOT1 |
| Hgene | PTOV1 | chr19:50361371 | chr19:6746462 | ENST00000391842 | + | 9 | 12 | 184_416 | 312.0 | 417.0 | FLOT1 |
| Hgene | PTOV1 | chr19:50361371 | chr19:6746462 | ENST00000599732 | + | 9 | 13 | 184_416 | 312.0 | 357.6666666666667 | FLOT1 |
| Hgene | PTOV1 | chr19:50361371 | chr19:6746462 | ENST00000600603 | + | 9 | 13 | 184_416 | 280.0 | 365.6666666666667 | FLOT1 |
| Hgene | PTOV1 | chr19:50361371 | chr19:6746462 | ENST00000601675 | + | 9 | 13 | 184_416 | 312.0 | 383.6666666666667 | FLOT1 |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 293_537 | 384.0 | 602.0 | CDC42 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 293_537 | 328.0 | 546.0 | CDC42 | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313244 | 9 | 15 | 293_601 | 384.0 | 602.0 | PDE6G | |
| Tgene | TRIP10 | chr19:50361371 | chr19:6746462 | ENST00000313285 | 8 | 14 | 293_601 | 328.0 | 546.0 | PDE6G |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PTOV1-TRIP10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PTOV1-TRIP10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
