
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:PTPN14-MARK3 (FusionGDB2 ID:70219) |
Fusion Gene Summary for PTPN14-MARK3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: PTPN14-MARK3 | Fusion gene ID: 70219 | Hgene | Tgene | Gene symbol | PTPN14 | MARK3 | Gene ID | 5784 | 4140 |
| Gene name | protein tyrosine phosphatase non-receptor type 14 | microtubule affinity regulating kinase 3 | |
| Synonyms | CATLPH|PEZ|PTP36 | CTAK1|KP78|PAR1A|Par-1a|VIPB | |
| Cytomap | 1q32.3-q41 | 14q32.32-q32.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tyrosine-protein phosphatase non-receptor type 14cytoskeletal-associated protein tyrosine phosphataseprotein-tyrosine phosphatase pez | MAP/microtubule affinity-regulating kinase 3C-TAK1ELKL motif kinase 2EMK-2cdc25C-associated protein kinase 1protein kinase STK10ser/Thr protein kinase PAR-1serine/threonine-protein kinase p78 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | P27448 | |
| Ensembl transtripts involved in fusion gene | ENST00000366956, ENST00000543945, ENST00000491277, | ENST00000561071, ENST00000216288, ENST00000303622, ENST00000335102, ENST00000416682, ENST00000429436, ENST00000440884, ENST00000553942, | |
| Fusion gene scores | * DoF score | 9 X 11 X 5=495 | 16 X 19 X 9=2736 |
| # samples | 11 | 21 | |
| ** MAII score | log2(11/495*10)=-2.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(21/2736*10)=-3.70360699721978 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: PTPN14 [Title/Abstract] AND MARK3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | PTPN14(214637972)-MARK3(103928741), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | PTPN14-MARK3 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. PTPN14-MARK3 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. PTPN14-MARK3 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. PTPN14-MARK3 seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | PTPN14 | GO:0046825 | regulation of protein export from nucleus | 22948661 |
| Tgene | MARK3 | GO:0018105 | peptidyl-serine phosphorylation | 9543386 |
| Tgene | MARK3 | GO:0032092 | positive regulation of protein binding | 9543386 |
| Tgene | MARK3 | GO:0035331 | negative regulation of hippo signaling | 28087714 |
| Tgene | MARK3 | GO:0036289 | peptidyl-serine autophosphorylation | 9543386 |
 Fusion gene breakpoints across PTPN14 (5'-gene) Fusion gene breakpoints across PTPN14 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
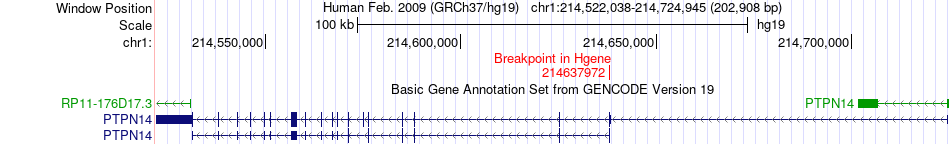 |
 Fusion gene breakpoints across MARK3 (3'-gene) Fusion gene breakpoints across MARK3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
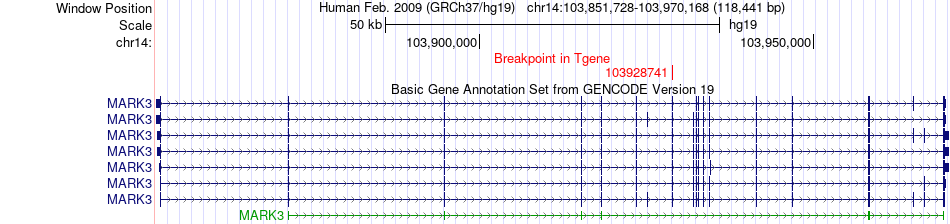 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-2297 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
Top |
Fusion Gene ORF analysis for PTPN14-MARK3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000366956 | ENST00000561071 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| 5CDS-intron | ENST00000543945 | ENST00000561071 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000216288 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000303622 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000335102 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000416682 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000429436 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000440884 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| Frame-shift | ENST00000366956 | ENST00000553942 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000216288 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000303622 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000335102 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000416682 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000429436 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000440884 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| In-frame | ENST00000543945 | ENST00000553942 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000216288 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000303622 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000335102 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000416682 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000429436 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000440884 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-3CDS | ENST00000491277 | ENST00000553942 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
| intron-intron | ENST00000491277 | ENST00000561071 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000440884 | MARK3 | chr14 | 103928741 | + | 1696 | 174 | 0 | 1670 | 556 |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000416682 | MARK3 | chr14 | 103928741 | + | 2014 | 174 | 0 | 1880 | 626 |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000429436 | MARK3 | chr14 | 103928741 | + | 2552 | 174 | 0 | 1952 | 650 |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000303622 | MARK3 | chr14 | 103928741 | + | 2485 | 174 | 0 | 1880 | 626 |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000216288 | MARK3 | chr14 | 103928741 | + | 2432 | 174 | 0 | 1832 | 610 |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000553942 | MARK3 | chr14 | 103928741 | + | 1944 | 174 | 0 | 1925 | 641 |
| ENST00000543945 | PTPN14 | chr1 | 214637972 | - | ENST00000335102 | MARK3 | chr14 | 103928741 | + | 1953 | 174 | 0 | 1952 | 650 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000543945 | ENST00000440884 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.004527659 | 0.9954724 |
| ENST00000543945 | ENST00000416682 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.007230932 | 0.9927691 |
| ENST00000543945 | ENST00000429436 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.002687376 | 0.9973126 |
| ENST00000543945 | ENST00000303622 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.002588115 | 0.9974119 |
| ENST00000543945 | ENST00000216288 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.002722023 | 0.99727803 |
| ENST00000543945 | ENST00000553942 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.009804621 | 0.99019533 |
| ENST00000543945 | ENST00000335102 | PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 0.009097172 | 0.99090284 |
Top |
Fusion Genomic Features for PTPN14-MARK3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 1.01E-07 | 0.9999999 |
| PTPN14 | chr1 | 214637972 | - | MARK3 | chr14 | 103928741 | + | 1.01E-07 | 0.9999999 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
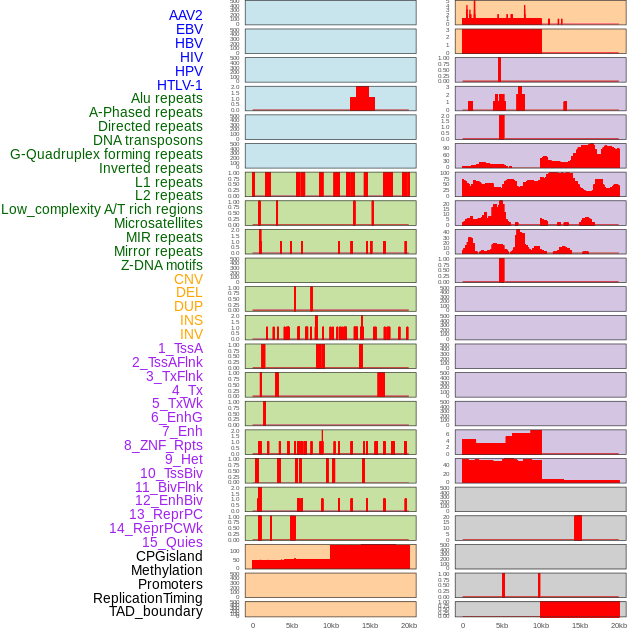 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
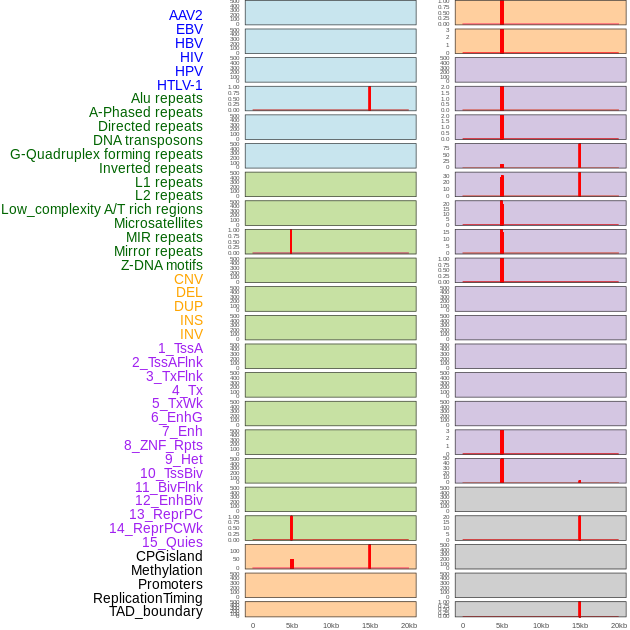 |
Top |
Fusion Protein Features for PTPN14-MARK3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:214637972/chr14:103928741) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MARK3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Serine/threonine-protein kinase (PubMed:23666762). Involved in the specific phosphorylation of microtubule-associated proteins for MAP2 and MAP4. Phosphorylates the microtubule-associated protein MAPT/TAU (PubMed:23666762). Phosphorylates CDC25C on 'Ser-216'. Regulates localization and activity of some histone deacetylases by mediating phosphorylation of HDAC7, promoting subsequent interaction between HDAC7 and 14-3-3 and export from the nucleus (PubMed:16980613). Negatively regulates the Hippo signaling pathway and antagonizes the phosphorylation of LATS1. Cooperates with DLG5 to inhibit the kinase activity of STK3/MST2 toward LATS1 (PubMed:28087714). {ECO:0000269|PubMed:16980613, ECO:0000269|PubMed:23666762, ECO:0000269|PubMed:28087714}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000216288 | 5 | 16 | 326_365 | 161 | 714.0 | Domain | UBA | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000216288 | 5 | 16 | 704_753 | 161 | 714.0 | Domain | KA1 | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000303622 | 5 | 16 | 326_365 | 161 | 730.0 | Domain | UBA | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000303622 | 5 | 16 | 704_753 | 161 | 730.0 | Domain | KA1 | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000416682 | 6 | 17 | 326_365 | 184 | 753.0 | Domain | UBA | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000416682 | 6 | 17 | 704_753 | 184 | 753.0 | Domain | KA1 | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000429436 | 5 | 18 | 326_365 | 161 | 754.0 | Domain | UBA | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000429436 | 5 | 18 | 704_753 | 161 | 754.0 | Domain | KA1 | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000440884 | 5 | 16 | 326_365 | 161 | 660.0 | Domain | UBA | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000440884 | 5 | 16 | 704_753 | 161 | 660.0 | Domain | KA1 | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000553942 | 5 | 17 | 326_365 | 161 | 745.0 | Domain | UBA | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000553942 | 5 | 17 | 704_753 | 161 | 745.0 | Domain | KA1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | PTPN14 | chr1:214637972 | chr14:103928741 | ENST00000366956 | - | 2 | 19 | 566_573 | 58 | 1188.0 | Compositional bias | Note=Poly-Pro |
| Hgene | PTPN14 | chr1:214637972 | chr14:103928741 | ENST00000366956 | - | 2 | 19 | 709_716 | 58 | 1188.0 | Compositional bias | Note=Poly-Glu |
| Hgene | PTPN14 | chr1:214637972 | chr14:103928741 | ENST00000366956 | - | 2 | 19 | 21_306 | 58 | 1188.0 | Domain | FERM |
| Hgene | PTPN14 | chr1:214637972 | chr14:103928741 | ENST00000366956 | - | 2 | 19 | 909_1180 | 58 | 1188.0 | Domain | Tyrosine-protein phosphatase |
| Hgene | PTPN14 | chr1:214637972 | chr14:103928741 | ENST00000366956 | - | 2 | 19 | 1121_1127 | 58 | 1188.0 | Region | Substrate binding |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000216288 | 5 | 16 | 56_307 | 161 | 714.0 | Domain | Protein kinase | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000303622 | 5 | 16 | 56_307 | 161 | 730.0 | Domain | Protein kinase | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000416682 | 6 | 17 | 56_307 | 184 | 753.0 | Domain | Protein kinase | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000429436 | 5 | 18 | 56_307 | 161 | 754.0 | Domain | Protein kinase | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000440884 | 5 | 16 | 56_307 | 161 | 660.0 | Domain | Protein kinase | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000553942 | 5 | 17 | 56_307 | 161 | 745.0 | Domain | Protein kinase | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000216288 | 5 | 16 | 62_70 | 161 | 714.0 | Nucleotide binding | ATP | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000303622 | 5 | 16 | 62_70 | 161 | 730.0 | Nucleotide binding | ATP | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000416682 | 6 | 17 | 62_70 | 184 | 753.0 | Nucleotide binding | ATP | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000429436 | 5 | 18 | 62_70 | 161 | 754.0 | Nucleotide binding | ATP | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000440884 | 5 | 16 | 62_70 | 161 | 660.0 | Nucleotide binding | ATP | |
| Tgene | MARK3 | chr1:214637972 | chr14:103928741 | ENST00000553942 | 5 | 17 | 62_70 | 161 | 745.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for PTPN14-MARK3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >70219_70219_1_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000216288_length(transcript)=2432nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGCTGAAAATCTATTGTTAGATGCCGATATGAACATTAAA ATAGCAGATTTCGGTTTTAGCAATGAATTTACTGTTGGCGGTAAACTCGACACGTTTTGTGGCAGTCCTCCATACGCAGCACCTGAGCTC TTCCAGGGCAAGAAATATGACGGGCCAGAAGTGGATGTGTGGAGTCTGGGGGTCATTTTATACACACTAGTCAGTGGCTCACTTCCCTTT GATGGGCAAAACCTAAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATTCCCTTCTACATGTCTACAGACTGTGAAAACCTT CTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATGAAGGACAGGTGGATCAATGCAGGGCATGAAGAA GATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATAGATATTATGGTGGGAATGGGATATTCACAAGAA GAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTGTTATTGGGGAGAAAATCTTCAGAGGTTAGGCCG AGCAGTGATCTCAACAACAGTACTGGCCAGTCTCCTCACCACAAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGT GACCATGCTGGACCAGCTATTCCTTCTGTTGTGGCGTATCCGAAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGA ATTTCCTCCCGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAGGGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAAT AAGGCGGATATTCCTGAACGCAAGAAAAGCTCCACTGTCCCTAGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTT TGCAGTGAGAGAACTACAGCTGATAGACACTCAGTGATTCAGAATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCT TCAACACACAGTATCAGTAGTGCAGCCACCCCAGATCGAATCCGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCC CGGGAACGGCGAACCGCAACATATAATGGCCCTCCTGCCTCTCCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGA GGCTCCACTAATCTCTTTAGTAAATTAACTTCAAAACTCACAAGGAGTCGCAATGTATCTGCTGAGCAAAAAGATGAAAACAAAGAAGCA AAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACTAGTTCAATGGATCCCGGGGACATGATGCGGGAAATCCGCAAAGTGTTG GACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTGCTCTTCTGCGTCCACGGAGATGGGCACGCGGAGAACCTCGTGCAGTGG GAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGGGTCCGGTTTAAGCGGATATCGGGGACATCCATAGCCTTCAAAAATATT GCTTCCAAAATTGCCAATGAGCTAAAGCTGTAACCCAGTGATTATGATGTAAATTAAGTAGCAATTAAAGTGTTTTCCTGAACACTGATG GAAATGTATAGAATAATATTTAGGCAATAACGTCTGCATCTTCTAAATCATGAAATTAAAGTCTGAGGACGAGAGCACGCCTGGGAGCGA AAGCTGGCCTTTTTTCTACGAATGCACTACATTAAAGATGTGCAACCTATGCGCCCCCTGCCCTACTTCCGTTACCCTGAGAGTCGGTGT GTGGCCCCATCTCCATGTGCCTCCCGTCTGGGTGGGTGTGAGAGTGGACGGTATGTGTGTGAAGTGGTGTATATGGAAGCATCTCCCTAC ACTGGCAGCCAGTCATTACTAGTACCTCTGCGGGAGATCATCCGGTGCTAAAACATTACAGTTGCCAAGGAGGAAAATACTGAATGACTG CTAAGAATTAACCTTAAGACCAGTTCATAGTTAATACAGGTTTACAGTTCATGCCTGTGGTTTTGTGTTTGTTGTTTTGTGTTTTTTTAG TGCAAAAGGTTTAAATTTATAGTTGTGAACATTGCTTGTGTGTGTTTTTCTAAGTAGATTCACAAGATAATTAAAAATTCACTTTTTCTC >70219_70219_1_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000216288_length(amino acids)=610AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKAENLLLDADMNIK IADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENL LKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSEVRP SSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGKGIAPASPMLGNASNPN KADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRIRFPRGTASRSTFHGQP RERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRSRNVSAEQKDENKEAKPRSLRFTWSMKTTSSMDPGDMMREIRKVL -------------------------------------------------------------- >70219_70219_2_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000303622_length(transcript)=2485nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGCTGAAAATCTATTGTTAGATGCCGATATGAACATTAAA ATAGCAGATTTCGGTTTTAGCAATGAATTTACTGTTGGCGGTAAACTCGACACGTTTTGTGGCAGTCCTCCATACGCAGCACCTGAGCTC TTCCAGGGCAAGAAATATGACGGGCCAGAAGTGGATGTGTGGAGTCTGGGGGTCATTTTATACACACTAGTCAGTGGCTCACTTCCCTTT GATGGGCAAAACCTAAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATTCCCTTCTACATGTCTACAGACTGTGAAAACCTT CTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATGAAGGACAGGTGGATCAATGCAGGGCATGAAGAA GATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATAGATATTATGGTGGGAATGGGATATTCACAAGAA GAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTGTTATTGGGGAGAAAATCTTCAGAGCTGGATGCT AGTGATTCCAGTTCTAGCAGCAATCTTTCACTTGCTAAGGTTAGGCCGAGCAGTGATCTCAACAACAGTACTGGCCAGTCTCCTCACCAC AAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGTGACCATGCTGGACCAGCTATTCCTTCTGTTGTGGCGTATCCG AAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGAATTTCCTCCCGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAG GGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAATAAGGCGGATATTCCTGAACGCAAGAAAAGCTCCACTGTCCCT AGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTTTGCAGTGAGAGAACTACAGCTGATAGACACTCAGTGATTCAG AATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCTTCAACACACAGTATCAGTAGTGCAGCCACCCCAGATCGAATC CGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCCCGGGAACGGCGAACCGCAACATATAATGGCCCTCCTGCCTCT CCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGAGGCTCCACTAATCTCTTTAGTAAATTAACTTCAAAACTCACA AGGAGTCGCAATGTATCTGCTGAGCAAAAAGATGAAAACAAAGAAGCAAAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACT AGTTCAATGGATCCCGGGGACATGATGCGGGAAATCCGCAAAGTGTTGGACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTG CTCTTCTGCGTCCACGGAGATGGGCACGCGGAGAACCTCGTGCAGTGGGAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGG GTCCGGTTTAAGCGGATATCGGGGACATCCATAGCCTTCAAAAATATTGCTTCCAAAATTGCCAATGAGCTAAAGCTGTAACCCAGTGAT TATGATGTAAATTAAGTAGCAATTAAAGTGTTTTCCTGAACACTGATGGAAATGTATAGAATAATATTTAGGCAATAACGTCTGCATCTT CTAAATCATGAAATTAAAGTCTGAGGACGAGAGCACGCCTGGGAGCGAAAGCTGGCCTTTTTTCTACGAATGCACTACATTAAAGATGTG CAACCTATGCGCCCCCTGCCCTACTTCCGTTACCCTGAGAGTCGGTGTGTGGCCCCATCTCCATGTGCCTCCCGTCTGGGTGGGTGTGAG AGTGGACGGTATGTGTGTGAAGTGGTGTATATGGAAGCATCTCCCTACACTGGCAGCCAGTCATTACTAGTACCTCTGCGGGAGATCATC CGGTGCTAAAACATTACAGTTGCCAAGGAGGAAAATACTGAATGACTGCTAAGAATTAACCTTAAGACCAGTTCATAGTTAATACAGGTT TACAGTTCATGCCTGTGGTTTTGTGTTTGTTGTTTTGTGTTTTTTTAGTGCAAAAGGTTTAAATTTATAGTTGTGAACATTGCTTGTGTG >70219_70219_2_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000303622_length(amino acids)=626AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKAENLLLDADMNIK IADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENL LKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSELDA SDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGK GIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRI RFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRSRNVSAEQKDENKEAKPRSLRFTWSMKTT -------------------------------------------------------------- >70219_70219_3_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000335102_length(transcript)=1953nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGCTGAAAATCTATTGTTAGATGCCGATATGAACATTAAA ATAGCAGATTTCGGTTTTAGCAATGAATTTACTGTTGGCGGTAAACTCGACACGTTTTGTGGCAGTCCTCCATACGCAGCACCTGAGCTC TTCCAGGGCAAGAAATATGACGGGCCAGAAGTGGATGTGTGGAGTCTGGGGGTCATTTTATACACACTAGTCAGTGGCTCACTTCCCTTT GATGGGCAAAACCTAAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATTCCCTTCTACATGTCTACAGACTGTGAAAACCTT CTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATGAAGGACAGGTGGATCAATGCAGGGCATGAAGAA GATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATAGATATTATGGTGGGAATGGGATATTCACAAGAA GAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTGTTATTGGGGAGAAAATCTTCAGAGCTGGATGCT AGTGATTCCAGTTCTAGCAGCAATCTTTCACTTGCTAAGGTTAGGCCGAGCAGTGATCTCAACAACAGTACTGGCCAGTCTCCTCACCAC AAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGTGACCATGCTGGACCAGCTATTCCTTCTGTTGTGGCGTATCCG AAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGAATTTCCTCCCGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAG GGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAATAAGGCGGATATTCCTGAACGCAAGAAAAGCTCCACTGTCCCT AGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTTTGCAGTGAGAGAACTACAGCTGATAGACACTCAGTGATTCAG AATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCTTCAACACACAGTATCAGTAGTGCAGCCACCCCAGATCGAATC CGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCCCGGGAACGGCGAACCGCAACATATAATGGCCCTCCTGCCTCT CCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGAGGCTCCACTAATCTCTTTAGTAAATTAACTTCAAAACTCACA AGGAGAAACATGTCATTCAGGTTTATCAAAAGGCTTCCAACTGAATATGAGAGGAACGGGAGATATGAGGGCTCAAGTCGCAATGTATCT GCTGAGCAAAAAGATGAAAACAAAGAAGCAAAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACTAGTTCAATGGATCCCGGG GACATGATGCGGGAAATCCGCAAAGTGTTGGACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTGCTCTTCTGCGTCCACGGA GATGGGCACGCGGAGAACCTCGTGCAGTGGGAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGGGTCCGGTTTAAGCGGATA >70219_70219_3_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000335102_length(amino acids)=650AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKAENLLLDADMNIK IADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENL LKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSELDA SDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGK GIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRI RFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRRNMSFRFIKRLPTEYERNGRYEGSSRNVS AEQKDENKEAKPRSLRFTWSMKTTSSMDPGDMMREIRKVLDANNCDYEQRERFLLFCVHGDGHAENLVQWEMEVCKLPRLSLNGVRFKRI -------------------------------------------------------------- >70219_70219_4_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000416682_length(transcript)=2014nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGCTGAAAATCTATTGTTAGATGCCGATATGAACATTAAA ATAGCAGATTTCGGTTTTAGCAATGAATTTACTGTTGGCGGTAAACTCGACACGTTTTGTGGCAGTCCTCCATACGCAGCACCTGAGCTC TTCCAGGGCAAGAAATATGACGGGCCAGAAGTGGATGTGTGGAGTCTGGGGGTCATTTTATACACACTAGTCAGTGGCTCACTTCCCTTT GATGGGCAAAACCTAAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATTCCCTTCTACATGTCTACAGACTGTGAAAACCTT CTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATGAAGGACAGGTGGATCAATGCAGGGCATGAAGAA GATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATAGATATTATGGTGGGAATGGGATATTCACAAGAA GAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTGTTATTGGGGAGAAAATCTTCAGAGCTGGATGCT AGTGATTCCAGTTCTAGCAGCAATCTTTCACTTGCTAAGGTTAGGCCGAGCAGTGATCTCAACAACAGTACTGGCCAGTCTCCTCACCAC AAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGTGACCATGCTGGACCAGCTATTCCTTCTGTTGTGGCGTATCCG AAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGAATTTCCTCCCGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAG GGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAATAAGGCGGATATTCCTGAACGCAAGAAAAGCTCCACTGTCCCT AGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTTTGCAGTGAGAGAACTACAGCTGATAGACACTCAGTGATTCAG AATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCTTCAACACACAGTATCAGTAGTGCAGCCACCCCAGATCGAATC CGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCCCGGGAACGGCGAACCGCAACATATAATGGCCCTCCTGCCTCT CCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGAGGCTCCACTAATCTCTTTAGTAAATTAACTTCAAAACTCACA AGGAGTCGCAATGTATCTGCTGAGCAAAAAGATGAAAACAAAGAAGCAAAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACT AGTTCAATGGATCCCGGGGACATGATGCGGGAAATCCGCAAAGTGTTGGACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTG CTCTTCTGCGTCCACGGAGATGGGCACGCGGAGAACCTCGTGCAGTGGGAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGG GTCCGGTTTAAGCGGATATCGGGGACATCCATAGCCTTCAAAAATATTGCTTCCAAAATTGCCAATGAGCTAAAGCTGTAACCCAGTGAT TATGATGTAAATTAAGTAGCAATTAAAGTGTTTTCCTGAACACTGATGGAAATGTATAGAATAATATTTAGGCAATAACGTCTGCATCTT >70219_70219_4_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000416682_length(amino acids)=626AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKAENLLLDADMNIK IADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENL LKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSELDA SDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGK GIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRI RFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRSRNVSAEQKDENKEAKPRSLRFTWSMKTT -------------------------------------------------------------- >70219_70219_5_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000429436_length(transcript)=2552nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGCTGAAAATCTATTGTTAGATGCCGATATGAACATTAAA ATAGCAGATTTCGGTTTTAGCAATGAATTTACTGTTGGCGGTAAACTCGACACGTTTTGTGGCAGTCCTCCATACGCAGCACCTGAGCTC TTCCAGGGCAAGAAATATGACGGGCCAGAAGTGGATGTGTGGAGTCTGGGGGTCATTTTATACACACTAGTCAGTGGCTCACTTCCCTTT GATGGGCAAAACCTAAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATTCCCTTCTACATGTCTACAGACTGTGAAAACCTT CTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATGAAGGACAGGTGGATCAATGCAGGGCATGAAGAA GATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATAGATATTATGGTGGGAATGGGATATTCACAAGAA GAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTGTTATTGGGGAGAAAATCTTCAGAGCTGGATGCT AGTGATTCCAGTTCTAGCAGCAATCTTTCACTTGCTAAGGTTAGGCCGAGCAGTGATCTCAACAACAGTACTGGCCAGTCTCCTCACCAC AAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGTGACCATGCTGGACCAGCTATTCCTTCTGTTGTGGCGTATCCG AAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGAATTTCCTCCCGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAG GGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAATAAGGCGGATATTCCTGAACGCAAGAAAAGCTCCACTGTCCCT AGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTTTGCAGTGAGAGAACTACAGCTGATAGACACTCAGTGATTCAG AATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCTTCAACACACAGTATCAGTAGTGCAGCCACCCCAGATCGAATC CGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCCCGGGAACGGCGAACCGCAACATATAATGGCCCTCCTGCCTCT CCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGAGGCTCCACTAATCTCTTTAGTAAATTAACTTCAAAACTCACA AGGAGAAACATGTCATTCAGGTTTATCAAAAGGCTTCCAACTGAATATGAGAGGAACGGGAGATATGAGGGCTCAAGTCGCAATGTATCT GCTGAGCAAAAAGATGAAAACAAAGAAGCAAAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACTAGTTCAATGGATCCCGGG GACATGATGCGGGAAATCCGCAAAGTGTTGGACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTGCTCTTCTGCGTCCACGGA GATGGGCACGCGGAGAACCTCGTGCAGTGGGAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGGGTCCGGTTTAAGCGGATA TCGGGGACATCCATAGCCTTCAAAAATATTGCTTCCAAAATTGCCAATGAGCTAAAGCTGTAACCCAGTGATTATGATGTAAATTAAGTA GCAATTAAAGTGTTTTCCTGAACACTGATGGAAATGTATAGAATAATATTTAGGCAATAACGTCTGCATCTTCTAAATCATGAAATTAAA GTCTGAGGACGAGAGCACGCCTGGGAGCGAAAGCTGGCCTTTTTTCTACGAATGCACTACATTAAAGATGTGCAACCTATGCGCCCCCTG CCCTACTTCCGTTACCCTGAGAGTCGGTGTGTGGCCCCATCTCCATGTGCCTCCCGTCTGGGTGGGTGTGAGAGTGGACGGTATGTGTGT GAAGTGGTGTATATGGAAGCATCTCCCTACACTGGCAGCCAGTCATTACTAGTACCTCTGCGGGAGATCATCCGGTGCTAAAACATTACA GTTGCCAAGGAGGAAAATACTGAATGACTGCTAAGAATTAACCTTAAGACCAGTTCATAGTTAATACAGGTTTACAGTTCATGCCTGTGG TTTTGTGTTTGTTGTTTTGTGTTTTTTTAGTGCAAAAGGTTTAAATTTATAGTTGTGAACATTGCTTGTGTGTGTTTTTCTAAGTAGATT >70219_70219_5_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000429436_length(amino acids)=650AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKAENLLLDADMNIK IADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENL LKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSELDA SDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGK GIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRI RFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRRNMSFRFIKRLPTEYERNGRYEGSSRNVS AEQKDENKEAKPRSLRFTWSMKTTSSMDPGDMMREIRKVLDANNCDYEQRERFLLFCVHGDGHAENLVQWEMEVCKLPRLSLNGVRFKRI -------------------------------------------------------------- >70219_70219_6_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000440884_length(transcript)=1696nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATT CCCTTCTACATGTCTACAGACTGTGAAAACCTTCTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATG AAGGACAGGTGGATCAATGCAGGGCATGAAGAAGATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATA GATATTATGGTGGGAATGGGATATTCACAAGAAGAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTG TTATTGGGGAGAAAATCTTCAGAGCTGGATGCTAGTGATTCCAGTTCTAGCAGCAATCTTTCACTTGCTAAGGTTAGGCCGAGCAGTGAT CTCAACAACAGTACTGGCCAGTCTCCTCACCACAAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGTGACCATGCT GGACCAGCTATTCCTTCTGTTGTGGCGTATCCGAAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGAATTTCCTCC CGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAGGGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAATAAGGCGGAT ATTCCTGAACGCAAGAAAAGCTCCACTGTCCCTAGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTTTGCAGTGAG AGAACTACAGCTGATAGACACTCAGTGATTCAGAATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCTTCAACACAC AGTATCAGTAGTGCAGCCACCCCAGATCGAATCCGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCCCGGGAACGG CGAACCGCAACATATAATGGCCCTCCTGCCTCTCCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGAGGCTCCACT AATCTCTTTAGTAAATTAACTTCAAAACTCACAAGGAGAAACATGTCATTCAGGTTTATCAAAAGTCGCAATGTATCTGCTGAGCAAAAA GATGAAAACAAAGAAGCAAAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACTAGTTCAATGGATCCCGGGGACATGATGCGG GAAATCCGCAAAGTGTTGGACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTGCTCTTCTGCGTCCACGGAGATGGGCACGCG GAGAACCTCGTGCAGTGGGAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGGGTCCGGTTTAAGCGGATATCGGGGACATCC >70219_70219_6_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000440884_length(amino acids)=556AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKELRERVLRGKYRI PFYMSTDCENLLKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYL LLGRKSSELDASDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISS RKSSGSAVGGKGIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTH SISSAATPDRIRFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRRNMSFRFIKSRNVSAEQK DENKEAKPRSLRFTWSMKTTSSMDPGDMMREIRKVLDANNCDYEQRERFLLFCVHGDGHAENLVQWEMEVCKLPRLSLNGVRFKRISGTS -------------------------------------------------------------- >70219_70219_7_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000553942_length(transcript)=1944nt_BP=174nt ATGCCTTTTGGTCTGAAGCTCCGCCGGACACGGCGCTACAACGTCCTGAGCAAGAACTGCTTTGTCACACGGATTCGCCTGCTGGACAGC AATGTTATCGAGTGCACGCTGTCGGTGGAAAGCACAGGGCAAGAATGCCTGGAGGCTGTGGCCCAGAGGCTGGAGCTGCGAGAGATTGTG TCTGCAGTTCAATACTGCCATCAGAAACGGATCGTACATCGAGACCTCAAGGCTGAAAATCTATTGTTAGATGCCGATATGAACATTAAA ATAGCAGATTTCGGTTTTAGCAATGAATTTACTGTTGGCGGTAAACTCGACACGTTTTGTGGCAGTCCTCCATACGCAGCACCTGAGCTC TTCCAGGGCAAGAAATATGACGGGCCAGAAGTGGATGTGTGGAGTCTGGGGGTCATTTTATACACACTAGTCAGTGGCTCACTTCCCTTT GATGGGCAAAACCTAAAGGAACTGAGAGAGAGAGTATTAAGAGGGAAATACAGAATTCCCTTCTACATGTCTACAGACTGTGAAAACCTT CTCAAACGTTTCCTGGTGCTAAATCCAATTAAACGCGGCACTCTAGAGCAAATCATGAAGGACAGGTGGATCAATGCAGGGCATGAAGAA GATGAACTCAAACCATTTGTTGAACCAGAGCTAGACATCTCAGACCAAAAAAGAATAGATATTATGGTGGGAATGGGATATTCACAAGAA GAAATTCAAGAATCTCTTAGTAAGATGAAATACGATGAAATCACAGCTACATATTTGTTATTGGGGAGAAAATCTTCAGAGCTGGATGCT AGTGATTCCAGTTCTAGCAGCAATCTTTCACTTGCTAAGGTTAGGCCGAGCAGTGATCTCAACAACAGTACTGGCCAGTCTCCTCACCAC AAAGTGCAGAGAAGTGTTTTTTCAAGCCAAAAGCAAAGACGCTACAGTGACCATGCTGGACCAGCTATTCCTTCTGTTGTGGCGTATCCG AAAAGGAGTCAGACCAGCACTGCAGATAGTGACCTCAAAGAAGATGGAATTTCCTCCCGGAAATCAAGTGGCAGTGCTGTTGGAGGAAAG GGAATTGCTCCAGCCAGTCCCATGCTTGGGAATGCAAGTAATCCTAATAAGGCGGATATTCCTGAACGCAAGAAAAGCTCCACTGTCCCT AGTAGTAACACAGCATCTGGTGGAATGACACGACGAAATACTTATGTTTGCAGTGAGAGAACTACAGCTGATAGACACTCAGTGATTCAG AATGGCAAAGAAAACAGCACTATTCCTGATCAGAGAACTCCAGTTGCTTCAACACACAGTATCAGTAGTGCAGCCACCCCAGATCGAATC CGCTTCCCAAGAGGCACTGCCAGTCGTAGCACTTTCCACGGCCAGCCCCGGGAACGGCGAACCGCAACATATAATGGCCCTCCTGCCTCT CCCAGCCTGTCCCATGAAGCCACACCATTGTCCCAGACTCGAAGCCGAGGCTCCACTAATCTCTTTAGTAAATTAACTTCAAAACTCACA AGGAGGCTTCCAACTGAATATGAGAGGAACGGGAGATATGAGGGCTCAAGTCGCAATGTATCTGCTGAGCAAAAAGATGAAAACAAAGAA GCAAAGCCTCGATCCCTACGCTTCACCTGGAGCATGAAAACCACTAGTTCAATGGATCCCGGGGACATGATGCGGGAAATCCGCAAAGTG TTGGACGCCAATAACTGCGACTATGAGCAGAGGGAGCGCTTCTTGCTCTTCTGCGTCCACGGAGATGGGCACGCGGAGAACCTCGTGCAG TGGGAAATGGAAGTGTGCAAGCTGCCAAGACTGTCTCTGAACGGGGTCCGGTTTAAGCGGATATCGGGGACATCCATAGCCTTCAAAAAT >70219_70219_7_PTPN14-MARK3_PTPN14_chr1_214637972_ENST00000543945_MARK3_chr14_103928741_ENST00000553942_length(amino acids)=641AA_BP=58 MPFGLKLRRTRRYNVLSKNCFVTRIRLLDSNVIECTLSVESTGQECLEAVAQRLELREIVSAVQYCHQKRIVHRDLKAENLLLDADMNIK IADFGFSNEFTVGGKLDTFCGSPPYAAPELFQGKKYDGPEVDVWSLGVILYTLVSGSLPFDGQNLKELRERVLRGKYRIPFYMSTDCENL LKRFLVLNPIKRGTLEQIMKDRWINAGHEEDELKPFVEPELDISDQKRIDIMVGMGYSQEEIQESLSKMKYDEITATYLLLGRKSSELDA SDSSSSSNLSLAKVRPSSDLNNSTGQSPHHKVQRSVFSSQKQRRYSDHAGPAIPSVVAYPKRSQTSTADSDLKEDGISSRKSSGSAVGGK GIAPASPMLGNASNPNKADIPERKKSSTVPSSNTASGGMTRRNTYVCSERTTADRHSVIQNGKENSTIPDQRTPVASTHSISSAATPDRI RFPRGTASRSTFHGQPRERRTATYNGPPASPSLSHEATPLSQTRSRGSTNLFSKLTSKLTRRLPTEYERNGRYEGSSRNVSAEQKDENKE AKPRSLRFTWSMKTTSSMDPGDMMREIRKVLDANNCDYEQRERFLLFCVHGDGHAENLVQWEMEVCKLPRLSLNGVRFKRISGTSIAFKN -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for PTPN14-MARK3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for PTPN14-MARK3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for PTPN14-MARK3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
