
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:R3HDM4-TMEM259 (FusionGDB2 ID:71107) |
Fusion Gene Summary for R3HDM4-TMEM259 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: R3HDM4-TMEM259 | Fusion gene ID: 71107 | Hgene | Tgene | Gene symbol | R3HDM4 | TMEM259 | Gene ID | 91300 | 91304 |
| Gene name | R3H domain containing 4 | transmembrane protein 259 | |
| Synonyms | C19orf22 | ASBABP1|C19orf6|MBRL|MEMBRALIN|R32184_3 | |
| Cytomap | 19p13.3 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | R3H domain-containing protein 4R3H domain-containing protein C19orf22 | membralinaspecific BCL2 ARE-binding protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q96D70 | Q4ZIN3 | |
| Ensembl transtripts involved in fusion gene | ENST00000361574, ENST00000587975, | ENST00000356663, ENST00000333175, | |
| Fusion gene scores | * DoF score | 4 X 3 X 5=60 | 6 X 6 X 5=180 |
| # samples | 7 | 8 | |
| ** MAII score | log2(7/60*10)=0.222392421336448 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(8/180*10)=-1.16992500144231 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: R3HDM4 [Title/Abstract] AND TMEM259 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | R3HDM4(913087)-TMEM259(1014472), # samples:1 R3HDM4(913087)-TMEM259(1010894), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across R3HDM4 (5'-gene) Fusion gene breakpoints across R3HDM4 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
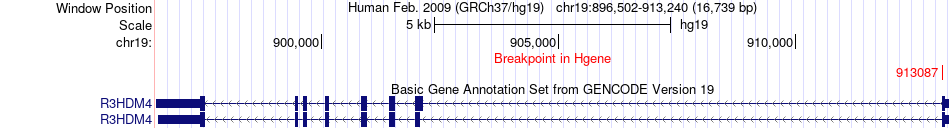 |
 Fusion gene breakpoints across TMEM259 (3'-gene) Fusion gene breakpoints across TMEM259 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
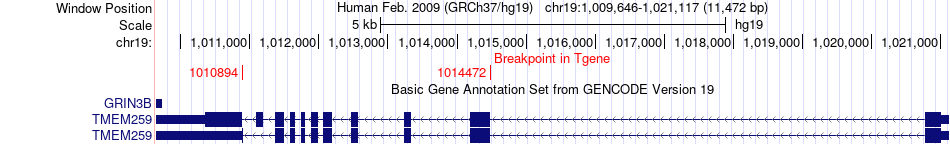 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-ZF-AA51-01A | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - |
| ChimerDB4 | GBM | TCGA-14-1402-02A | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1010894 | - |
Top |
Fusion Gene ORF analysis for R3HDM4-TMEM259 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| Frame-shift | ENST00000361574 | ENST00000356663 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1010894 | - |
| Frame-shift | ENST00000587975 | ENST00000356663 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1010894 | - |
| In-frame | ENST00000361574 | ENST00000333175 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - |
| In-frame | ENST00000361574 | ENST00000333175 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1010894 | - |
| In-frame | ENST00000361574 | ENST00000356663 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - |
| In-frame | ENST00000587975 | ENST00000333175 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - |
| In-frame | ENST00000587975 | ENST00000333175 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1010894 | - |
| In-frame | ENST00000587975 | ENST00000356663 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000361574 | R3HDM4 | chr19 | 913087 | - | ENST00000356663 | TMEM259 | chr19 | 1014472 | - | 2485 | 145 | 148 | 1782 | 544 |
| ENST00000361574 | R3HDM4 | chr19 | 913087 | - | ENST00000333175 | TMEM259 | chr19 | 1014472 | - | 2382 | 145 | 148 | 1146 | 332 |
| ENST00000587975 | R3HDM4 | chr19 | 913087 | - | ENST00000356663 | TMEM259 | chr19 | 1014472 | - | 2494 | 154 | 157 | 1791 | 544 |
| ENST00000587975 | R3HDM4 | chr19 | 913087 | - | ENST00000333175 | TMEM259 | chr19 | 1014472 | - | 2391 | 154 | 157 | 1155 | 332 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000361574 | ENST00000356663 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - | 0.005486656 | 0.9945134 |
| ENST00000361574 | ENST00000333175 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - | 0.005047562 | 0.99495244 |
| ENST00000587975 | ENST00000356663 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - | 0.00545833 | 0.9945417 |
| ENST00000587975 | ENST00000333175 | R3HDM4 | chr19 | 913087 | - | TMEM259 | chr19 | 1014472 | - | 0.00448713 | 0.9955129 |
Top |
Fusion Genomic Features for R3HDM4-TMEM259 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
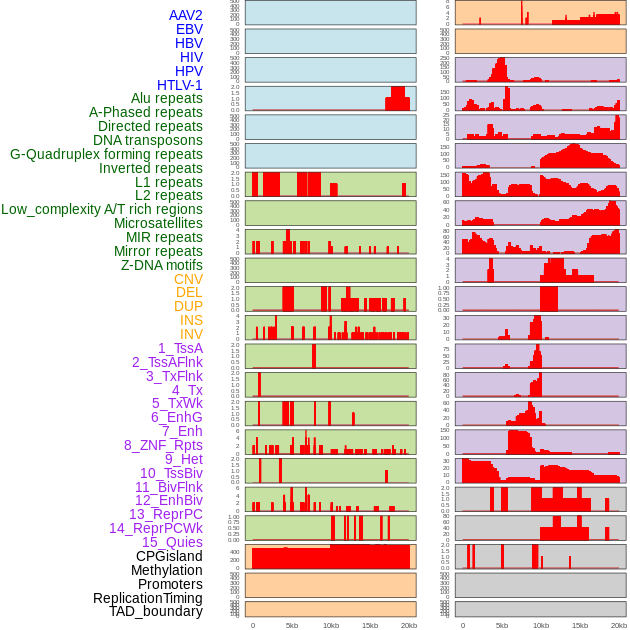 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for R3HDM4-TMEM259 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:913087/chr19:1014472) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| R3HDM4 | TMEM259 |
| FUNCTION: May have a role in the ERAD pathway required for clearance of misfolded proteins in the endoplasmic reticulum (ER). Promotes survival of motor neurons, probably by protecting against ER stress. {ECO:0000250|UniProtKB:Q8CIV2}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000333175 | 8 | 10 | 426_446 | 405 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000333175 | 0 | 10 | 302_322 | 75 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000333175 | 0 | 10 | 346_366 | 75 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000333175 | 0 | 10 | 426_446 | 75 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000356663 | 0 | 11 | 302_322 | 75 | 621.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000356663 | 0 | 11 | 346_366 | 75 | 621.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000356663 | 0 | 11 | 426_446 | 75 | 621.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | R3HDM4 | chr19:913087 | chr19:1010894 | ENST00000361574 | - | 1 | 8 | 132_184 | 23 | 269.0 | Compositional bias | Note=Arg-rich |
| Hgene | R3HDM4 | chr19:913087 | chr19:1014472 | ENST00000361574 | - | 1 | 8 | 132_184 | 23 | 269.0 | Compositional bias | Note=Arg-rich |
| Hgene | R3HDM4 | chr19:913087 | chr19:1010894 | ENST00000361574 | - | 1 | 8 | 187_250 | 23 | 269.0 | Domain | R3H |
| Hgene | R3HDM4 | chr19:913087 | chr19:1014472 | ENST00000361574 | - | 1 | 8 | 187_250 | 23 | 269.0 | Domain | R3H |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000333175 | 8 | 10 | 302_322 | 405 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000333175 | 8 | 10 | 346_366 | 405 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000333175 | 8 | 10 | 70_90 | 405 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000356663 | 9 | 11 | 302_322 | 439 | 621.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000356663 | 9 | 11 | 346_366 | 439 | 621.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000356663 | 9 | 11 | 426_446 | 439 | 621.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1010894 | ENST00000356663 | 9 | 11 | 70_90 | 439 | 621.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000333175 | 0 | 10 | 70_90 | 75 | 409.0 | Transmembrane | Helical | |
| Tgene | TMEM259 | chr19:913087 | chr19:1014472 | ENST00000356663 | 0 | 11 | 70_90 | 75 | 621.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for R3HDM4-TMEM259 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >71107_71107_1_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000361574_TMEM259_chr19_1014472_ENST00000333175_length(transcript)=2382nt_BP=145nt CGGGCGCCCGTCACTTCCTGCCCGGCCCGGTGAAGGCCCTGCCGGGCGGCGGCGGCGGCGACAGCGTGCGAGCCATGGTCGCGCTGGAGA ACCCCGAGTGCGGCCCGGAGGCGGCGGAGGGCACCCCGGGCGGGCGGCGGCTGCTGCCCTGTTTGTGCTCTTCGTCCTGGCCTACATCCA CATCGTCTTCTCCCGCTCGCCCATCAACTGCCTGGAGCATGTGCGTGACAAGTGGCCGCGTGAGGGCATCCTGCGTGTGGAAGTGCGGCA CAACTCGAGCCGCGCGCCCGTCTTCCTACAGTTCTGTGACAGCGGCGGCCGCGGGAGCTTCCCGGGCCTGGCCGTGGAACCAGGCAGCAA CCTGGACATGGAAGATGAGGAGGAGGAAGAGCTGACCATGGAGATGTTTGGGAACAGCTCCATCAAGTTTGAGCTGGACATCGAGCCCAA GGTGTTCAAGCCGCCGAGTAGCACAGAGGCCCTGAATGACAGCCAGGAGTTCCCCTTCCCCGAGACGCCCACCAAAGTGTGGCCGCAGGA CGAGTACATCGTGGAGTACTCACTAGAGTATGGCTTCCTTCGCCTGTCGCAGGCCACCCGCCAGCGCCTGAGCATCCCCGTCATGGTGGT CACCCTGGACCCCACGCGGGACCAGTGCTTCGGGGACCGCTTCAGCCGCCTGCTGCTGGATGAGTTCCTGGGCTACGATGACATCCTCAT GTCCAGCGTGAAGGGCCTGGCCGAGAACGAGGAGAACAAGGGCTTCCTGCGGAATGTGGTGTCGGGCGAGCACTACCGCTTTGTGAGCAT GTGGATGGCGCGGACGTCCTACCTGGCCGCCTTCGCCATCATGGTCATCTTCACGCTGAGCGTGTCCATGCTGCTGCGGTACTCACACCA CCAGATCTTCGTCTTCATCGTGGACCTGCTGCAGATGCTGGAGATGAACATGGCCATCGCCTTCCCCGCAGCGCCCCTGCTGACCGTCAT CCTGGCCCTCGTCGGGATGGAGGCCATCATGTCGGAGTTCTTCAACGACACCACCACCGCCTTCTACATCATCCTCATCGTGTGGCTCGC GGACCAGTATGACGCCATCTGCTGCCACACCAGCACCAGCAAGCGGCATTGGCTGCGCATTCCATGATCTACTTCTTCCACCACTACGAG CTGCCTGCCATCCTGCAGCAGGTCCGCATCCAGGAGATGCTGCTTCAGGCGCCGCCACTGGGCCCCGGGACCCCCACGGCGCTGCCCGAT GACATGAACAACAACTCGGGCGCCCCGGCTACAGCCCCTGACTCTGCCGGCCAGCCCCCCGCCCTGGGCCCCGTCTCGCCTGGGGCCAGC GGGAGTCCCGGGCCTGTGGCAGCGGCGCCCAGCTCCCTGGTGGCCGCGGCAGCCTCAGTGGCAGCAGCTGCCGGTGGTGACCTGGGTTGG ATGGCAGAGACCGCTGCCATCATCACAGACGCCTCCTTCCTGTCCGGCCTGAGCGCCTCCCTCCTGGAGCGGCGTCCAGCCAGCCCGCTG GGCCCTGCTGGGGGCCTCCCCCACGCCCCCCAGGACAGTGTCCCCCCGAGTGACTCCGCAGCTTCTGACACAACTCCCCTGGGGGCTGCG GTAGGCGGGCCTAGCCCGGCCTCCATGGCCCCAACGGAGGCGCCCTCGGAGGTGGGGTCCTGAGCCGCACAGCTGAGCCGCCTCTGACCC CTGCTGGCTGGGCCTGACCTTCCCGAGCCCGTGGGGGTGGGGGAGGCCAGCCACCTCCTTCCTCTGGGACTGCCCAGCCTGTGTCGGGGG CTTTCAGGGTTTCGTGGGGTTTGCCCGGAAGGCGGCTTTCCTCCCCCTGGTGTGAGGTCGCGCCCGAGGCTTGTACCCGCTAGTGAGGTG TTTGAGCTGGTCAGCAAGGAGAGGGGGTGGGGTTCCGCGGAAGGTTCTGGAGGGGTCTTGGTAGGTCTGCAGTGAACCGTCCTGAGGATG GAGTGGGGTCCCATGGTGCAGGTCTCTGAGCAAGGCGGAGGTGTGGAGGAGAGGCCGGCTTGGGGTGGGGCCTCGCGCCCTAGTGCCGGC CGGCCTCAGCCCGGCTCTGCCTGGTGCTCCCTGCAGTGCCTTCTCCATGGCCCCGCCCTCCCCGCGTGTGCGCCAGGCTTGGGGTCCCCG GGAGAGCAGAGCTTGCGCCTCGGGCATAGGGACGTGGGGTGCAGGCGCCAACATCAGTGGCAGCAGCCAGGGCCGTGGTCCAGTCCCACT CGGGGATGGAGTGGGCCGGCGGCCAAACCAGTCACTCGGGGAGGAATGCGGAGGAGCGCTCATTCCATTCTATTTAATTGCAGTGTACAA >71107_71107_1_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000361574_TMEM259_chr19_1014472_ENST00000333175_length(amino acids)=332AA_BP=1 MFVLFVLAYIHIVFSRSPINCLEHVRDKWPREGILRVEVRHNSSRAPVFLQFCDSGGRGSFPGLAVEPGSNLDMEDEEEEELTMEMFGNS SIKFELDIEPKVFKPPSSTEALNDSQEFPFPETPTKVWPQDEYIVEYSLEYGFLRLSQATRQRLSIPVMVVTLDPTRDQCFGDRFSRLLL DEFLGYDDILMSSVKGLAENEENKGFLRNVVSGEHYRFVSMWMARTSYLAAFAIMVIFTLSVSMLLRYSHHQIFVFIVDLLQMLEMNMAI -------------------------------------------------------------- >71107_71107_2_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000361574_TMEM259_chr19_1014472_ENST00000356663_length(transcript)=2485nt_BP=145nt CGGGCGCCCGTCACTTCCTGCCCGGCCCGGTGAAGGCCCTGCCGGGCGGCGGCGGCGGCGACAGCGTGCGAGCCATGGTCGCGCTGGAGA ACCCCGAGTGCGGCCCGGAGGCGGCGGAGGGCACCCCGGGCGGGCGGCGGCTGCTGCCCTGTTTGTGCTCTTCGTCCTGGCCTACATCCA CATCGTCTTCTCCCGCTCGCCCATCAACTGCCTGGAGCATGTGCGTGACAAGTGGCCGCGTGAGGGCATCCTGCGTGTGGAAGTGCGGCA CAACTCGAGCCGCGCGCCCGTCTTCCTACAGTTCTGTGACAGCGGCGGCCGCGGGAGCTTCCCGGGCCTGGCCGTGGAACCAGGCAGCAA CCTGGACATGGAAGATGAGGAGGAGGAAGAGCTGACCATGGAGATGTTTGGGAACAGCTCCATCAAGTTTGAGCTGGACATCGAGCCCAA GGTGTTCAAGCCGCCGAGTAGCACAGAGGCCCTGAATGACAGCCAGGAGTTCCCCTTCCCCGAGACGCCCACCAAAGTGTGGCCGCAGGA CGAGTACATCGTGGAGTACTCACTAGAGTATGGCTTCCTTCGCCTGTCGCAGGCCACCCGCCAGCGCCTGAGCATCCCCGTCATGGTGGT CACCCTGGACCCCACGCGGGACCAGTGCTTCGGGGACCGCTTCAGCCGCCTGCTGCTGGATGAGTTCCTGGGCTACGATGACATCCTCAT GTCCAGCGTGAAGGGCCTGGCCGAGAACGAGGAGAACAAGGGCTTCCTGCGGAATGTGGTGTCGGGCGAGCACTACCGCTTTGTGAGCAT GTGGATGGCGCGGACGTCCTACCTGGCCGCCTTCGCCATCATGGTCATCTTCACGCTGAGCGTGTCCATGCTGCTGCGGTACTCACACCA CCAGATCTTCGTCTTCATCGTGGACCTGCTGCAGATGCTGGAGATGAACATGGCCATCGCCTTCCCCGCAGCGCCCCTGCTGACCGTCAT CCTGGCCCTCGTCGGGATGGAGGCCATCATGTCGGAGTTCTTCAACGACACCACCACCGCCTTCTACATCATCCTCATCGTGTGGCTCGC GGACCAGTATGACGCCATCTGCTGCCACACCAGCACCAGCAAGCGGCATTGGCTGCGGTTCTTCTATCTCTACCACTTCGCCTTCTATGC CTATCACTACCGCTTCAATGGGCAGTATAGCAGCCTGGCCCTGGTCACCTCCTGGCTCTTCATCCAGCATTCCATGATCTACTTCTTCCA CCACTACGAGCTGCCTGCCATCCTGCAGCAGGTCCGCATCCAGGAGATGCTGCTTCAGGCGCCGCCACTGGGCCCCGGGACCCCCACGGC GCTGCCCGATGACATGAACAACAACTCGGGCGCCCCGGCTACAGCCCCTGACTCTGCCGGCCAGCCCCCCGCCCTGGGCCCCGTCTCGCC TGGGGCCAGCGGGAGTCCCGGGCCTGTGGCAGCGGCGCCCAGCTCCCTGGTGGCCGCGGCAGCCTCAGTGGCAGCAGCTGCCGGTGGTGA CCTGGGTTGGATGGCAGAGACCGCTGCCATCATCACAGACGCCTCCTTCCTGTCCGGCCTGAGCGCCTCCCTCCTGGAGCGGCGTCCAGC CAGCCCGCTGGGCCCTGCTGGGGGCCTCCCCCACGCCCCCCAGGACAGTGTCCCCCCGAGTGACTCCGCAGCTTCTGACACAACTCCCCT GGGGGCTGCGGTAGGCGGGCCTAGCCCGGCCTCCATGGCCCCAACGGAGGCGCCCTCGGAGGTGGGGTCCTGAGCCGCACAGCTGAGCCG CCTCTGACCCCTGCTGGCTGGGCCTGACCTTCCCGAGCCCGTGGGGGTGGGGGAGGCCAGCCACCTCCTTCCTCTGGGACTGCCCAGCCT GTGTCGGGGGCTTTCAGGGTTTCGTGGGGTTTGCCCGGAAGGCGGCTTTCCTCCCCCTGGTGTGAGGTCGCGCCCGAGGCTTGTACCCGC TAGTGAGGTGTTTGAGCTGGTCAGCAAGGAGAGGGGGTGGGGTTCCGCGGAAGGTTCTGGAGGGGTCTTGGTAGGTCTGCAGTGAACCGT CCTGAGGATGGAGTGGGGTCCCATGGTGCAGGTCTCTGAGCAAGGCGGAGGTGTGGAGGAGAGGCCGGCTTGGGGTGGGGCCTCGCGCCC TAGTGCCGGCCGGCCTCAGCCCGGCTCTGCCTGGTGCTCCCTGCAGTGCCTTCTCCATGGCCCCGCCCTCCCCGCGTGTGCGCCAGGCTT GGGGTCCCCGGGAGAGCAGAGCTTGCGCCTCGGGCATAGGGACGTGGGGTGCAGGCGCCAACATCAGTGGCAGCAGCCAGGGCCGTGGTC CAGTCCCACTCGGGGATGGAGTGGGCCGGCGGCCAAACCAGTCACTCGGGGAGGAATGCGGAGGAGCGCTCATTCCATTCTATTTAATTG >71107_71107_2_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000361574_TMEM259_chr19_1014472_ENST00000356663_length(amino acids)=544AA_BP=1 MFVLFVLAYIHIVFSRSPINCLEHVRDKWPREGILRVEVRHNSSRAPVFLQFCDSGGRGSFPGLAVEPGSNLDMEDEEEEELTMEMFGNS SIKFELDIEPKVFKPPSSTEALNDSQEFPFPETPTKVWPQDEYIVEYSLEYGFLRLSQATRQRLSIPVMVVTLDPTRDQCFGDRFSRLLL DEFLGYDDILMSSVKGLAENEENKGFLRNVVSGEHYRFVSMWMARTSYLAAFAIMVIFTLSVSMLLRYSHHQIFVFIVDLLQMLEMNMAI AFPAAPLLTVILALVGMEAIMSEFFNDTTTAFYIILIVWLADQYDAICCHTSTSKRHWLRFFYLYHFAFYAYHYRFNGQYSSLALVTSWL FIQHSMIYFFHHYELPAILQQVRIQEMLLQAPPLGPGTPTALPDDMNNNSGAPATAPDSAGQPPALGPVSPGASGSPGPVAAAPSSLVAA AASVAAAAGGDLGWMAETAAIITDASFLSGLSASLLERRPASPLGPAGGLPHAPQDSVPPSDSAASDTTPLGAAVGGPSPASMAPTEAPS -------------------------------------------------------------- >71107_71107_3_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000587975_TMEM259_chr19_1014472_ENST00000333175_length(transcript)=2391nt_BP=154nt GCCCGCCTCCGGGCGCCCGTCACTTCCTGCCCGGCCCGGTGAAGGCCCTGCCGGGCGGCGGCGGCGGCGACAGCGTGCGAGCCATGGTCG CGCTGGAGAACCCCGAGTGCGGCCCGGAGGCGGCGGAGGGCACCCCGGGCGGGCGGCGGCTGCTGCCCTGTTTGTGCTCTTCGTCCTGGC CTACATCCACATCGTCTTCTCCCGCTCGCCCATCAACTGCCTGGAGCATGTGCGTGACAAGTGGCCGCGTGAGGGCATCCTGCGTGTGGA AGTGCGGCACAACTCGAGCCGCGCGCCCGTCTTCCTACAGTTCTGTGACAGCGGCGGCCGCGGGAGCTTCCCGGGCCTGGCCGTGGAACC AGGCAGCAACCTGGACATGGAAGATGAGGAGGAGGAAGAGCTGACCATGGAGATGTTTGGGAACAGCTCCATCAAGTTTGAGCTGGACAT CGAGCCCAAGGTGTTCAAGCCGCCGAGTAGCACAGAGGCCCTGAATGACAGCCAGGAGTTCCCCTTCCCCGAGACGCCCACCAAAGTGTG GCCGCAGGACGAGTACATCGTGGAGTACTCACTAGAGTATGGCTTCCTTCGCCTGTCGCAGGCCACCCGCCAGCGCCTGAGCATCCCCGT CATGGTGGTCACCCTGGACCCCACGCGGGACCAGTGCTTCGGGGACCGCTTCAGCCGCCTGCTGCTGGATGAGTTCCTGGGCTACGATGA CATCCTCATGTCCAGCGTGAAGGGCCTGGCCGAGAACGAGGAGAACAAGGGCTTCCTGCGGAATGTGGTGTCGGGCGAGCACTACCGCTT TGTGAGCATGTGGATGGCGCGGACGTCCTACCTGGCCGCCTTCGCCATCATGGTCATCTTCACGCTGAGCGTGTCCATGCTGCTGCGGTA CTCACACCACCAGATCTTCGTCTTCATCGTGGACCTGCTGCAGATGCTGGAGATGAACATGGCCATCGCCTTCCCCGCAGCGCCCCTGCT GACCGTCATCCTGGCCCTCGTCGGGATGGAGGCCATCATGTCGGAGTTCTTCAACGACACCACCACCGCCTTCTACATCATCCTCATCGT GTGGCTCGCGGACCAGTATGACGCCATCTGCTGCCACACCAGCACCAGCAAGCGGCATTGGCTGCGCATTCCATGATCTACTTCTTCCAC CACTACGAGCTGCCTGCCATCCTGCAGCAGGTCCGCATCCAGGAGATGCTGCTTCAGGCGCCGCCACTGGGCCCCGGGACCCCCACGGCG CTGCCCGATGACATGAACAACAACTCGGGCGCCCCGGCTACAGCCCCTGACTCTGCCGGCCAGCCCCCCGCCCTGGGCCCCGTCTCGCCT GGGGCCAGCGGGAGTCCCGGGCCTGTGGCAGCGGCGCCCAGCTCCCTGGTGGCCGCGGCAGCCTCAGTGGCAGCAGCTGCCGGTGGTGAC CTGGGTTGGATGGCAGAGACCGCTGCCATCATCACAGACGCCTCCTTCCTGTCCGGCCTGAGCGCCTCCCTCCTGGAGCGGCGTCCAGCC AGCCCGCTGGGCCCTGCTGGGGGCCTCCCCCACGCCCCCCAGGACAGTGTCCCCCCGAGTGACTCCGCAGCTTCTGACACAACTCCCCTG GGGGCTGCGGTAGGCGGGCCTAGCCCGGCCTCCATGGCCCCAACGGAGGCGCCCTCGGAGGTGGGGTCCTGAGCCGCACAGCTGAGCCGC CTCTGACCCCTGCTGGCTGGGCCTGACCTTCCCGAGCCCGTGGGGGTGGGGGAGGCCAGCCACCTCCTTCCTCTGGGACTGCCCAGCCTG TGTCGGGGGCTTTCAGGGTTTCGTGGGGTTTGCCCGGAAGGCGGCTTTCCTCCCCCTGGTGTGAGGTCGCGCCCGAGGCTTGTACCCGCT AGTGAGGTGTTTGAGCTGGTCAGCAAGGAGAGGGGGTGGGGTTCCGCGGAAGGTTCTGGAGGGGTCTTGGTAGGTCTGCAGTGAACCGTC CTGAGGATGGAGTGGGGTCCCATGGTGCAGGTCTCTGAGCAAGGCGGAGGTGTGGAGGAGAGGCCGGCTTGGGGTGGGGCCTCGCGCCCT AGTGCCGGCCGGCCTCAGCCCGGCTCTGCCTGGTGCTCCCTGCAGTGCCTTCTCCATGGCCCCGCCCTCCCCGCGTGTGCGCCAGGCTTG GGGTCCCCGGGAGAGCAGAGCTTGCGCCTCGGGCATAGGGACGTGGGGTGCAGGCGCCAACATCAGTGGCAGCAGCCAGGGCCGTGGTCC AGTCCCACTCGGGGATGGAGTGGGCCGGCGGCCAAACCAGTCACTCGGGGAGGAATGCGGAGGAGCGCTCATTCCATTCTATTTAATTGC >71107_71107_3_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000587975_TMEM259_chr19_1014472_ENST00000333175_length(amino acids)=332AA_BP=1 MFVLFVLAYIHIVFSRSPINCLEHVRDKWPREGILRVEVRHNSSRAPVFLQFCDSGGRGSFPGLAVEPGSNLDMEDEEEEELTMEMFGNS SIKFELDIEPKVFKPPSSTEALNDSQEFPFPETPTKVWPQDEYIVEYSLEYGFLRLSQATRQRLSIPVMVVTLDPTRDQCFGDRFSRLLL DEFLGYDDILMSSVKGLAENEENKGFLRNVVSGEHYRFVSMWMARTSYLAAFAIMVIFTLSVSMLLRYSHHQIFVFIVDLLQMLEMNMAI -------------------------------------------------------------- >71107_71107_4_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000587975_TMEM259_chr19_1014472_ENST00000356663_length(transcript)=2494nt_BP=154nt GCCCGCCTCCGGGCGCCCGTCACTTCCTGCCCGGCCCGGTGAAGGCCCTGCCGGGCGGCGGCGGCGGCGACAGCGTGCGAGCCATGGTCG CGCTGGAGAACCCCGAGTGCGGCCCGGAGGCGGCGGAGGGCACCCCGGGCGGGCGGCGGCTGCTGCCCTGTTTGTGCTCTTCGTCCTGGC CTACATCCACATCGTCTTCTCCCGCTCGCCCATCAACTGCCTGGAGCATGTGCGTGACAAGTGGCCGCGTGAGGGCATCCTGCGTGTGGA AGTGCGGCACAACTCGAGCCGCGCGCCCGTCTTCCTACAGTTCTGTGACAGCGGCGGCCGCGGGAGCTTCCCGGGCCTGGCCGTGGAACC AGGCAGCAACCTGGACATGGAAGATGAGGAGGAGGAAGAGCTGACCATGGAGATGTTTGGGAACAGCTCCATCAAGTTTGAGCTGGACAT CGAGCCCAAGGTGTTCAAGCCGCCGAGTAGCACAGAGGCCCTGAATGACAGCCAGGAGTTCCCCTTCCCCGAGACGCCCACCAAAGTGTG GCCGCAGGACGAGTACATCGTGGAGTACTCACTAGAGTATGGCTTCCTTCGCCTGTCGCAGGCCACCCGCCAGCGCCTGAGCATCCCCGT CATGGTGGTCACCCTGGACCCCACGCGGGACCAGTGCTTCGGGGACCGCTTCAGCCGCCTGCTGCTGGATGAGTTCCTGGGCTACGATGA CATCCTCATGTCCAGCGTGAAGGGCCTGGCCGAGAACGAGGAGAACAAGGGCTTCCTGCGGAATGTGGTGTCGGGCGAGCACTACCGCTT TGTGAGCATGTGGATGGCGCGGACGTCCTACCTGGCCGCCTTCGCCATCATGGTCATCTTCACGCTGAGCGTGTCCATGCTGCTGCGGTA CTCACACCACCAGATCTTCGTCTTCATCGTGGACCTGCTGCAGATGCTGGAGATGAACATGGCCATCGCCTTCCCCGCAGCGCCCCTGCT GACCGTCATCCTGGCCCTCGTCGGGATGGAGGCCATCATGTCGGAGTTCTTCAACGACACCACCACCGCCTTCTACATCATCCTCATCGT GTGGCTCGCGGACCAGTATGACGCCATCTGCTGCCACACCAGCACCAGCAAGCGGCATTGGCTGCGGTTCTTCTATCTCTACCACTTCGC CTTCTATGCCTATCACTACCGCTTCAATGGGCAGTATAGCAGCCTGGCCCTGGTCACCTCCTGGCTCTTCATCCAGCATTCCATGATCTA CTTCTTCCACCACTACGAGCTGCCTGCCATCCTGCAGCAGGTCCGCATCCAGGAGATGCTGCTTCAGGCGCCGCCACTGGGCCCCGGGAC CCCCACGGCGCTGCCCGATGACATGAACAACAACTCGGGCGCCCCGGCTACAGCCCCTGACTCTGCCGGCCAGCCCCCCGCCCTGGGCCC CGTCTCGCCTGGGGCCAGCGGGAGTCCCGGGCCTGTGGCAGCGGCGCCCAGCTCCCTGGTGGCCGCGGCAGCCTCAGTGGCAGCAGCTGC CGGTGGTGACCTGGGTTGGATGGCAGAGACCGCTGCCATCATCACAGACGCCTCCTTCCTGTCCGGCCTGAGCGCCTCCCTCCTGGAGCG GCGTCCAGCCAGCCCGCTGGGCCCTGCTGGGGGCCTCCCCCACGCCCCCCAGGACAGTGTCCCCCCGAGTGACTCCGCAGCTTCTGACAC AACTCCCCTGGGGGCTGCGGTAGGCGGGCCTAGCCCGGCCTCCATGGCCCCAACGGAGGCGCCCTCGGAGGTGGGGTCCTGAGCCGCACA GCTGAGCCGCCTCTGACCCCTGCTGGCTGGGCCTGACCTTCCCGAGCCCGTGGGGGTGGGGGAGGCCAGCCACCTCCTTCCTCTGGGACT GCCCAGCCTGTGTCGGGGGCTTTCAGGGTTTCGTGGGGTTTGCCCGGAAGGCGGCTTTCCTCCCCCTGGTGTGAGGTCGCGCCCGAGGCT TGTACCCGCTAGTGAGGTGTTTGAGCTGGTCAGCAAGGAGAGGGGGTGGGGTTCCGCGGAAGGTTCTGGAGGGGTCTTGGTAGGTCTGCA GTGAACCGTCCTGAGGATGGAGTGGGGTCCCATGGTGCAGGTCTCTGAGCAAGGCGGAGGTGTGGAGGAGAGGCCGGCTTGGGGTGGGGC CTCGCGCCCTAGTGCCGGCCGGCCTCAGCCCGGCTCTGCCTGGTGCTCCCTGCAGTGCCTTCTCCATGGCCCCGCCCTCCCCGCGTGTGC GCCAGGCTTGGGGTCCCCGGGAGAGCAGAGCTTGCGCCTCGGGCATAGGGACGTGGGGTGCAGGCGCCAACATCAGTGGCAGCAGCCAGG GCCGTGGTCCAGTCCCACTCGGGGATGGAGTGGGCCGGCGGCCAAACCAGTCACTCGGGGAGGAATGCGGAGGAGCGCTCATTCCATTCT >71107_71107_4_R3HDM4-TMEM259_R3HDM4_chr19_913087_ENST00000587975_TMEM259_chr19_1014472_ENST00000356663_length(amino acids)=544AA_BP=1 MFVLFVLAYIHIVFSRSPINCLEHVRDKWPREGILRVEVRHNSSRAPVFLQFCDSGGRGSFPGLAVEPGSNLDMEDEEEEELTMEMFGNS SIKFELDIEPKVFKPPSSTEALNDSQEFPFPETPTKVWPQDEYIVEYSLEYGFLRLSQATRQRLSIPVMVVTLDPTRDQCFGDRFSRLLL DEFLGYDDILMSSVKGLAENEENKGFLRNVVSGEHYRFVSMWMARTSYLAAFAIMVIFTLSVSMLLRYSHHQIFVFIVDLLQMLEMNMAI AFPAAPLLTVILALVGMEAIMSEFFNDTTTAFYIILIVWLADQYDAICCHTSTSKRHWLRFFYLYHFAFYAYHYRFNGQYSSLALVTSWL FIQHSMIYFFHHYELPAILQQVRIQEMLLQAPPLGPGTPTALPDDMNNNSGAPATAPDSAGQPPALGPVSPGASGSPGPVAAAPSSLVAA AASVAAAAGGDLGWMAETAAIITDASFLSGLSASLLERRPASPLGPAGGLPHAPQDSVPPSDSAASDTTPLGAAVGGPSPASMAPTEAPS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for R3HDM4-TMEM259 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for R3HDM4-TMEM259 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for R3HDM4-TMEM259 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
