
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ASH2L-C8orf86 (FusionGDB2 ID:7124) |
Fusion Gene Summary for ASH2L-C8orf86 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ASH2L-C8orf86 | Fusion gene ID: 7124 | Hgene | Tgene | Gene symbol | ASH2L | C8orf86 | Gene ID | 9070 | 389649 |
| Gene name | ASH2 like, histone lysine methyltransferase complex subunit | chromosome 8 open reading frame 86 | |
| Synonyms | ASH2|ASH2L1|ASH2L2|Bre2 | - | |
| Cytomap | 8p11.23 | 8p11.22 | |
| Type of gene | protein-coding | protein-coding | |
| Description | set1/Ash2 histone methyltransferase complex subunit ASH2ASH2-like proteinash2 (absent, small, or homeotic)-like | uncharacterized protein C8orf86 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | Q9UBL3 | . | |
| Ensembl transtripts involved in fusion gene | ENST00000524263, ENST00000250635, ENST00000343823, ENST00000428278, ENST00000521652, ENST00000545394, | ENST00000358138, ENST00000437935, | |
| Fusion gene scores | * DoF score | 13 X 10 X 6=780 | 3 X 2 X 2=12 |
| # samples | 14 | 2 | |
| ** MAII score | log2(14/780*10)=-2.47804729680464 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(2/12*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: ASH2L [Title/Abstract] AND C8orf86 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ASH2L(37968351)-C8orf86(38374030), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ASH2L | GO:0006974 | cellular response to DNA damage stimulus | 17500065 |
| Hgene | ASH2L | GO:0043627 | response to estrogen | 16603732 |
| Hgene | ASH2L | GO:0051568 | histone H3-K4 methylation | 17355966|19556245 |
 Fusion gene breakpoints across ASH2L (5'-gene) Fusion gene breakpoints across ASH2L (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
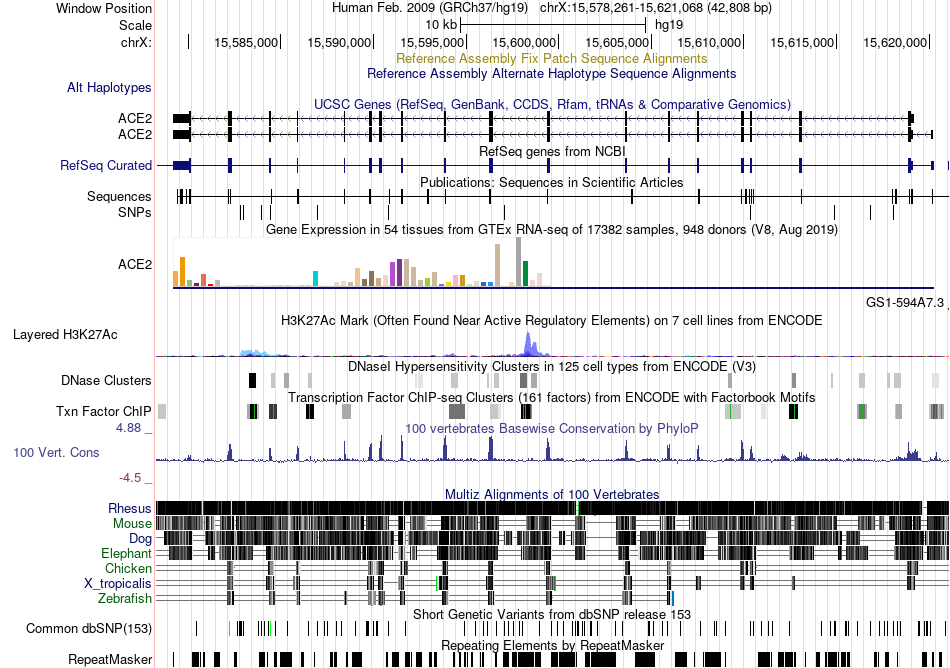 |
 Fusion gene breakpoints across C8orf86 (3'-gene) Fusion gene breakpoints across C8orf86 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
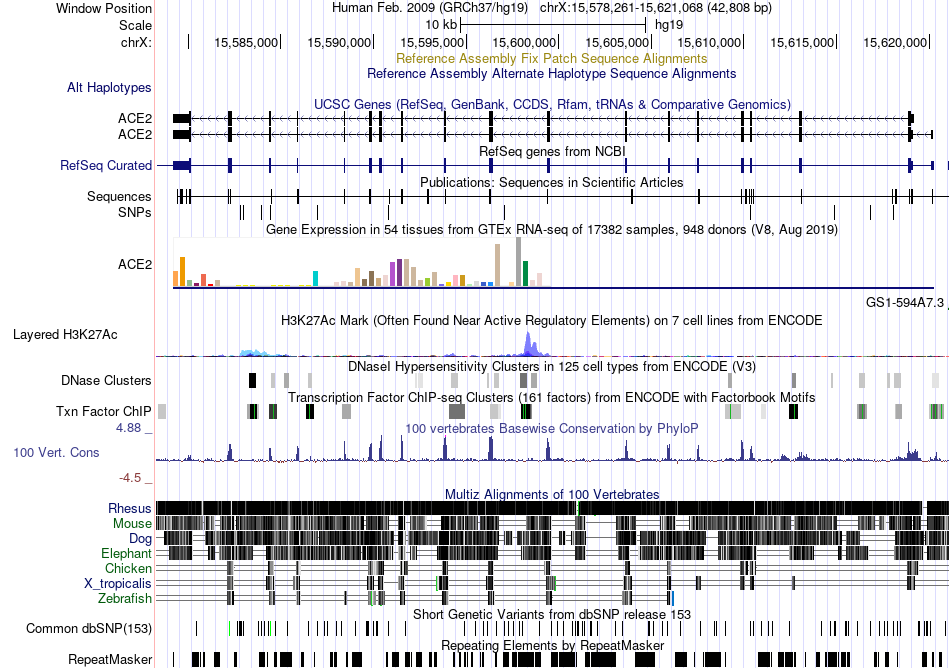 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CR-7364-01A | ASH2L | chr8 | 37968351 | - | C8orf86 | chr8 | 38374030 | - |
| ChimerDB4 | HNSC | TCGA-CR-7364-01A | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| ChimerDB4 | HNSC | TCGA-CR-7364 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
Top |
Fusion Gene ORF analysis for ASH2L-C8orf86 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000524263 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| 3UTR-intron | ENST00000524263 | ENST00000437935 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| 5CDS-intron | ENST00000250635 | ENST00000437935 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| 5CDS-intron | ENST00000343823 | ENST00000437935 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| 5CDS-intron | ENST00000428278 | ENST00000437935 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| 5CDS-intron | ENST00000521652 | ENST00000437935 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| 5CDS-intron | ENST00000545394 | ENST00000437935 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| In-frame | ENST00000250635 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| In-frame | ENST00000343823 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| In-frame | ENST00000428278 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| In-frame | ENST00000521652 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
| In-frame | ENST00000545394 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000343823 | ASH2L | chr8 | 37968351 | + | ENST00000358138 | C8orf86 | chr8 | 38374030 | - | 2804 | 894 | 309 | 1250 | 313 |
| ENST00000250635 | ASH2L | chr8 | 37968351 | + | ENST00000358138 | C8orf86 | chr8 | 38374030 | - | 2553 | 643 | 58 | 999 | 313 |
| ENST00000545394 | ASH2L | chr8 | 37968351 | + | ENST00000358138 | C8orf86 | chr8 | 38374030 | - | 2350 | 440 | 272 | 796 | 174 |
| ENST00000428278 | ASH2L | chr8 | 37968351 | + | ENST00000358138 | C8orf86 | chr8 | 38374030 | - | 2654 | 744 | 357 | 1100 | 247 |
| ENST00000521652 | ASH2L | chr8 | 37968351 | + | ENST00000358138 | C8orf86 | chr8 | 38374030 | - | 2508 | 598 | 211 | 954 | 247 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000343823 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - | 0.00689695 | 0.993103 |
| ENST00000250635 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - | 0.007010733 | 0.9929892 |
| ENST00000545394 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - | 0.15229848 | 0.84770155 |
| ENST00000428278 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - | 0.14255592 | 0.8574441 |
| ENST00000521652 | ENST00000358138 | ASH2L | chr8 | 37968351 | + | C8orf86 | chr8 | 38374030 | - | 0.15703891 | 0.84296113 |
Top |
Fusion Genomic Features for ASH2L-C8orf86 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
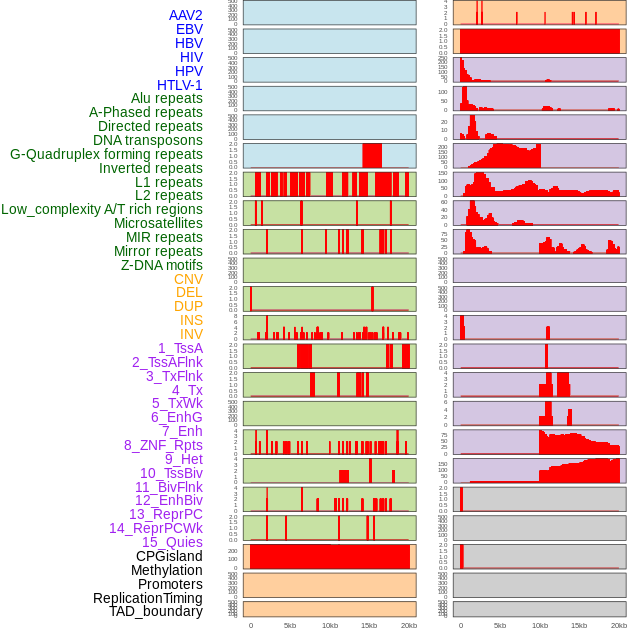 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ASH2L-C8orf86 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:37968351/chr8:38374030) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ASH2L | . |
| FUNCTION: Transcriptional regulator (PubMed:12670868). Component or associated component of some histone methyltransferase complexes which regulates transcription through recruitment of those complexes to gene promoters (PubMed:19131338). Component of the Set1/Ash2 histone methyltransferase (HMT) complex, a complex that specifically methylates 'Lys-4' of histone H3, but not if the neighboring 'Lys-9' residue is already methylated (PubMed:19556245). As part of the MLL1/MLL complex it is involved in methylation and dimethylation at 'Lys-4' of histone H3 (PubMed:19556245). May play a role in hematopoiesis (PubMed:12670868). In association with RBBP5 and WDR5, stimulates the histone methyltransferase activities of KMT2A, KMT2B, KMT2C, KMT2D, SETD1A and SETD1B (PubMed:21220120, PubMed:22266653). {ECO:0000269|PubMed:12670868, ECO:0000269|PubMed:19131338, ECO:0000269|PubMed:19556245, ECO:0000269|PubMed:21220120, ECO:0000269|PubMed:22266653}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000343823 | + | 5 | 16 | 67_177 | 195 | 629.0 | Region | Note=DNA-binding |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000250635 | + | 5 | 15 | 1_66 | 101 | 502.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000343823 | + | 5 | 16 | 117_150 | 195 | 629.0 | Zinc finger | Note=C4-type |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000343823 | + | 5 | 16 | 1_66 | 195 | 629.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000428278 | + | 5 | 16 | 1_66 | 101 | 535.0 | Zinc finger | Note=PHD-type%3B atypical |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000521652 | + | 5 | 15 | 1_66 | 101 | 502.0 | Zinc finger | Note=PHD-type%3B atypical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000250635 | + | 5 | 15 | 360_583 | 101 | 502.0 | Domain | B30.2/SPRY |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000343823 | + | 5 | 16 | 360_583 | 195 | 629.0 | Domain | B30.2/SPRY |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000428278 | + | 5 | 16 | 360_583 | 101 | 535.0 | Domain | B30.2/SPRY |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000521652 | + | 5 | 15 | 360_583 | 101 | 502.0 | Domain | B30.2/SPRY |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000250635 | + | 5 | 15 | 67_177 | 101 | 502.0 | Region | Note=DNA-binding |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000428278 | + | 5 | 16 | 67_177 | 101 | 535.0 | Region | Note=DNA-binding |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000521652 | + | 5 | 15 | 67_177 | 101 | 502.0 | Region | Note=DNA-binding |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000250635 | + | 5 | 15 | 117_150 | 101 | 502.0 | Zinc finger | Note=C4-type |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000428278 | + | 5 | 16 | 117_150 | 101 | 535.0 | Zinc finger | Note=C4-type |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000521652 | + | 5 | 15 | 117_150 | 101 | 502.0 | Zinc finger | Note=C4-type |
Top |
Fusion Gene Sequence for ASH2L-C8orf86 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >7124_7124_1_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000250635_C8orf86_chr8_38374030_ENST00000358138_length(transcript)=2553nt_BP=643nt CACAGCAACGCGCGCGAGAGAAGAGAGTATTCTCGCGAGAAGTCCAGGGGTGGCCGTGATGGCGGCGGCAGGAGCAGGACCTGGCCAGGA AGCGGGTGCCGGGCCTGGCCCAGGAGCGGTCGCAAATGCAACAGGGGCAGAAGAGGGGGAGATGAAGCCGGTGGCAGCGGGAGCAGCCGC TCCTCCTGGAGAGGGGATCTCTGCTGCTCCGACAGTTGAGCCCAGTTCCGGGGAGGCTGAAGGCGGGGAGGCAAACTTGGTCGATGTAAG CGGTGGCTTGGAGACAGAATCATCTAATGGAAAAGATACACTAGAAGGTGCTGGGGATACATCAGAGGTGATGGATACTCAGGCGGGCTC CGTGGATGAAGAGAATGGCCGACAGTTGGGTGAGGTAGAGCTGCAATGTGGGATTTGTACAAAATGGTTCACGGCTGACACATTTGGCAT AGATACCTCATCCTGTCTACCTTTCATGACCAACTACAGTTTTCATTGCAACGTCTGCCATCACAGTGGGAATACCTATTTCCTCCGGAA GCAAGCAAACTTGAAGGAAATGTGCCTTAGTGCTTTGGCCAACCTGACATGGCAGTCCCGAACACAGGATGAACATCCGAAGACAATGTT CTCCAAAGATAAGGCTTATGAGAATACAGGCCACTGGGAGAGACTCGCATCTGTGGTTTCTTCAAAAACACAGCAGCCCACAGTGATCTC TCATTCTTCCATTTCTATCACATTCAGTCATTACCCTCCAGCCACACTGGACTCCTTTCTTGTCCTGGAACCTATCAAACTCTTTCCTGT CTCAAGCCTCCGCAGTCCTCTCTGCTTGAACTGTGGCTCCTGCAGAGAAAGCATCAGAATCTCCGGGGAACTGATTGGAAATGCACATTC TCCAGCCCCGCCCAGAACTCCTGAATTAGAAACCCTGGGGTGGGACAAGCAAGCCGTGCTTTCTGGGGCACAGGTGATTCTGGTGTGTGC TGAAGTTTAAGAACCACTGGCCGAGAACATTCTTAGGTCTGCTCTTCTTTTGCCCGGCCCCTCCCTCGCGGAGGAATTCCTTCTTATTCC TCTCTGAAGAGTGGCTGCTGCCAAAAAACGTTTGTGAGATGGCCTGGGTTTTCTTTGTTGATTTATCATTTAGTTTGGAAGAAATCAGAA GTCTCTTTAAGAAGCCAATTTGAAACATTCACCCCATGGGAACAGTTCTGGATGAAGTCAGAAGATCTGGAGGCAGCGCAGTAACACACG TAGGTTTTCTGGCCATATGGACATTTCAGAGAAAACAACGCACAGAGGCCTGGAGCAGGTGAACTGGCTTAAGTAGAGAGAAACTAAGTC ATTTGGGGATATTTAGCACCTAATGTCAAGGCAGAAATGTCTAAGATGTAATTAACAGTTATATTCTAATCTCAATAGTAGCTAAGTACA GACTTAAACATAAGCCTGTATATAACAAAATAACCCCAGGAGAACCAAAGAAAATCTAGAAGTTGCTGCTAAAAACAGTTATGTTAGTGA TACCTAGGAAAGTTTTTTTTCTTTTAACATGTCATTGTGGTTTACAAATGAAAATTGAGGCCGGGCGTGGTGGCTCACGCCTGTAATCCC AGCACTTTTGGAGGCCAAGGTGGGCGAATCACAAGGTCAGGAGTTCAAGACCAGTCTGGCCAACATGGTGAAACCCCATCTTTACTAAAA ATACAAAAAATTAGCTGGGCGTGGTGGTGGGCGCCTGTAATCTCAGCTACTAGGGAGGCTGAGGAAGGAGAATCGCTTGAACCTGGGAGG CGGAGGTTGCAGTGAGCCAAGATCATGCCACCGCACTCCAGCCTGGGCAACAGTGTGAGACTCCATCTCAAAACAAACAAACAAACAAAT GAAACAAATGAAAATTGAAACTTCACCCATTTATGGCTATTGCCTAAAGAATTTATAAATGCCTGGGTCATTGCAAGCATATTGCTGACA TGTCTCTCGGTCTGCGTTACCCTGGTGGACATCACGACACTCACCTGACAGGCAGCAGCTTCCTTCCAGTAAAAGCAAAGAATCTGAAAG GAATGGAAAAGGCTCCACACAGTGCCATTTTATAGAAGGAAATGCAACAAGGTCACAGACCAGAAGGACAGCAGCCCAGGCCGGCTGGGC ATGGAGGAAGTCCCAAGATGCTGCTGGGCATGAACAGACCTCCTCATACAGTGTGCCTCTGAAGAAATAATGCAATTGTGTGGGGCCAGA GGAGCCACAAATAGAACAAAGGGAGGAAAGGAAAAATCAATATGCAGTAAAGAGGAGGAAGGGAGCCCGGCGCGATGTCTGAATCTCGCT GGCAAGAAAAGGAAACAGGTGGTCTAAGCAGGCAACCCTTCACCCCACATTGTAATGCTGGGGCATGGACGTGTTCCATAGATCACTCAC TGAGAAGTCTTCACAAGAACACTCCAAGGCAGACACTACTATCCATCTTCCACACACTGGGGGATTGAGAGTCAGAAGGATTATACAGCT >7124_7124_1_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000250635_C8orf86_chr8_38374030_ENST00000358138_length(amino acids)=313AA_BP=195 MAAAGAGPGQEAGAGPGPGAVANATGAEEGEMKPVAAGAAAPPGEGISAAPTVEPSSGEAEGGEANLVDVSGGLETESSNGKDTLEGAGD TSEVMDTQAGSVDEENGRQLGEVELQCGICTKWFTADTFGIDTSSCLPFMTNYSFHCNVCHHSGNTYFLRKQANLKEMCLSALANLTWQS RTQDEHPKTMFSKDKAYENTGHWERLASVVSSKTQQPTVISHSSISITFSHYPPATLDSFLVLEPIKLFPVSSLRSPLCLNCGSCRESIR -------------------------------------------------------------- >7124_7124_2_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000343823_C8orf86_chr8_38374030_ENST00000358138_length(transcript)=2804nt_BP=894nt GCAGAATATCCAGTCGTAGCCACGAAAATACGCCGCGTCAGTCCCTGACTCGAAAGGAAGTCCTCCAGAGAGCCGCACTAACCATCTAGC TTCCTCCACCTTCAGCCTGCCAGGCCCGCGGCCACCGCCGGCGCGGGAGGCCGGCCCACCTCTGCCCGCGCGCCTGGCCTTAAAGCGGTC GTGGAATTATGCTTCCTCAGGCAGGCGGTGGACGCACCGCGGGCTTTTTCTGTTTGAAGACTACAGCGTCACACAGCAACGCGCGCGAGA GAAGAGAGTATTCTCGCGAGAAGTCCAGGGGTGGCCGTGATGGCGGCGGCAGGAGCAGGACCTGGCCAGGAAGCGGGTGCCGGGCCTGGC CCAGGAGCGGTCGCAAATGCAACAGGGGCAGAAGAGGGGGAGATGAAGCCGGTGGCAGCGGGAGCAGCCGCTCCTCCTGGAGAGGGGATC TCTGCTGCTCCGACAGTTGAGCCCAGTTCCGGGGAGGCTGAAGGCGGGGAGGCAAACTTGGTCGATGTAAGCGGTGGCTTGGAGACAGAA TCATCTAATGGAAAAGATACACTAGAAGGTGCTGGGGATACATCAGAGGTGATGGATACTCAGGCGGGCTCCGTGGATGAAGAGAATGGC CGACAGTTGGGTGAGGTAGAGCTGCAATGTGGGATTTGTACAAAATGGTTCACGGCTGACACATTTGGCATAGATACCTCATCCTGTCTA CCTTTCATGACCAACTACAGTTTTCATTGCAACGTCTGCCATCACAGTGGGAATACCTATTTCCTCCGGAAGCAAGCAAACTTGAAGGAA ATGTGCCTTAGTGCTTTGGCCAACCTGACATGGCAGTCCCGAACACAGGATGAACATCCGAAGACAATGTTCTCCAAAGATAAGGCTTAT GAGAATACAGGCCACTGGGAGAGACTCGCATCTGTGGTTTCTTCAAAAACACAGCAGCCCACAGTGATCTCTCATTCTTCCATTTCTATC ACATTCAGTCATTACCCTCCAGCCACACTGGACTCCTTTCTTGTCCTGGAACCTATCAAACTCTTTCCTGTCTCAAGCCTCCGCAGTCCT CTCTGCTTGAACTGTGGCTCCTGCAGAGAAAGCATCAGAATCTCCGGGGAACTGATTGGAAATGCACATTCTCCAGCCCCGCCCAGAACT CCTGAATTAGAAACCCTGGGGTGGGACAAGCAAGCCGTGCTTTCTGGGGCACAGGTGATTCTGGTGTGTGCTGAAGTTTAAGAACCACTG GCCGAGAACATTCTTAGGTCTGCTCTTCTTTTGCCCGGCCCCTCCCTCGCGGAGGAATTCCTTCTTATTCCTCTCTGAAGAGTGGCTGCT GCCAAAAAACGTTTGTGAGATGGCCTGGGTTTTCTTTGTTGATTTATCATTTAGTTTGGAAGAAATCAGAAGTCTCTTTAAGAAGCCAAT TTGAAACATTCACCCCATGGGAACAGTTCTGGATGAAGTCAGAAGATCTGGAGGCAGCGCAGTAACACACGTAGGTTTTCTGGCCATATG GACATTTCAGAGAAAACAACGCACAGAGGCCTGGAGCAGGTGAACTGGCTTAAGTAGAGAGAAACTAAGTCATTTGGGGATATTTAGCAC CTAATGTCAAGGCAGAAATGTCTAAGATGTAATTAACAGTTATATTCTAATCTCAATAGTAGCTAAGTACAGACTTAAACATAAGCCTGT ATATAACAAAATAACCCCAGGAGAACCAAAGAAAATCTAGAAGTTGCTGCTAAAAACAGTTATGTTAGTGATACCTAGGAAAGTTTTTTT TCTTTTAACATGTCATTGTGGTTTACAAATGAAAATTGAGGCCGGGCGTGGTGGCTCACGCCTGTAATCCCAGCACTTTTGGAGGCCAAG GTGGGCGAATCACAAGGTCAGGAGTTCAAGACCAGTCTGGCCAACATGGTGAAACCCCATCTTTACTAAAAATACAAAAAATTAGCTGGG CGTGGTGGTGGGCGCCTGTAATCTCAGCTACTAGGGAGGCTGAGGAAGGAGAATCGCTTGAACCTGGGAGGCGGAGGTTGCAGTGAGCCA AGATCATGCCACCGCACTCCAGCCTGGGCAACAGTGTGAGACTCCATCTCAAAACAAACAAACAAACAAATGAAACAAATGAAAATTGAA ACTTCACCCATTTATGGCTATTGCCTAAAGAATTTATAAATGCCTGGGTCATTGCAAGCATATTGCTGACATGTCTCTCGGTCTGCGTTA CCCTGGTGGACATCACGACACTCACCTGACAGGCAGCAGCTTCCTTCCAGTAAAAGCAAAGAATCTGAAAGGAATGGAAAAGGCTCCACA CAGTGCCATTTTATAGAAGGAAATGCAACAAGGTCACAGACCAGAAGGACAGCAGCCCAGGCCGGCTGGGCATGGAGGAAGTCCCAAGAT GCTGCTGGGCATGAACAGACCTCCTCATACAGTGTGCCTCTGAAGAAATAATGCAATTGTGTGGGGCCAGAGGAGCCACAAATAGAACAA AGGGAGGAAAGGAAAAATCAATATGCAGTAAAGAGGAGGAAGGGAGCCCGGCGCGATGTCTGAATCTCGCTGGCAAGAAAAGGAAACAGG TGGTCTAAGCAGGCAACCCTTCACCCCACATTGTAATGCTGGGGCATGGACGTGTTCCATAGATCACTCACTGAGAAGTCTTCACAAGAA CACTCCAAGGCAGACACTACTATCCATCTTCCACACACTGGGGGATTGAGAGTCAGAAGGATTATACAGCTTGTTCAAATTATAAATAAA >7124_7124_2_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000343823_C8orf86_chr8_38374030_ENST00000358138_length(amino acids)=313AA_BP=195 MAAAGAGPGQEAGAGPGPGAVANATGAEEGEMKPVAAGAAAPPGEGISAAPTVEPSSGEAEGGEANLVDVSGGLETESSNGKDTLEGAGD TSEVMDTQAGSVDEENGRQLGEVELQCGICTKWFTADTFGIDTSSCLPFMTNYSFHCNVCHHSGNTYFLRKQANLKEMCLSALANLTWQS RTQDEHPKTMFSKDKAYENTGHWERLASVVSSKTQQPTVISHSSISITFSHYPPATLDSFLVLEPIKLFPVSSLRSPLCLNCGSCRESIR -------------------------------------------------------------- >7124_7124_3_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000428278_C8orf86_chr8_38374030_ENST00000358138_length(transcript)=2654nt_BP=744nt ATCCCGCGGCTCCTGGAGCCCTGCCGCCCGCCCCCTGCGAACCCAGGCCCCGCCGCCAATGGCTCGCCCTGCCTCTGCGCCGCTTGGCCC GTCCCCTCTCAAGCATATCTCGGATAACGCCCCTTCCGCACCTTTCACGGGCGGTGGGAGCTGAGGCTCCTGTCGTTATCTCTGATCCTT GCACCCTGGCAGGAAGCTGGTAGCTCACACTTTAACGGGAGGCCTTCACATATTCCAGAAAAGAAACCACTTTGCAGTGCCAGACTGGAA GAAGTAACGGTCACTCTGAAAACAGGGTGGGAGAGCTGCCTCTCTTTGAACCTCTCCCAGGACCAACTCTAACCCAGGGAGGCAAACTTG GTCGATGTAAGCGGTGGCTTGGAGACAGAATCATCTAATGGAAAAGATACACTAGAAGGTGCTGGGGATACATCAGAGGTGATGGATACT CAGGCGGGCTCCGTGGATGAAGAGAATGGCCGACAGTTGGGTGAGGTAGAGCTGCAATGTGGGATTTGTACAAAATGGTTCACGGCTGAC ACATTTGGCATAGATACCTCATCCTGTCTACCTTTCATGACCAACTACAGTTTTCATTGCAACGTCTGCCATCACAGTGGGAATACCTAT TTCCTCCGGAAGCAAGCAAACTTGAAGGAAATGTGCCTTAGTGCTTTGGCCAACCTGACATGGCAGTCCCGAACACAGGATGAACATCCG AAGACAATGTTCTCCAAAGATAAGGCTTATGAGAATACAGGCCACTGGGAGAGACTCGCATCTGTGGTTTCTTCAAAAACACAGCAGCCC ACAGTGATCTCTCATTCTTCCATTTCTATCACATTCAGTCATTACCCTCCAGCCACACTGGACTCCTTTCTTGTCCTGGAACCTATCAAA CTCTTTCCTGTCTCAAGCCTCCGCAGTCCTCTCTGCTTGAACTGTGGCTCCTGCAGAGAAAGCATCAGAATCTCCGGGGAACTGATTGGA AATGCACATTCTCCAGCCCCGCCCAGAACTCCTGAATTAGAAACCCTGGGGTGGGACAAGCAAGCCGTGCTTTCTGGGGCACAGGTGATT CTGGTGTGTGCTGAAGTTTAAGAACCACTGGCCGAGAACATTCTTAGGTCTGCTCTTCTTTTGCCCGGCCCCTCCCTCGCGGAGGAATTC CTTCTTATTCCTCTCTGAAGAGTGGCTGCTGCCAAAAAACGTTTGTGAGATGGCCTGGGTTTTCTTTGTTGATTTATCATTTAGTTTGGA AGAAATCAGAAGTCTCTTTAAGAAGCCAATTTGAAACATTCACCCCATGGGAACAGTTCTGGATGAAGTCAGAAGATCTGGAGGCAGCGC AGTAACACACGTAGGTTTTCTGGCCATATGGACATTTCAGAGAAAACAACGCACAGAGGCCTGGAGCAGGTGAACTGGCTTAAGTAGAGA GAAACTAAGTCATTTGGGGATATTTAGCACCTAATGTCAAGGCAGAAATGTCTAAGATGTAATTAACAGTTATATTCTAATCTCAATAGT AGCTAAGTACAGACTTAAACATAAGCCTGTATATAACAAAATAACCCCAGGAGAACCAAAGAAAATCTAGAAGTTGCTGCTAAAAACAGT TATGTTAGTGATACCTAGGAAAGTTTTTTTTCTTTTAACATGTCATTGTGGTTTACAAATGAAAATTGAGGCCGGGCGTGGTGGCTCACG CCTGTAATCCCAGCACTTTTGGAGGCCAAGGTGGGCGAATCACAAGGTCAGGAGTTCAAGACCAGTCTGGCCAACATGGTGAAACCCCAT CTTTACTAAAAATACAAAAAATTAGCTGGGCGTGGTGGTGGGCGCCTGTAATCTCAGCTACTAGGGAGGCTGAGGAAGGAGAATCGCTTG AACCTGGGAGGCGGAGGTTGCAGTGAGCCAAGATCATGCCACCGCACTCCAGCCTGGGCAACAGTGTGAGACTCCATCTCAAAACAAACA AACAAACAAATGAAACAAATGAAAATTGAAACTTCACCCATTTATGGCTATTGCCTAAAGAATTTATAAATGCCTGGGTCATTGCAAGCA TATTGCTGACATGTCTCTCGGTCTGCGTTACCCTGGTGGACATCACGACACTCACCTGACAGGCAGCAGCTTCCTTCCAGTAAAAGCAAA GAATCTGAAAGGAATGGAAAAGGCTCCACACAGTGCCATTTTATAGAAGGAAATGCAACAAGGTCACAGACCAGAAGGACAGCAGCCCAG GCCGGCTGGGCATGGAGGAAGTCCCAAGATGCTGCTGGGCATGAACAGACCTCCTCATACAGTGTGCCTCTGAAGAAATAATGCAATTGT GTGGGGCCAGAGGAGCCACAAATAGAACAAAGGGAGGAAAGGAAAAATCAATATGCAGTAAAGAGGAGGAAGGGAGCCCGGCGCGATGTC TGAATCTCGCTGGCAAGAAAAGGAAACAGGTGGTCTAAGCAGGCAACCCTTCACCCCACATTGTAATGCTGGGGCATGGACGTGTTCCAT AGATCACTCACTGAGAAGTCTTCACAAGAACACTCCAAGGCAGACACTACTATCCATCTTCCACACACTGGGGGATTGAGAGTCAGAAGG >7124_7124_3_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000428278_C8orf86_chr8_38374030_ENST00000358138_length(amino acids)=247AA_BP=129 MVDVSGGLETESSNGKDTLEGAGDTSEVMDTQAGSVDEENGRQLGEVELQCGICTKWFTADTFGIDTSSCLPFMTNYSFHCNVCHHSGNT YFLRKQANLKEMCLSALANLTWQSRTQDEHPKTMFSKDKAYENTGHWERLASVVSSKTQQPTVISHSSISITFSHYPPATLDSFLVLEPI -------------------------------------------------------------- >7124_7124_4_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000521652_C8orf86_chr8_38374030_ENST00000358138_length(transcript)=2508nt_BP=598nt GGAGCTGAGGCTCCTGTCGTTATCTCTGATCCTTGCACCCTGGCAGGAAGCTGGTAGCTCACACTTTAACGGGAGGCCTTCACATATTCC AGAAAAGAAACCACTTTGCAGTGCCAGACTGGAAGAAGTAACGGTCACTCTGAAAACAGGGTGGGAGAGCTGCCTCTCTTTGAACCTCTC CCAGGACCAACTCTAACCCAGGGAGGCAAACTTGGTCGATGTAAGCGGTGGCTTGGAGACAGAATCATCTAATGGAAAAGATACACTAGA AGGTGCTGGGGATACATCAGAGGTGATGGATACTCAGGCGGGCTCCGTGGATGAAGAGAATGGCCGACAGTTGGGTGAGGTAGAGCTGCA ATGTGGGATTTGTACAAAATGGTTCACGGCTGACACATTTGGCATAGATACCTCATCCTGTCTACCTTTCATGACCAACTACAGTTTTCA TTGCAACGTCTGCCATCACAGTGGGAATACCTATTTCCTCCGGAAGCAAGCAAACTTGAAGGAAATGTGCCTTAGTGCTTTGGCCAACCT GACATGGCAGTCCCGAACACAGGATGAACATCCGAAGACAATGTTCTCCAAAGATAAGGCTTATGAGAATACAGGCCACTGGGAGAGACT CGCATCTGTGGTTTCTTCAAAAACACAGCAGCCCACAGTGATCTCTCATTCTTCCATTTCTATCACATTCAGTCATTACCCTCCAGCCAC ACTGGACTCCTTTCTTGTCCTGGAACCTATCAAACTCTTTCCTGTCTCAAGCCTCCGCAGTCCTCTCTGCTTGAACTGTGGCTCCTGCAG AGAAAGCATCAGAATCTCCGGGGAACTGATTGGAAATGCACATTCTCCAGCCCCGCCCAGAACTCCTGAATTAGAAACCCTGGGGTGGGA CAAGCAAGCCGTGCTTTCTGGGGCACAGGTGATTCTGGTGTGTGCTGAAGTTTAAGAACCACTGGCCGAGAACATTCTTAGGTCTGCTCT TCTTTTGCCCGGCCCCTCCCTCGCGGAGGAATTCCTTCTTATTCCTCTCTGAAGAGTGGCTGCTGCCAAAAAACGTTTGTGAGATGGCCT GGGTTTTCTTTGTTGATTTATCATTTAGTTTGGAAGAAATCAGAAGTCTCTTTAAGAAGCCAATTTGAAACATTCACCCCATGGGAACAG TTCTGGATGAAGTCAGAAGATCTGGAGGCAGCGCAGTAACACACGTAGGTTTTCTGGCCATATGGACATTTCAGAGAAAACAACGCACAG AGGCCTGGAGCAGGTGAACTGGCTTAAGTAGAGAGAAACTAAGTCATTTGGGGATATTTAGCACCTAATGTCAAGGCAGAAATGTCTAAG ATGTAATTAACAGTTATATTCTAATCTCAATAGTAGCTAAGTACAGACTTAAACATAAGCCTGTATATAACAAAATAACCCCAGGAGAAC CAAAGAAAATCTAGAAGTTGCTGCTAAAAACAGTTATGTTAGTGATACCTAGGAAAGTTTTTTTTCTTTTAACATGTCATTGTGGTTTAC AAATGAAAATTGAGGCCGGGCGTGGTGGCTCACGCCTGTAATCCCAGCACTTTTGGAGGCCAAGGTGGGCGAATCACAAGGTCAGGAGTT CAAGACCAGTCTGGCCAACATGGTGAAACCCCATCTTTACTAAAAATACAAAAAATTAGCTGGGCGTGGTGGTGGGCGCCTGTAATCTCA GCTACTAGGGAGGCTGAGGAAGGAGAATCGCTTGAACCTGGGAGGCGGAGGTTGCAGTGAGCCAAGATCATGCCACCGCACTCCAGCCTG GGCAACAGTGTGAGACTCCATCTCAAAACAAACAAACAAACAAATGAAACAAATGAAAATTGAAACTTCACCCATTTATGGCTATTGCCT AAAGAATTTATAAATGCCTGGGTCATTGCAAGCATATTGCTGACATGTCTCTCGGTCTGCGTTACCCTGGTGGACATCACGACACTCACC TGACAGGCAGCAGCTTCCTTCCAGTAAAAGCAAAGAATCTGAAAGGAATGGAAAAGGCTCCACACAGTGCCATTTTATAGAAGGAAATGC AACAAGGTCACAGACCAGAAGGACAGCAGCCCAGGCCGGCTGGGCATGGAGGAAGTCCCAAGATGCTGCTGGGCATGAACAGACCTCCTC ATACAGTGTGCCTCTGAAGAAATAATGCAATTGTGTGGGGCCAGAGGAGCCACAAATAGAACAAAGGGAGGAAAGGAAAAATCAATATGC AGTAAAGAGGAGGAAGGGAGCCCGGCGCGATGTCTGAATCTCGCTGGCAAGAAAAGGAAACAGGTGGTCTAAGCAGGCAACCCTTCACCC CACATTGTAATGCTGGGGCATGGACGTGTTCCATAGATCACTCACTGAGAAGTCTTCACAAGAACACTCCAAGGCAGACACTACTATCCA >7124_7124_4_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000521652_C8orf86_chr8_38374030_ENST00000358138_length(amino acids)=247AA_BP=129 MVDVSGGLETESSNGKDTLEGAGDTSEVMDTQAGSVDEENGRQLGEVELQCGICTKWFTADTFGIDTSSCLPFMTNYSFHCNVCHHSGNT YFLRKQANLKEMCLSALANLTWQSRTQDEHPKTMFSKDKAYENTGHWERLASVVSSKTQQPTVISHSSISITFSHYPPATLDSFLVLEPI -------------------------------------------------------------- >7124_7124_5_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000545394_C8orf86_chr8_38374030_ENST00000358138_length(transcript)=2350nt_BP=440nt GATGGCGGCGGCAGGAGCAGGACCTGGCCAGGAAGCGGGTGCCGGGCCTGGCCCAGGAGCGGTCGCAAATGCAACAGGGGCAGAAGAGGG GGAGATGAAGCCGGTGGCAGCGGGAGCAGCCGCTCCTCCTGGAGAGGGGATCTCTGCTGCTCCGACAGTTGAGCCCAGTTCCGGGGAGGC TGAAGGCGGGGAGGCAAACTTGGTCGATGTAAGCGGTGGCTTGGAGACAGAATCATCTAATGGAAAAGATACACTAATCCTGTCTACCTT TCATGACCAACTACAGTTTTCATTGCAACGTCTGCCATCACAGTGGGAATACCTATTTCCTCCGGAAGCAAGCAAACTTGAAGGAAATGT GCCTTAGTGCTTTGGCCAACCTGACATGGCAGTCCCGAACACAGGATGAACATCCGAAGACAATGTTCTCCAAAGATAAGGCTTATGAGA ATACAGGCCACTGGGAGAGACTCGCATCTGTGGTTTCTTCAAAAACACAGCAGCCCACAGTGATCTCTCATTCTTCCATTTCTATCACAT TCAGTCATTACCCTCCAGCCACACTGGACTCCTTTCTTGTCCTGGAACCTATCAAACTCTTTCCTGTCTCAAGCCTCCGCAGTCCTCTCT GCTTGAACTGTGGCTCCTGCAGAGAAAGCATCAGAATCTCCGGGGAACTGATTGGAAATGCACATTCTCCAGCCCCGCCCAGAACTCCTG AATTAGAAACCCTGGGGTGGGACAAGCAAGCCGTGCTTTCTGGGGCACAGGTGATTCTGGTGTGTGCTGAAGTTTAAGAACCACTGGCCG AGAACATTCTTAGGTCTGCTCTTCTTTTGCCCGGCCCCTCCCTCGCGGAGGAATTCCTTCTTATTCCTCTCTGAAGAGTGGCTGCTGCCA AAAAACGTTTGTGAGATGGCCTGGGTTTTCTTTGTTGATTTATCATTTAGTTTGGAAGAAATCAGAAGTCTCTTTAAGAAGCCAATTTGA AACATTCACCCCATGGGAACAGTTCTGGATGAAGTCAGAAGATCTGGAGGCAGCGCAGTAACACACGTAGGTTTTCTGGCCATATGGACA TTTCAGAGAAAACAACGCACAGAGGCCTGGAGCAGGTGAACTGGCTTAAGTAGAGAGAAACTAAGTCATTTGGGGATATTTAGCACCTAA TGTCAAGGCAGAAATGTCTAAGATGTAATTAACAGTTATATTCTAATCTCAATAGTAGCTAAGTACAGACTTAAACATAAGCCTGTATAT AACAAAATAACCCCAGGAGAACCAAAGAAAATCTAGAAGTTGCTGCTAAAAACAGTTATGTTAGTGATACCTAGGAAAGTTTTTTTTCTT TTAACATGTCATTGTGGTTTACAAATGAAAATTGAGGCCGGGCGTGGTGGCTCACGCCTGTAATCCCAGCACTTTTGGAGGCCAAGGTGG GCGAATCACAAGGTCAGGAGTTCAAGACCAGTCTGGCCAACATGGTGAAACCCCATCTTTACTAAAAATACAAAAAATTAGCTGGGCGTG GTGGTGGGCGCCTGTAATCTCAGCTACTAGGGAGGCTGAGGAAGGAGAATCGCTTGAACCTGGGAGGCGGAGGTTGCAGTGAGCCAAGAT CATGCCACCGCACTCCAGCCTGGGCAACAGTGTGAGACTCCATCTCAAAACAAACAAACAAACAAATGAAACAAATGAAAATTGAAACTT CACCCATTTATGGCTATTGCCTAAAGAATTTATAAATGCCTGGGTCATTGCAAGCATATTGCTGACATGTCTCTCGGTCTGCGTTACCCT GGTGGACATCACGACACTCACCTGACAGGCAGCAGCTTCCTTCCAGTAAAAGCAAAGAATCTGAAAGGAATGGAAAAGGCTCCACACAGT GCCATTTTATAGAAGGAAATGCAACAAGGTCACAGACCAGAAGGACAGCAGCCCAGGCCGGCTGGGCATGGAGGAAGTCCCAAGATGCTG CTGGGCATGAACAGACCTCCTCATACAGTGTGCCTCTGAAGAAATAATGCAATTGTGTGGGGCCAGAGGAGCCACAAATAGAACAAAGGG AGGAAAGGAAAAATCAATATGCAGTAAAGAGGAGGAAGGGAGCCCGGCGCGATGTCTGAATCTCGCTGGCAAGAAAAGGAAACAGGTGGT CTAAGCAGGCAACCCTTCACCCCACATTGTAATGCTGGGGCATGGACGTGTTCCATAGATCACTCACTGAGAAGTCTTCACAAGAACACT CCAAGGCAGACACTACTATCCATCTTCCACACACTGGGGGATTGAGAGTCAGAAGGATTATACAGCTTGTTCAAATTATAAATAAAAGCC >7124_7124_5_ASH2L-C8orf86_ASH2L_chr8_37968351_ENST00000545394_C8orf86_chr8_38374030_ENST00000358138_length(amino acids)=174AA_BP=56 MTNYSFHCNVCHHSGNTYFLRKQANLKEMCLSALANLTWQSRTQDEHPKTMFSKDKAYENTGHWERLASVVSSKTQQPTVISHSSISITF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ASH2L-C8orf86 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000250635 | + | 5 | 15 | 316_628 | 101.0 | 502.0 | RBBP5 |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000343823 | + | 5 | 16 | 316_628 | 195.0 | 629.0 | RBBP5 |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000428278 | + | 5 | 16 | 316_628 | 101.0 | 535.0 | RBBP5 |
| Hgene | ASH2L | chr8:37968351 | chr8:38374030 | ENST00000521652 | + | 5 | 15 | 316_628 | 101.0 | 502.0 | RBBP5 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ASH2L-C8orf86 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ASH2L-C8orf86 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
