
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RASA1-XRCC4 (FusionGDB2 ID:72378) |
Fusion Gene Summary for RASA1-XRCC4 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RASA1-XRCC4 | Fusion gene ID: 72378 | Hgene | Tgene | Gene symbol | RASA1 | XRCC4 | Gene ID | 8831 | 7518 |
| Gene name | synaptic Ras GTPase activating protein 1 | X-ray repair cross complementing 4 | |
| Synonyms | MRD5|RASA1|RASA5|SYNGAP | SSMED | |
| Cytomap | 6p21.32 | 5q14.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ras/Rap GTPase-activating protein SynGAPRas GTPase-activating protein SynGAPneuronal RasGAPsynaptic Ras GTPase activating protein 1 homologsynaptic Ras GTPase activating protein, 135kDa | DNA repair protein XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000274376, ENST00000456692, ENST00000506290, ENST00000512763, | ENST00000509268, ENST00000282268, ENST00000511817, ENST00000338635, ENST00000396027, | |
| Fusion gene scores | * DoF score | 9 X 7 X 6=378 | 9 X 8 X 4=288 |
| # samples | 9 | 9 | |
| ** MAII score | log2(9/378*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/288*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: RASA1 [Title/Abstract] AND XRCC4 [Title/Abstract] AND fusion [Title/Abstract] | ||
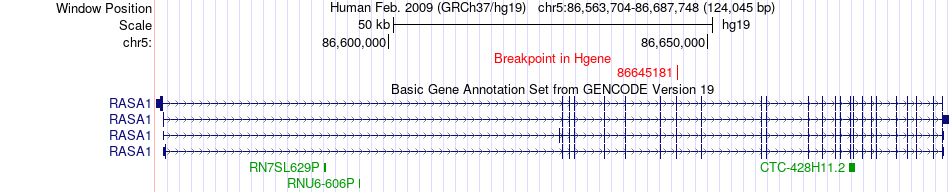
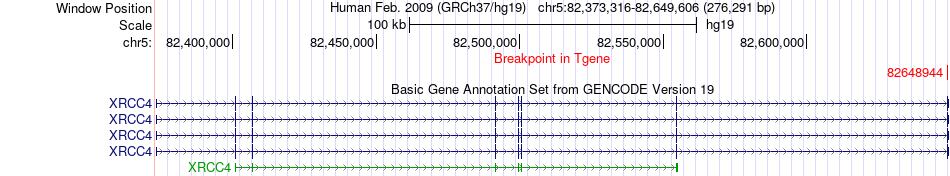
| Most frequent breakpoint | RASA1(86645181)-XRCC4(82648944), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | XRCC4 | GO:0006302 | double-strand break repair | 9242410 |
| Tgene | XRCC4 | GO:0006303 | double-strand break repair via nonhomologous end joining | 12517771 |
| Tgene | XRCC4 | GO:0010165 | response to X-ray | 9242410 |
| Tgene | XRCC4 | GO:0051103 | DNA ligation involved in DNA repair | 12517771 |
| Tgene | XRCC4 | GO:0051351 | positive regulation of ligase activity | 9242410 |
 Fusion gene breakpoints across RASA1 (5'-gene) Fusion gene breakpoints across RASA1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across XRCC4 (3'-gene) Fusion gene breakpoints across XRCC4 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-K1-A6RV-01A | RASA1 | chr5 | 86645181 | - | XRCC4 | chr5 | 82648944 | + |
| ChimerDB4 | SARC | TCGA-K1-A6RV-01A | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
Top |
Fusion Gene ORF analysis for RASA1-XRCC4 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000274376 | ENST00000509268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| 5CDS-intron | ENST00000456692 | ENST00000509268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| 5CDS-intron | ENST00000506290 | ENST00000509268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| 5CDS-intron | ENST00000512763 | ENST00000509268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000274376 | ENST00000282268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000274376 | ENST00000511817 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000456692 | ENST00000282268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000456692 | ENST00000511817 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000506290 | ENST00000282268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000506290 | ENST00000511817 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000512763 | ENST00000282268 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| Frame-shift | ENST00000512763 | ENST00000511817 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000274376 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000274376 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000456692 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000456692 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000506290 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000506290 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000512763 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
| In-frame | ENST00000512763 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000274376 | RASA1 | chr5 | 86645181 | + | ENST00000338635 | XRCC4 | chr5 | 82648944 | + | 2404 | 1817 | 564 | 1934 | 456 |
| ENST00000274376 | RASA1 | chr5 | 86645181 | + | ENST00000396027 | XRCC4 | chr5 | 82648944 | + | 2398 | 1817 | 564 | 1928 | 454 |
| ENST00000456692 | RASA1 | chr5 | 86645181 | + | ENST00000338635 | XRCC4 | chr5 | 82648944 | + | 1424 | 837 | 115 | 954 | 279 |
| ENST00000456692 | RASA1 | chr5 | 86645181 | + | ENST00000396027 | XRCC4 | chr5 | 82648944 | + | 1418 | 837 | 115 | 948 | 277 |
| ENST00000512763 | RASA1 | chr5 | 86645181 | + | ENST00000338635 | XRCC4 | chr5 | 82648944 | + | 1540 | 953 | 201 | 1070 | 289 |
| ENST00000512763 | RASA1 | chr5 | 86645181 | + | ENST00000396027 | XRCC4 | chr5 | 82648944 | + | 1534 | 953 | 201 | 1064 | 287 |
| ENST00000506290 | RASA1 | chr5 | 86645181 | + | ENST00000338635 | XRCC4 | chr5 | 82648944 | + | 1459 | 872 | 117 | 989 | 290 |
| ENST00000506290 | RASA1 | chr5 | 86645181 | + | ENST00000396027 | XRCC4 | chr5 | 82648944 | + | 1453 | 872 | 117 | 983 | 288 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000274376 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000354077 | 0.99964595 |
| ENST00000274376 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000354288 | 0.9996457 |
| ENST00000456692 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000997447 | 0.9990025 |
| ENST00000456692 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000999474 | 0.9990005 |
| ENST00000512763 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000254487 | 0.99974555 |
| ENST00000512763 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000328093 | 0.99967194 |
| ENST00000506290 | ENST00000338635 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.000263561 | 0.9997365 |
| ENST00000506290 | ENST00000396027 | RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648944 | + | 0.00025304 | 0.999747 |
Top |
Fusion Genomic Features for RASA1-XRCC4 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648943 | + | 0.001255627 | 0.99874437 |
| RASA1 | chr5 | 86645181 | + | XRCC4 | chr5 | 82648943 | + | 0.001255627 | 0.99874437 |
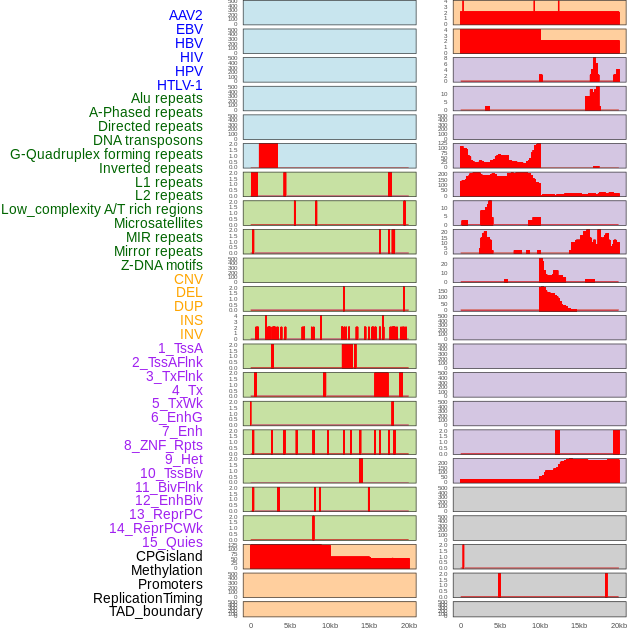
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
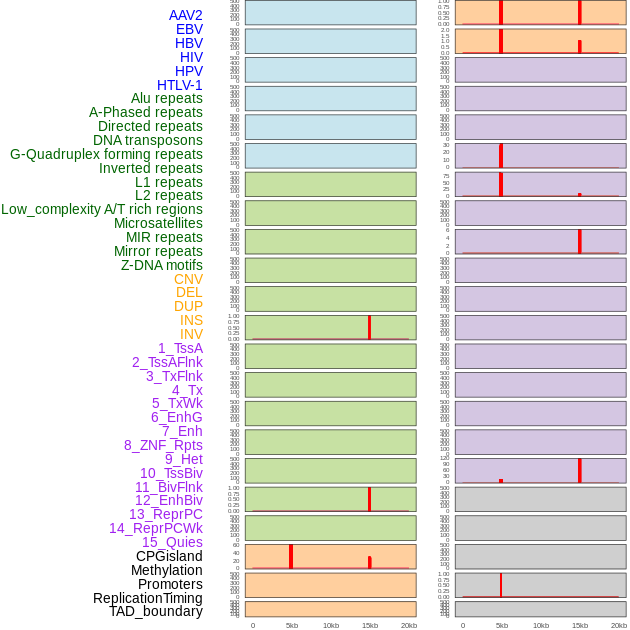
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for RASA1-XRCC4 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr5:86645181/chr5:82648944) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 135_141 | 417 | 1048.0 | Compositional bias | Note=Poly-Pro |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 163_168 | 417 | 1048.0 | Compositional bias | Note=Poly-Glu |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 17_22 | 417 | 1048.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 135_141 | 240 | 871.0 | Compositional bias | Note=Poly-Pro |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 163_168 | 240 | 871.0 | Compositional bias | Note=Poly-Glu |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 17_22 | 240 | 871.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 181_272 | 417 | 1048.0 | Domain | SH2 1 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 279_341 | 417 | 1048.0 | Domain | SH3 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 351_441 | 417 | 1048.0 | Domain | SH2 2 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 474_577 | 417 | 1048.0 | Domain | PH |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 577_690 | 417 | 1048.0 | Domain | C2 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000274376 | + | 8 | 25 | 748_942 | 417 | 1048.0 | Domain | Ras-GAP |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 181_272 | 240 | 871.0 | Domain | SH2 1 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 279_341 | 240 | 871.0 | Domain | SH3 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 351_441 | 240 | 871.0 | Domain | SH2 2 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 474_577 | 240 | 871.0 | Domain | PH |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 577_690 | 240 | 871.0 | Domain | C2 |
| Hgene | RASA1 | chr5:86645181 | chr5:82648944 | ENST00000456692 | + | 8 | 25 | 748_942 | 240 | 871.0 | Domain | Ras-GAP |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000282268 | 6 | 8 | 131_165 | 297 | 335.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000282268 | 6 | 8 | 184_212 | 297 | 335.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000338635 | 6 | 8 | 131_165 | 297 | 337.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000338635 | 6 | 8 | 184_212 | 297 | 337.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000396027 | 6 | 8 | 131_165 | 297 | 335.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000396027 | 6 | 8 | 184_212 | 297 | 335.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000511817 | 6 | 8 | 131_165 | 297 | 337.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000511817 | 6 | 8 | 184_212 | 297 | 337.0 | Coiled coil | Ontology_term=ECO:0000255 |
Top |
Fusion Gene Sequence for RASA1-XRCC4 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >72378_72378_1_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000274376_XRCC4_chr5_82648944_ENST00000338635_length(transcript)=2404nt_BP=1817nt CTGAGAGCTCCAGGTAGTGAGCAGTTCAGTCGATTTCCTCGTTACCCCGCCCCCCTTTCTCTTGCCCCCCCACCCCTCTCATCTGCCTGG TGGAGGATGAAGCGGCTGCAGTGGCCCCAGCCTCAGCAGCGGCACCGGCGGTGGCTGCGGTGTGGGTGGCCGGAACTGGGGTGGTGAAGA AGCGGTGGTGGCGGCTGAAGGAGCGGTAGCAGCCTCAGCCTCTTTCCCTGTGCTTCCTTTCCTCTTTAGTGCAGCGAGGAAGTTTTCGCT TCTGTACATGTGTTTGTGTGCGTGAGTGTGGGTGTGTGCGGTGAGGTTTGGGTGGCGTTTGTGCAGGCGTTGGGTTTTTTGCCCACTTGG CTTCCCGTAACCCAGGCAGCTGGGGAGCCTGGGCTGTGGCCCTAGGAGGGGGCGCGGCGGCGGGCTCTCTCCTTTTGTTGTTGTTTCCTC AGCCTGGGGAGCTGAAGGGGAGACGCGTCTGGGTGGGGCTGCTCGGAGCCCGGGCCTGGTGGCCCCTGGGGCTCCCGGGCGGGCAGGGTA GGGCAGAGTAGAGCGGGCTTCAACATGATGGCGGCCGAGGCCGGCAGTGAGGAGGGCGGCCCGGTAACAGCCGGAGCTGGAGGAGGCGGC GCGGCAGCGGGCTCCAGTGCCTATCCCGCAGTGTGTCGGGTGAAGATACCCGCGGCCCTGCCTGTGGCAGCCGCCCCCTATCCTGGGCTG GTGGAGACCGGAGTGGCTGGAACTCTGGGTGGCGGAGCCGCTTTGGGGTCAGAGTTCCTAGGAGCCGGGTCTGTGGCAGGGGCACTGGGG GGAGCTGGACTGACAGGGGGAGGTACTGCTGCTGGCGTAGCTGGTGCTGCTGCTGGCGTGGCCGGTGCTGCTGTTGCTGGACCTAGTGGA GACATGGCTCTCACCAAACTGCCCACTTCGTTGCTTGCTGAGACTCTCGGGCCAGGCGGCGGTTTTCCCCCTCTGCCCCCTCCCCCTTAC CTGCCCCCTTTGGGGGCGGGCCTCGGGACAGTGGACGAAGGTGACTCTCTGGATGGACCAGAATACGAGGAGGAAGAGGTGGCCATACCG TTGACCGCTCCTCCAACTAACCAGTGGTATCACGGAAAACTTGACAGAACGATAGCAGAAGAACGCCTCAGGCAGGCAGGGAAGTCTGGC AGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTACTTTCATTTCTTAGCCAGATGAATGTTGTCAACCATTTTAGGATT ATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCTTCACTGTCAGACCTAATAGGTTATTACAGTCATGTTTCTTGTTTG CTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCAGTAGAAGATAGAAGGCGTGTACGAGCTATTCTACCTTACACAAAA GTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTCATTGTTCATAATGAATTAGAAGATGGATGGATGTGGGTTACAAAT TTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAAGAGGTGGGCCGGGAAGAAGATCCACATGAAGGAAAAATATGGTTC CATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACAGTTGGTCAAGTCTGCAGTTTTCTTGTGAGGCCCTCAGATAATACT CCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAGCGATTTAAAATATGTCCAACGCCAAACAATCAGTTTATGATGGGA GGCCGGTATTATAACAGTTCTAGGCCTGATTCTTCACTACCTGAGACGTCTAAAAAGGAGCACATCTCAGCTGAAAACATGTCTTTAGAA ACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCTCAAAAAATACTTTGATGTTCACTAGACTATGTTTTCTAT TCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTACCGTATCTTACAATTTAATTACATACACAGTGAATTGAAA CCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATGACACTGGCACATTATTCTAAACTATTCATTCAGCATGCC TATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATCATATTCATTCACACATATTATATGTGATAGCTGTCCAAC ATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAAATGTCTTCAGCTTTTTTTGAATAGACGTATTCAAACATA >72378_72378_1_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000274376_XRCC4_chr5_82648944_ENST00000338635_length(amino acids)=456AA_BP=417 MMAAEAGSEEGGPVTAGAGGGGAAAGSSAYPAVCRVKIPAALPVAAAPYPGLVETGVAGTLGGGAALGSEFLGAGSVAGALGGAGLTGGG TAAGVAGAAAGVAGAAVAGPSGDMALTKLPTSLLAETLGPGGGFPPLPPPPYLPPLGAGLGTVDEGDSLDGPEYEEEEVAIPLTAPPTNQ WYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYYSHVSCLLKGEKLLY PVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPHEGKIWFHGKISKQE AYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNSSRPDSSLPETSKKEHISAENMSLETLRNSSPE -------------------------------------------------------------- >72378_72378_2_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000274376_XRCC4_chr5_82648944_ENST00000396027_length(transcript)=2398nt_BP=1817nt CTGAGAGCTCCAGGTAGTGAGCAGTTCAGTCGATTTCCTCGTTACCCCGCCCCCCTTTCTCTTGCCCCCCCACCCCTCTCATCTGCCTGG TGGAGGATGAAGCGGCTGCAGTGGCCCCAGCCTCAGCAGCGGCACCGGCGGTGGCTGCGGTGTGGGTGGCCGGAACTGGGGTGGTGAAGA AGCGGTGGTGGCGGCTGAAGGAGCGGTAGCAGCCTCAGCCTCTTTCCCTGTGCTTCCTTTCCTCTTTAGTGCAGCGAGGAAGTTTTCGCT TCTGTACATGTGTTTGTGTGCGTGAGTGTGGGTGTGTGCGGTGAGGTTTGGGTGGCGTTTGTGCAGGCGTTGGGTTTTTTGCCCACTTGG CTTCCCGTAACCCAGGCAGCTGGGGAGCCTGGGCTGTGGCCCTAGGAGGGGGCGCGGCGGCGGGCTCTCTCCTTTTGTTGTTGTTTCCTC AGCCTGGGGAGCTGAAGGGGAGACGCGTCTGGGTGGGGCTGCTCGGAGCCCGGGCCTGGTGGCCCCTGGGGCTCCCGGGCGGGCAGGGTA GGGCAGAGTAGAGCGGGCTTCAACATGATGGCGGCCGAGGCCGGCAGTGAGGAGGGCGGCCCGGTAACAGCCGGAGCTGGAGGAGGCGGC GCGGCAGCGGGCTCCAGTGCCTATCCCGCAGTGTGTCGGGTGAAGATACCCGCGGCCCTGCCTGTGGCAGCCGCCCCCTATCCTGGGCTG GTGGAGACCGGAGTGGCTGGAACTCTGGGTGGCGGAGCCGCTTTGGGGTCAGAGTTCCTAGGAGCCGGGTCTGTGGCAGGGGCACTGGGG GGAGCTGGACTGACAGGGGGAGGTACTGCTGCTGGCGTAGCTGGTGCTGCTGCTGGCGTGGCCGGTGCTGCTGTTGCTGGACCTAGTGGA GACATGGCTCTCACCAAACTGCCCACTTCGTTGCTTGCTGAGACTCTCGGGCCAGGCGGCGGTTTTCCCCCTCTGCCCCCTCCCCCTTAC CTGCCCCCTTTGGGGGCGGGCCTCGGGACAGTGGACGAAGGTGACTCTCTGGATGGACCAGAATACGAGGAGGAAGAGGTGGCCATACCG TTGACCGCTCCTCCAACTAACCAGTGGTATCACGGAAAACTTGACAGAACGATAGCAGAAGAACGCCTCAGGCAGGCAGGGAAGTCTGGC AGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTACTTTCATTTCTTAGCCAGATGAATGTTGTCAACCATTTTAGGATT ATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCTTCACTGTCAGACCTAATAGGTTATTACAGTCATGTTTCTTGTTTG CTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCAGTAGAAGATAGAAGGCGTGTACGAGCTATTCTACCTTACACAAAA GTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTCATTGTTCATAATGAATTAGAAGATGGATGGATGTGGGTTACAAAT TTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAAGAGGTGGGCCGGGAAGAAGATCCACATGAAGGAAAAATATGGTTC CATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACAGTTGGTCAAGTCTGCAGTTTTCTTGTGAGGCCCTCAGATAATACT CCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAGCGATTTAAAATATGTCCAACGCCAAACAATCAGTTTATGATGGGA GGCCGGTATTATAACAGGCCTGATTCTTCACTACCTGAGACGTCTAAAAAGGAGCACATCTCAGCTGAAAACATGTCTTTAGAAACTCTG AGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCTCAAAAAATACTTTGATGTTCACTAGACTATGTTTTCTATTCATTT CTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTACCGTATCTTACAATTTAATTACATACACAGTGAATTGAAACCATTG TGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATGACACTGGCACATTATTCTAAACTATTCATTCAGCATGCCTATAAT TACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATCATATTCATTCACACATATTATATGTGATAGCTGTCCAACATCCTG TCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAAATGTCTTCAGCTTTTTTTGAATAGACGTATTCAAACATATTCTGA >72378_72378_2_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000274376_XRCC4_chr5_82648944_ENST00000396027_length(amino acids)=454AA_BP=417 MMAAEAGSEEGGPVTAGAGGGGAAAGSSAYPAVCRVKIPAALPVAAAPYPGLVETGVAGTLGGGAALGSEFLGAGSVAGALGGAGLTGGG TAAGVAGAAAGVAGAAVAGPSGDMALTKLPTSLLAETLGPGGGFPPLPPPPYLPPLGAGLGTVDEGDSLDGPEYEEEEVAIPLTAPPTNQ WYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYYSHVSCLLKGEKLLY PVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPHEGKIWFHGKISKQE AYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNRPDSSLPETSKKEHISAENMSLETLRNSSPEDL -------------------------------------------------------------- >72378_72378_3_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000456692_XRCC4_chr5_82648944_ENST00000338635_length(transcript)=1424nt_BP=837nt GACCAGAATACGAGGAGGAAGAGGTGGCCATACCGTTGACCGCTCCTCCAACTAACCAGTAAGTTAAGACTGCTGTTCAGGAATTTGGGA AGCTGGCTCCAGAAAAGAAGTGGAAATGAAGGGGTGGTATCACGGAAAACTTGACAGAACGATAGCAGAAGAACGCCTCAGGCAGGCAGG GAAGTCTGGCAGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTACTTTCATTTCTTAGCCAGATGAATGTTGTCAACCA TTTTAGGATTATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCTTCACTGTCAGACCTAATAGGTTATTACAGTCATGT TTCTTGTTTGCTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCAGTAGAAGATAGAAGGCGTGTACGAGCTATTCTACC TTACACAAAAGTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTCATTGTTCATAATGAATTAGAAGATGGATGGATGTG GGTTACAAATTTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAAGAGGTGGGCCGGGAAGAAGATCCACATGAAGGAAA AATATGGTTCCATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACAGTTGGTCAAGTCTGCAGTTTTCTTGTGAGGCCCTC AGATAATACTCCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAGCGATTTAAAATATGTCCAACGCCAAACAATCAGTT TATGATGGGAGGCCGGTATTATAACAGTTCTAGGCCTGATTCTTCACTACCTGAGACGTCTAAAAAGGAGCACATCTCAGCTGAAAACAT GTCTTTAGAAACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCTCAAAAAATACTTTGATGTTCACTAGACTA TGTTTTCTATTCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTACCGTATCTTACAATTTAATTACATACACAG TGAATTGAAACCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATGACACTGGCACATTATTCTAAACTATTCAT TCAGCATGCCTATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATCATATTCATTCACACATATTATATGTGATA GCTGTCCAACATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAAATGTCTTCAGCTTTTTTTGAATAGACGTA >72378_72378_3_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000456692_XRCC4_chr5_82648944_ENST00000338635_length(amino acids)=279AA_BP=240 MKGWYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYYSHVSCLLKGEK LLYPVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPHEGKIWFHGKIS KQEAYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNSSRPDSSLPETSKKEHISAENMSLETLRNS -------------------------------------------------------------- >72378_72378_4_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000456692_XRCC4_chr5_82648944_ENST00000396027_length(transcript)=1418nt_BP=837nt GACCAGAATACGAGGAGGAAGAGGTGGCCATACCGTTGACCGCTCCTCCAACTAACCAGTAAGTTAAGACTGCTGTTCAGGAATTTGGGA AGCTGGCTCCAGAAAAGAAGTGGAAATGAAGGGGTGGTATCACGGAAAACTTGACAGAACGATAGCAGAAGAACGCCTCAGGCAGGCAGG GAAGTCTGGCAGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTACTTTCATTTCTTAGCCAGATGAATGTTGTCAACCA TTTTAGGATTATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCTTCACTGTCAGACCTAATAGGTTATTACAGTCATGT TTCTTGTTTGCTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCAGTAGAAGATAGAAGGCGTGTACGAGCTATTCTACC TTACACAAAAGTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTCATTGTTCATAATGAATTAGAAGATGGATGGATGTG GGTTACAAATTTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAAGAGGTGGGCCGGGAAGAAGATCCACATGAAGGAAA AATATGGTTCCATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACAGTTGGTCAAGTCTGCAGTTTTCTTGTGAGGCCCTC AGATAATACTCCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAGCGATTTAAAATATGTCCAACGCCAAACAATCAGTT TATGATGGGAGGCCGGTATTATAACAGGCCTGATTCTTCACTACCTGAGACGTCTAAAAAGGAGCACATCTCAGCTGAAAACATGTCTTT AGAAACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCTCAAAAAATACTTTGATGTTCACTAGACTATGTTTT CTATTCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTACCGTATCTTACAATTTAATTACATACACAGTGAATT GAAACCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATGACACTGGCACATTATTCTAAACTATTCATTCAGCA TGCCTATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATCATATTCATTCACACATATTATATGTGATAGCTGTC CAACATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAAATGTCTTCAGCTTTTTTTGAATAGACGTATTCAAA >72378_72378_4_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000456692_XRCC4_chr5_82648944_ENST00000396027_length(amino acids)=277AA_BP=240 MKGWYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYYSHVSCLLKGEK LLYPVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPHEGKIWFHGKIS KQEAYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNRPDSSLPETSKKEHISAENMSLETLRNSSP -------------------------------------------------------------- >72378_72378_5_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000506290_XRCC4_chr5_82648944_ENST00000338635_length(transcript)=1459nt_BP=872nt GAGGAGGAAGAGGTGGCCATACCGTTGACCGCTCCTCCAACTAACCAGTAAGTTAAGACTGCTGTTCAGGAATTTGGGAAGCTGGCTCCA GAAAAGAAGTGGAAATGAAGGGGTTCCATGTGCCTTGTGAAGTCATGGTTCCTCAGTATAAGATCGAGGTGGTATCACGGAAAACTTGAC AGAACGATAGCAGAAGAACGCCTCAGGCAGGCAGGGAAGTCTGGCAGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTA CTTTCATTTCTTAGCCAGATGAATGTTGTCAACCATTTTAGGATTATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCT TCACTGTCAGACCTAATAGGTTATTACAGTCATGTTTCTTGTTTGCTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCA GTAGAAGATAGAAGGCGTGTACGAGCTATTCTACCTTACACAAAAGTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTC ATTGTTCATAATGAATTAGAAGATGGATGGATGTGGGTTACAAATTTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAA GAGGTGGGCCGGGAAGAAGATCCACATGAAGGAAAAATATGGTTCCATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACA GTTGGTCAAGTCTGCAGTTTTCTTGTGAGGCCCTCAGATAATACTCCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAG CGATTTAAAATATGTCCAACGCCAAACAATCAGTTTATGATGGGAGGCCGGTATTATAACAGTTCTAGGCCTGATTCTTCACTACCTGAG ACGTCTAAAAAGGAGCACATCTCAGCTGAAAACATGTCTTTAGAAACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAA CAGTCTCAAAAAATACTTTGATGTTCACTAGACTATGTTTTCTATTCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGC TATTACCGTATCTTACAATTTAATTACATACACAGTGAATTGAAACCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTG ATGATGACACTGGCACATTATTCTAAACTATTCATTCAGCATGCCTATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACA TTTATCATATTCATTCACACATATTATATGTGATAGCTGTCCAACATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCAT TTTAAAATGTCTTCAGCTTTTTTTGAATAGACGTATTCAAACATATTCTGAACATTGATGTTTGAACATTTTAATTTGTGTGATGATGTA >72378_72378_5_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000506290_XRCC4_chr5_82648944_ENST00000338635_length(amino acids)=290AA_BP=251 MCLVKSWFLSIRSRWYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYY SHVSCLLKGEKLLYPVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPH EGKIWFHGKISKQEAYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNSSRPDSSLPETSKKEHISA -------------------------------------------------------------- >72378_72378_6_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000506290_XRCC4_chr5_82648944_ENST00000396027_length(transcript)=1453nt_BP=872nt GAGGAGGAAGAGGTGGCCATACCGTTGACCGCTCCTCCAACTAACCAGTAAGTTAAGACTGCTGTTCAGGAATTTGGGAAGCTGGCTCCA GAAAAGAAGTGGAAATGAAGGGGTTCCATGTGCCTTGTGAAGTCATGGTTCCTCAGTATAAGATCGAGGTGGTATCACGGAAAACTTGAC AGAACGATAGCAGAAGAACGCCTCAGGCAGGCAGGGAAGTCTGGCAGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTA CTTTCATTTCTTAGCCAGATGAATGTTGTCAACCATTTTAGGATTATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCT TCACTGTCAGACCTAATAGGTTATTACAGTCATGTTTCTTGTTTGCTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCA GTAGAAGATAGAAGGCGTGTACGAGCTATTCTACCTTACACAAAAGTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTC ATTGTTCATAATGAATTAGAAGATGGATGGATGTGGGTTACAAATTTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAA GAGGTGGGCCGGGAAGAAGATCCACATGAAGGAAAAATATGGTTCCATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACA GTTGGTCAAGTCTGCAGTTTTCTTGTGAGGCCCTCAGATAATACTCCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAG CGATTTAAAATATGTCCAACGCCAAACAATCAGTTTATGATGGGAGGCCGGTATTATAACAGGCCTGATTCTTCACTACCTGAGACGTCT AAAAAGGAGCACATCTCAGCTGAAAACATGTCTTTAGAAACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCT CAAAAAATACTTTGATGTTCACTAGACTATGTTTTCTATTCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTAC CGTATCTTACAATTTAATTACATACACAGTGAATTGAAACCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATG ACACTGGCACATTATTCTAAACTATTCATTCAGCATGCCTATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATC ATATTCATTCACACATATTATATGTGATAGCTGTCCAACATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAA ATGTCTTCAGCTTTTTTTGAATAGACGTATTCAAACATATTCTGAACATTGATGTTTGAACATTTTAATTTGTGTGATGATGTAGAAAAT >72378_72378_6_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000506290_XRCC4_chr5_82648944_ENST00000396027_length(amino acids)=288AA_BP=251 MCLVKSWFLSIRSRWYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYY SHVSCLLKGEKLLYPVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPH EGKIWFHGKISKQEAYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNRPDSSLPETSKKEHISAEN -------------------------------------------------------------- >72378_72378_7_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000512763_XRCC4_chr5_82648944_ENST00000338635_length(transcript)=1540nt_BP=953nt GAGGAGGAAGAGGTGGCCATACCGTTGACCGCTCCTCCAACTAACCAGTAAGTTAAGACTGCTGTTCAGGAATTTGGGAAGCTGGCTCCA GAAAAGAAGTGGAAATGAAGGGGCATACTACCTGGCGTCTTCCTACATTTATTGCTCCTCTGGGAAAGTGTAAAGTCTTTAAGATGGAAA GCATGAGAAAATGTGTATCTAATGAGAACCGGATATAGTTCTGTCCCTTCCAAGTTGAGGTGGTATCACGGAAAACTTGACAGAACGATA GCAGAAGAACGCCTCAGGCAGGCAGGGAAGTCTGGCAGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTACTTTCATTT CTTAGCCAGATGAATGTTGTCAACCATTTTAGGATTATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCTTCACTGTCA GACCTAATAGGTTATTACAGTCATGTTTCTTGTTTGCTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCAGTAGAAGAT AGAAGGCGTGTACGAGCTATTCTACCTTACACAAAAGTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTCATTGTTCAT AATGAATTAGAAGATGGATGGATGTGGGTTACAAATTTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAAGAGGTGGGC CGGGAAGAAGATCCACATGAAGGAAAAATATGGTTCCATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACAGTTGGTCAA GTCTGCAGTTTTCTTGTGAGGCCCTCAGATAATACTCCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAGCGATTTAAA ATATGTCCAACGCCAAACAATCAGTTTATGATGGGAGGCCGGTATTATAACAGTTCTAGGCCTGATTCTTCACTACCTGAGACGTCTAAA AAGGAGCACATCTCAGCTGAAAACATGTCTTTAGAAACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCTCAA AAAATACTTTGATGTTCACTAGACTATGTTTTCTATTCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTACCGT ATCTTACAATTTAATTACATACACAGTGAATTGAAACCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATGACA CTGGCACATTATTCTAAACTATTCATTCAGCATGCCTATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATCATA TTCATTCACACATATTATATGTGATAGCTGTCCAACATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAAATG TCTTCAGCTTTTTTTGAATAGACGTATTCAAACATATTCTGAACATTGATGTTTGAACATTTTAATTTGTGTGATGATGTAGAAAATATA >72378_72378_7_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000512763_XRCC4_chr5_82648944_ENST00000338635_length(amino acids)=289AA_BP=250 MRTGYSSVPSKLRWYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYYS HVSCLLKGEKLLYPVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPHE GKIWFHGKISKQEAYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNSSRPDSSLPETSKKEHISAE -------------------------------------------------------------- >72378_72378_8_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000512763_XRCC4_chr5_82648944_ENST00000396027_length(transcript)=1534nt_BP=953nt GAGGAGGAAGAGGTGGCCATACCGTTGACCGCTCCTCCAACTAACCAGTAAGTTAAGACTGCTGTTCAGGAATTTGGGAAGCTGGCTCCA GAAAAGAAGTGGAAATGAAGGGGCATACTACCTGGCGTCTTCCTACATTTATTGCTCCTCTGGGAAAGTGTAAAGTCTTTAAGATGGAAA GCATGAGAAAATGTGTATCTAATGAGAACCGGATATAGTTCTGTCCCTTCCAAGTTGAGGTGGTATCACGGAAAACTTGACAGAACGATA GCAGAAGAACGCCTCAGGCAGGCAGGGAAGTCTGGCAGTTATCTTATAAGAGAGAGTGATCGGAGGCCAGGGTCCTTTGTACTTTCATTT CTTAGCCAGATGAATGTTGTCAACCATTTTAGGATTATTGCTATGTGTGGAGATTACTACATTGGTGGAAGACGTTTTTCTTCACTGTCA GACCTAATAGGTTATTACAGTCATGTTTCTTGTTTGCTTAAAGGAGAAAAATTACTTTACCCAGTTGCACCACCAGAGCCAGTAGAAGAT AGAAGGCGTGTACGAGCTATTCTACCTTACACAAAAGTACCAGACACTGATGAAATAAGTTTCTTAAAAGGAGATATGTTCATTGTTCAT AATGAATTAGAAGATGGATGGATGTGGGTTACAAATTTAAGAACAGATGAACAAGGCCTTATTGTTGAAGACCTAGTAGAAGAGGTGGGC CGGGAAGAAGATCCACATGAAGGAAAAATATGGTTCCATGGGAAGATTTCCAAACAGGAAGCTTATAATTTACTAATGACAGTTGGTCAA GTCTGCAGTTTTCTTGTGAGGCCCTCAGATAATACTCCTGGCGATTATTCACTTTATTTCCGGACCAATGAAAATATTCAGCGATTTAAA ATATGTCCAACGCCAAACAATCAGTTTATGATGGGAGGCCGGTATTATAACAGGCCTGATTCTTCACTACCTGAGACGTCTAAAAAGGAG CACATCTCAGCTGAAAACATGTCTTTAGAAACTCTGAGAAACAGCAGCCCAGAAGACCTCTTTGATGAGATTTAACAGTCTCAAAAAATA CTTTGATGTTCACTAGACTATGTTTTCTATTCATTTCTTTAAAATGAAAAAGGAGAATTTCAAGTCAGCAGCCGCTATTACCGTATCTTA CAATTTAATTACATACACAGTGAATTGAAACCATTGTGCAAAATGGATTACACATGTATACAAAGATACGATTTGATGATGACACTGGCA CATTATTCTAAACTATTCATTCAGCATGCCTATAATTACATAAATTGTATGAGACTTTTTGTTGCAAAGGACACATTTATCATATTCATT CACACATATTATATGTGATAGCTGTCCAACATCCTGTCTGGGAAGATTTTGAAAACAGGACAAAGAAAACATCATTTTAAAATGTCTTCA GCTTTTTTTGAATAGACGTATTCAAACATATTCTGAACATTGATGTTTGAACATTTTAATTTGTGTGATGATGTAGAAAATATAATTTTA >72378_72378_8_RASA1-XRCC4_RASA1_chr5_86645181_ENST00000512763_XRCC4_chr5_82648944_ENST00000396027_length(amino acids)=287AA_BP=250 MRTGYSSVPSKLRWYHGKLDRTIAEERLRQAGKSGSYLIRESDRRPGSFVLSFLSQMNVVNHFRIIAMCGDYYIGGRRFSSLSDLIGYYS HVSCLLKGEKLLYPVAPPEPVEDRRRVRAILPYTKVPDTDEISFLKGDMFIVHNELEDGWMWVTNLRTDEQGLIVEDLVEEVGREEDPHE GKIWFHGKISKQEAYNLLMTVGQVCSFLVRPSDNTPGDYSLYFRTNENIQRFKICPTPNNQFMMGGRYYNRPDSSLPETSKKEHISAENM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RASA1-XRCC4 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000282268 | 6 | 8 | 1_213 | 297.6666666666667 | 335.0 | IFFO1 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000338635 | 6 | 8 | 1_213 | 297.6666666666667 | 337.0 | IFFO1 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000396027 | 6 | 8 | 1_213 | 297.6666666666667 | 335.0 | IFFO1 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000511817 | 6 | 8 | 1_213 | 297.6666666666667 | 337.0 | IFFO1 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000282268 | 6 | 8 | 180_213 | 297.6666666666667 | 335.0 | LIG4 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000338635 | 6 | 8 | 180_213 | 297.6666666666667 | 337.0 | LIG4 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000396027 | 6 | 8 | 180_213 | 297.6666666666667 | 335.0 | LIG4 | |
| Tgene | XRCC4 | chr5:86645181 | chr5:82648944 | ENST00000511817 | 6 | 8 | 180_213 | 297.6666666666667 | 337.0 | LIG4 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RASA1-XRCC4 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RASA1-XRCC4 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
