
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RASAL2-KIF14 (FusionGDB2 ID:72423) |
Fusion Gene Summary for RASAL2-KIF14 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RASAL2-KIF14 | Fusion gene ID: 72423 | Hgene | Tgene | Gene symbol | RASAL2 | KIF14 | Gene ID | 9462 | 9928 |
| Gene name | RAS protein activator like 2 | kinesin family member 14 | |
| Synonyms | NGAP | MCPH20|MKS12 | |
| Cytomap | 1q25.2 | 1q32.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ras GTPase-activating protein nGAPRASAL2/ACVR1 fusionRas protein activator like 1 | kinesin-like protein KIF14epididymis secretory sperm binding protein | |
| Modification date | 20200313 | 20200315 | |
| UniProtAcc | . | Q15058 | |
| Ensembl transtripts involved in fusion gene | ENST00000465723, ENST00000367649, ENST00000448150, ENST00000462775, | ENST00000367350, | |
| Fusion gene scores | * DoF score | 19 X 13 X 10=2470 | 3 X 3 X 3=27 |
| # samples | 20 | 4 | |
| ** MAII score | log2(20/2470*10)=-3.62643913669732 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/27*10)=0.567040592723894 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: RASAL2 [Title/Abstract] AND KIF14 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RASAL2(178252826)-KIF14(200544819), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | KIF14 | GO:0008284 | positive regulation of cell proliferation | 24784001 |
| Tgene | KIF14 | GO:0034446 | substrate adhesion-dependent cell spreading | 23209302 |
| Tgene | KIF14 | GO:0045184 | establishment of protein localization | 23209302 |
 Fusion gene breakpoints across RASAL2 (5'-gene) Fusion gene breakpoints across RASAL2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
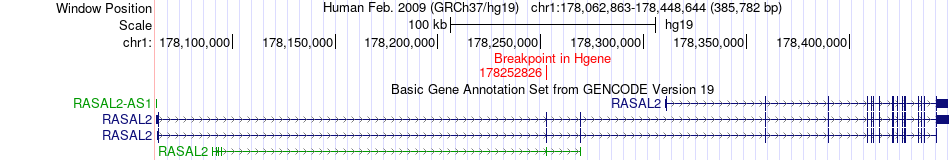 |
 Fusion gene breakpoints across KIF14 (3'-gene) Fusion gene breakpoints across KIF14 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
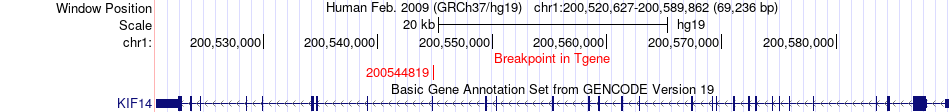 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SKCM | TCGA-BF-AAP4-01A | RASAL2 | chr1 | 178252826 | - | KIF14 | chr1 | 200544819 | - |
| ChimerDB4 | SKCM | TCGA-BF-AAP4-01A | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - |
Top |
Fusion Gene ORF analysis for RASAL2-KIF14 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000465723 | ENST00000367350 | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - |
| In-frame | ENST00000367649 | ENST00000367350 | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - |
| In-frame | ENST00000448150 | ENST00000367350 | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - |
| intron-3CDS | ENST00000462775 | ENST00000367350 | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000448150 | RASAL2 | chr1 | 178252826 | + | ENST00000367350 | KIF14 | chr1 | 200544819 | - | 4464 | 1094 | 410 | 2575 | 721 |
| ENST00000367649 | RASAL2 | chr1 | 178252826 | + | ENST00000367350 | KIF14 | chr1 | 200544819 | - | 4052 | 682 | 166 | 2163 | 665 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000448150 | ENST00000367350 | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - | 0.000447484 | 0.99955255 |
| ENST00000367649 | ENST00000367350 | RASAL2 | chr1 | 178252826 | + | KIF14 | chr1 | 200544819 | - | 0.000291167 | 0.99970883 |
Top |
Fusion Genomic Features for RASAL2-KIF14 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
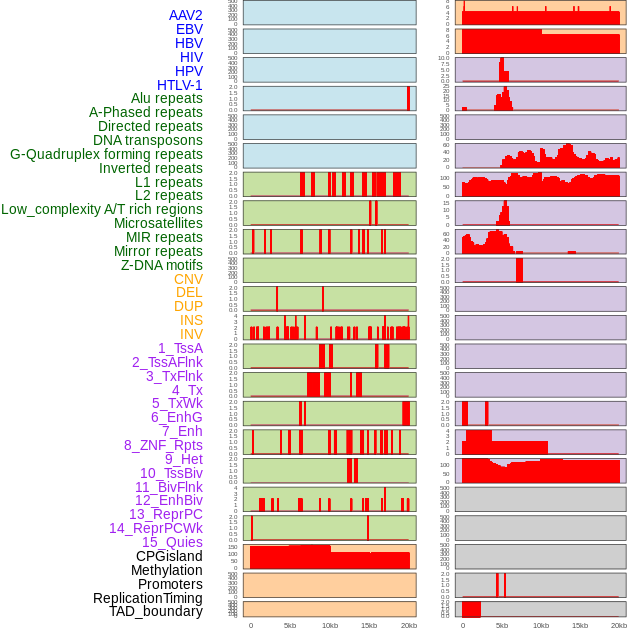 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for RASAL2-KIF14 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:178252826/chr1:200544819) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | KIF14 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Microtubule motor protein that binds to microtubules with high affinity through each tubulin heterodimer and has an ATPase activity (By similarity). Plays a role in many processes like cell division, cytokinesis and also in cell proliferation and apoptosis (PubMed:24784001, PubMed:16648480). During cytokinesis, targets to central spindle and midbody through its interaction with PRC1 and CIT respectively (PubMed:16431929). Regulates cell growth through regulation of cell cycle progression and cytokinesis (PubMed:24854087). During cell cycle progression acts through SCF-dependent proteasomal ubiquitin-dependent protein catabolic process which controls CDKN1B degradation, resulting in positive regulation of cyclins, including CCNE1, CCND1 and CCNB1 (PubMed:24854087). During late neurogenesis, regulates the cerebellar, cerebral cortex and olfactory bulb development through regulation of apoptosis, cell proliferation and cell division (By similarity). Also is required for chromosome congression and alignment during mitotic cell cycle process (PubMed:15843429). Regulates cell spreading, focal adhesion dynamics, and cell migration through its interaction with RADIL resulting in regulation of RAP1A-mediated inside-out integrin activation by tethering RADIL on microtubules (PubMed:23209302). {ECO:0000250|UniProtKB:L0N7N1, ECO:0000269|PubMed:15843429, ECO:0000269|PubMed:16431929, ECO:0000269|PubMed:16648480, ECO:0000269|PubMed:23209302, ECO:0000269|PubMed:24784001, ECO:0000269|PubMed:24854087}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 1332_1348 | 1155 | 1649.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 1468_1500 | 1155 | 1649.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000367649 | + | 2 | 18 | 237_243 | 110 | 1281.0 | Compositional bias | Note=Poly-Lys |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000367649 | + | 2 | 18 | 922_925 | 110 | 1281.0 | Compositional bias | Note=Poly-Ser |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000462775 | + | 1 | 16 | 237_243 | 0 | 1140.0 | Compositional bias | Note=Poly-Lys |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000462775 | + | 1 | 16 | 922_925 | 0 | 1140.0 | Compositional bias | Note=Poly-Ser |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000367649 | + | 2 | 18 | 149_267 | 110 | 1281.0 | Domain | C2 |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000367649 | + | 2 | 18 | 327_519 | 110 | 1281.0 | Domain | Ras-GAP |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000367649 | + | 2 | 18 | 41_158 | 110 | 1281.0 | Domain | Note=PH |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000462775 | + | 1 | 16 | 149_267 | 0 | 1140.0 | Domain | C2 |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000462775 | + | 1 | 16 | 327_519 | 0 | 1140.0 | Domain | Ras-GAP |
| Hgene | RASAL2 | chr1:178252826 | chr1:200544819 | ENST00000462775 | + | 1 | 16 | 41_158 | 0 | 1140.0 | Domain | Note=PH |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 705_791 | 1155 | 1649.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 922_1079 | 1155 | 1649.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 358_701 | 1155 | 1649.0 | Domain | Kinesin motor | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 825_891 | 1155 | 1649.0 | Domain | Note=FHA | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 447_454 | 1155 | 1649.0 | Nucleotide binding | ATP | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 1_356 | 1155 | 1649.0 | Region | Note=Required for PRC1-binding | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 356_737 | 1155 | 1649.0 | Region | Required for microtubule-binding with high affinity | |
| Tgene | KIF14 | chr1:178252826 | chr1:200544819 | ENST00000367350 | 20 | 30 | 901_1648 | 1155 | 1649.0 | Region | Note=Required for CIT-binding |
Top |
Fusion Gene Sequence for RASAL2-KIF14 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >72423_72423_1_RASAL2-KIF14_RASAL2_chr1_178252826_ENST00000367649_KIF14_chr1_200544819_ENST00000367350_length(transcript)=4052nt_BP=682nt GAAGAAGGCGGCTCCGAGGAGGTGGGAGGGCGAGCCTCCCCTCCCGGGTTCTTCTGCGTCCTCTCCCGGGAGGGACCCACACACACCTGA GCCCGGACCCACCCTTGGTCGGGGCCACGCTGGATCCTCCTCCCGGCCTGGGTCCCGCCCGCCGACTGCAGGTGGGCTGCATCGCCCGAG CCTCGGGCAGTGGGCGACGGGGAAGGAGGTGAGAGGTGTCCGCGCCGGCTGCCGCTCGGGTCCCTGCCCTCGCTGCGCGCTCTCCTCCTC CCCTTACCGCAGGCAGGGCGCGGAGCCGCGCGCCAGCCCGCCCCGAAGCCGCCGCCTCGTCCCCCTCCCGCCTCGGGGCACCATGGAGCT CTCTCCGTCGTCCGGAGGAGCCGCGGAGGCGCTGTCCTGGCCGGAGATGTTCCCGGCGCTGGAGTCCGACTCGCCGCTGCCCCCGGAGGA CCTGGACGCGGTTGTCCCAGTCAGTGGAGCCGTCGCCGGTGGCATGTTGGATCGGATCCTTCTGGAGTCCGTGTGCCAGCAACAGAGCTG GGTCCGGGTGTACGATGTGAAAGGACCACCCACCCACCGTCTGTCTTGTGGTCAGTCACCCTACACCGAGACAACAACGTGGGAGCGGAA GTATTGCATCCTCACAGACAGCCAGTTGGTATTGCTCAACAAGGAGAAGGAGAGTAATGGTAGTAACAGGGGTGAAGATGCCTTTTGTGA TCCTGAAGATGAATGGGAACCCGACATTACAGATGCACCAGTTTCTTCACTTTCTAGAAGGAGGAGTAGGAGTTTGATGAAGAACAGAAG AATTTCTGGTTGTTTACATGACATACAAGTCCATCCAATTAAGAATTTGCATTCTTCACATTCATCAGGTTTAATGGACAAATCAAGCAC TATTTACTCAAATTCAGCAGAGTCCTTTCTTCCTGGAATTTGCAAAGAATTGATTGGTTCTTCGTTAGATTTTTTTGGACAGAGTTATGA TGAAGAAAGAACTATAGCAGACAGCCTAATTAATAGTTTTCTTAAAATTTATAATGGGCTATTTGCCATTTCCAAGGCTCATGAAGAACA AGATGAAGAAAGTCAAGATAACTTGTTTTCTTCTGATCGAGCAATCCAGTCACTTACTATTCAGACTGCATGTGCTTTTGAGCAGCTAGT AGTGCTAATGAAACACTGGCTGAGTGATTTACTGCCTTGTACCAACATAGCAAGACTTGAGGATGAGTTGAGACAAGAAGTTAAAAAACT GGGAGGCTACTTACAGTTATTTTTGCAGGGATGCTGTTTGGATATTTCATCAATGATAAAAGAGGCTCAAAAGAATGCAATCCAAATTGT ACAACAAGCTGTAAAGTATGTGGGGCAGTTAGCAGTTCTGAAAGGGAGCAAGCTACATTTTCTAGAAAACGGTAACAATAAAGCTGCCAG TGTCCAGGAGGAATTCATGGATGCTGTTTGTGATGGTGTAGGCTTAGGAATGAAGATTTTATTAGATTCTGGACTGGAAAAAGCAAAAGA ACTTCAGCATGAACTCTTTAGGCAGTGTACAAAAAATGAGGTTACCAAAGAAATGAAAACTAATGCCATGGGATTGATTAGATCTCTTGA AAACATCTTTGCTGAATCGAAAATTAAAAGTTTCAGAAGGCAAGTACAAGAAGAAAACTTTGAATACCAAGATTTCAAGAGGATGGTTAA TCGTGCTCCAGAATTCTTAAAGTTAAAACATTGCTTAGAGAAAGCTATTGAAATTATTATTTCTGCACTGAAAGGATGCCATAGTGATAT AAATCTTCTCCAGACTTGTGTTGAAAGTATTCGCAACTTGGCCAGTGATTTTTACAGTGACTTCAGTGTGCCTTCTACTTCTGTTGGCAG CTATGAGAGTAGAGTAACTCACATTGTCCACCAGGAACTAGAATCTCTAGCTAAGTCTCTCCTCTTTTGTTTTGAATCTGAAGAAAGCCC TGATTTGTTGAAACCCTGGGAAACTTATAATCAAAATACCAAAGAAGAACACCAACAATCTAAATCAAGCGGGATTGACGGCAGTAAGAA TAAAGGTGTACCAAAGCGTGTCTATGAGCTCCATGGCTCATCCCCAGCAGTGAGCTCAGAGGAATGCACACCCAGTAGGATTCAGTGGGT GTGAATACTGATGTGTAGGCACTTTTATGACCACCCATGAAAGAAAAAGAACACTTGCTCGGTAATTTTCTTTATGCAGGAGAGTTTAAG AGAAATCAGCACAGATATTTCAAAAAAGTCCATGTCTTTTTATCTTTAAAATATCTATTTATCAAAGGCCAGACACAGTGGCTCACGCCT GTAATCCCAGCACTTTGGGAGGCGGGCAGATCACAAGGTCAGGAGTTTGAGACCGGCCTGGCCAACATGGCGAAACCCCGTCTCTACTAA AAATACAAAAATTTGCTGGGCATGGTGGCGCGTGCCTGTAATCCCAGCTACTAGGGGGGCTGAGGCAGGAGGATCGCTTGAACCTGAGAG GCAGAGGTTGCAGTGAGCCAAGATCATGCCACTTTACTCCAGTCTGAGCAACAGAACGAGACTTAGTCAAAATAAATAAATAAATAAATA AATAAATAAATAAATAAATAAATAAAATATATTTTTATCTTTAAAGTGTTTAACATTGGTATACTGTCTGTAGTTGGTTCATTAGTCGTT TATAAAGGGTTATTTTCTCATGAGTGGAAACCTGAACAATCAGTTACCTTTGTGCCTATGCCTTCTCTCTCCTCAGACAGCTGGGATGTT TATGGTGAAATGGCCTGTACAAGTTTAACTAAGACAACTTAACTTGCATTGTTAATCAAAAATTCTTTTCTCAAAGGGTTAACTGGTTGC CATTTTGAATAGTATGTTCAAGGGTGTAGCTTCCTGTTTCTTTCCAAATTATAAGTAGCTACCTAAATATAGTATAATTATATATTAATA ATATGGCTTGCTGGCACAGTAGTTTACCCTGTTATCTGTGTTTCATAATGGGGGCTGTATGAATATTATTTAAAACTAATAAAATGTTGC CAGAATTATACTAAACTGTTGGATGAGATTAGGAGATCAGAGGCTGGACCTTCTCTTGATAATGCTTGTTTTGTTAAAGGTATAATGAAA TAATTTGTATATGATTTGATGAAGATTAAAGACCCTTATTTTCCACAGCTTTAAAAAAAAACCTTTATTTATGATCAAGTAATAAAGATA ATATTCTACTTGTGGGATCTTACATTACGGAAATAGTTTGACGTTTTTGACCTCAAGAGTATGTATAATTTGAAGAGATACTTTGTAACT ATGCTTGGGTGATATTGAGCAGTTCCTAAAGAATAATTCATTTAAAAAAAAAGAAGAAAAAAAAAGAAGAATTCATTTAAATAACCTGAT CCTTTCATTTGCCCTTTTCGAATTTACAGATACTACTTGTACATTTGGCATAACTAGTTGAAATTGGCCATTCGTACCATGAATAAATCT GATAGTTTCCTTGTTAGGAAGAGATTGTAAGTAAATACAGTCATTGCAGTCAGAACAGTATTAGTGAACCTTGTGTGGTGTTTTCAAGCT CTTTAAAATGGTACAATGTAGCACATTTGCTTTCATTTCTTTTTTTATTTTTGGCATTTGACCTTGTATTCTTTCTGAAGCTCTATATGT GTTTTTATTAGTCAATAATCTGGCAAGTAGCACTTTGCCTGTGCAGTTTGCTGGAGTGTAGATGTACATATGAGGATTTCCCGGGAGGTG CACTTCTTTGAAGAACTTCCTAAAGTACCTGTATAGTAGTTTTCATCTTAATATTCAGTATTTAATCTTCAGTTTGTGCTTTGTAAACTC ATGACTTAATTGGTCAGAAACTTTTTAGTGTCTTTATAAAATTTTGTATACATATTTATACTAAACACATTGTGATACTGTATTTGAATG AATGGTGAAAAAATATTTGCTATTGGAATTATGTGCACTGACAAGAAATGTTATAAAGAGAATGCCTTTAATAAATCTTTTCAGCATTAG >72423_72423_1_RASAL2-KIF14_RASAL2_chr1_178252826_ENST00000367649_KIF14_chr1_200544819_ENST00000367350_length(amino acids)=665AA_BP=172 MHRPSLGQWATGKEVRGVRAGCRSGPCPRCALSSSPYRRQGAEPRASPPRSRRLVPLPPRGTMELSPSSGGAAEALSWPEMFPALESDSP LPPEDLDAVVPVSGAVAGGMLDRILLESVCQQQSWVRVYDVKGPPTHRLSCGQSPYTETTTWERKYCILTDSQLVLLNKEKESNGSNRGE DAFCDPEDEWEPDITDAPVSSLSRRRSRSLMKNRRISGCLHDIQVHPIKNLHSSHSSGLMDKSSTIYSNSAESFLPGICKELIGSSLDFF GQSYDEERTIADSLINSFLKIYNGLFAISKAHEEQDEESQDNLFSSDRAIQSLTIQTACAFEQLVVLMKHWLSDLLPCTNIARLEDELRQ EVKKLGGYLQLFLQGCCLDISSMIKEAQKNAIQIVQQAVKYVGQLAVLKGSKLHFLENGNNKAASVQEEFMDAVCDGVGLGMKILLDSGL EKAKELQHELFRQCTKNEVTKEMKTNAMGLIRSLENIFAESKIKSFRRQVQEENFEYQDFKRMVNRAPEFLKLKHCLEKAIEIIISALKG CHSDINLLQTCVESIRNLASDFYSDFSVPSTSVGSYESRVTHIVHQELESLAKSLLFCFESEESPDLLKPWETYNQNTKEEHQQSKSSGI -------------------------------------------------------------- >72423_72423_2_RASAL2-KIF14_RASAL2_chr1_178252826_ENST00000448150_KIF14_chr1_200544819_ENST00000367350_length(transcript)=4464nt_BP=1094nt CCCTCGATTGCTTCAGCCTATCACCCTGGGATGCCGCCTGCCGCTGCCGCCGCCGCCGCCGCCGCCACCGCCACCGCCACCGCCACCGCC ACCGCCGTGGCTGCCGGGTTTGTGGGATCCGCCGCGGAGCAGGAGCCAGAGCTGTGGCCGGAGCTGTGGCCTGAGAGTCAGGGGGCGGGC AGGCTCATTCCAGAAGTGGGGGCCGAGGACAGGCGGGCTAGAACCCGGGGCCACCGCCCCCTACCCCTCCTCAGGCTGAGGGAACGGGCA CTCGGAGCTTCTGAGGCGGCGGCGGCGGGAGCCGGTCAGAGGGGGCGGCCCCTGGCGGCGACGCCCCCAAGCCCAGCCCTGCCCGGCCCC GCTCCATCCCCAGCCAGAGCCTGGTCTGACCGCGAGAGACGCCGGCGGGGCTGAAGAAGGCGGCTCCGAGGAGGTGGGAGGGCGAGCCTC CCCTCCCGGGTTCTTCTGCGTCCTCTCCCGGGAGGGACCCACACACACCTGAGCCCGGACCCACCCTTGGTCGGGGCCACGCTGGATCCT CCTCCCGGCCTGGGTCCCGCCCGCCGACTGCAGGTGGGCTGCATCGCCCGAGCCTCGGGCAGTGGGCGACGGGGAAGGAGGTGAGAGGTG TCCGCGCCGGCTGCCGCTCGGGTCCCTGCCCTCGCTGCGCGCTCTCCTCCTCCCCTTACCGCAGGCAGGGCGCGGAGCCGCGCGCCAGCC CGCCCCGAAGCCGCCGCCTCGTCCCCCTCCCGCCTCGGGGCACCATGGAGCTCTCTCCGTCGTCCGGAGGAGCCGCGGAGGCGCTGTCCT GGCCGGAGATGTTCCCGGCGCTGGAGTCCGACTCGCCGCTGCCCCCGGAGGACCTGGACGCGGTTGTCCCAGTCAGTGGAGCCGTCGCCG GTGGCATGTTGGATCGGATCCTTCTGGAGTCCGTGTGCCAGCAACAGAGCTGGGTCCGGGTGTACGATGTGAAAGGACCACCCACCCACC GTCTGTCTTGTGGTCAGTCACCCTACACCGAGACAACAACGTGGGAGCGGAAGTATTGCATCCTCACAGACAGCCAGTTGGTATTGCTCA ACAAGGAGAAGGAGAGTAATGGTAGTAACAGGGGTGAAGATGCCTTTTGTGATCCTGAAGATGAATGGGAACCCGACATTACAGATGCAC CAGTTTCTTCACTTTCTAGAAGGAGGAGTAGGAGTTTGATGAAGAACAGAAGAATTTCTGGTTGTTTACATGACATACAAGTCCATCCAA TTAAGAATTTGCATTCTTCACATTCATCAGGTTTAATGGACAAATCAAGCACTATTTACTCAAATTCAGCAGAGTCCTTTCTTCCTGGAA TTTGCAAAGAATTGATTGGTTCTTCGTTAGATTTTTTTGGACAGAGTTATGATGAAGAAAGAACTATAGCAGACAGCCTAATTAATAGTT TTCTTAAAATTTATAATGGGCTATTTGCCATTTCCAAGGCTCATGAAGAACAAGATGAAGAAAGTCAAGATAACTTGTTTTCTTCTGATC GAGCAATCCAGTCACTTACTATTCAGACTGCATGTGCTTTTGAGCAGCTAGTAGTGCTAATGAAACACTGGCTGAGTGATTTACTGCCTT GTACCAACATAGCAAGACTTGAGGATGAGTTGAGACAAGAAGTTAAAAAACTGGGAGGCTACTTACAGTTATTTTTGCAGGGATGCTGTT TGGATATTTCATCAATGATAAAAGAGGCTCAAAAGAATGCAATCCAAATTGTACAACAAGCTGTAAAGTATGTGGGGCAGTTAGCAGTTC TGAAAGGGAGCAAGCTACATTTTCTAGAAAACGGTAACAATAAAGCTGCCAGTGTCCAGGAGGAATTCATGGATGCTGTTTGTGATGGTG TAGGCTTAGGAATGAAGATTTTATTAGATTCTGGACTGGAAAAAGCAAAAGAACTTCAGCATGAACTCTTTAGGCAGTGTACAAAAAATG AGGTTACCAAAGAAATGAAAACTAATGCCATGGGATTGATTAGATCTCTTGAAAACATCTTTGCTGAATCGAAAATTAAAAGTTTCAGAA GGCAAGTACAAGAAGAAAACTTTGAATACCAAGATTTCAAGAGGATGGTTAATCGTGCTCCAGAATTCTTAAAGTTAAAACATTGCTTAG AGAAAGCTATTGAAATTATTATTTCTGCACTGAAAGGATGCCATAGTGATATAAATCTTCTCCAGACTTGTGTTGAAAGTATTCGCAACT TGGCCAGTGATTTTTACAGTGACTTCAGTGTGCCTTCTACTTCTGTTGGCAGCTATGAGAGTAGAGTAACTCACATTGTCCACCAGGAAC TAGAATCTCTAGCTAAGTCTCTCCTCTTTTGTTTTGAATCTGAAGAAAGCCCTGATTTGTTGAAACCCTGGGAAACTTATAATCAAAATA CCAAAGAAGAACACCAACAATCTAAATCAAGCGGGATTGACGGCAGTAAGAATAAAGGTGTACCAAAGCGTGTCTATGAGCTCCATGGCT CATCCCCAGCAGTGAGCTCAGAGGAATGCACACCCAGTAGGATTCAGTGGGTGTGAATACTGATGTGTAGGCACTTTTATGACCACCCAT GAAAGAAAAAGAACACTTGCTCGGTAATTTTCTTTATGCAGGAGAGTTTAAGAGAAATCAGCACAGATATTTCAAAAAAGTCCATGTCTT TTTATCTTTAAAATATCTATTTATCAAAGGCCAGACACAGTGGCTCACGCCTGTAATCCCAGCACTTTGGGAGGCGGGCAGATCACAAGG TCAGGAGTTTGAGACCGGCCTGGCCAACATGGCGAAACCCCGTCTCTACTAAAAATACAAAAATTTGCTGGGCATGGTGGCGCGTGCCTG TAATCCCAGCTACTAGGGGGGCTGAGGCAGGAGGATCGCTTGAACCTGAGAGGCAGAGGTTGCAGTGAGCCAAGATCATGCCACTTTACT CCAGTCTGAGCAACAGAACGAGACTTAGTCAAAATAAATAAATAAATAAATAAATAAATAAATAAATAAATAAATAAAATATATTTTTAT CTTTAAAGTGTTTAACATTGGTATACTGTCTGTAGTTGGTTCATTAGTCGTTTATAAAGGGTTATTTTCTCATGAGTGGAAACCTGAACA ATCAGTTACCTTTGTGCCTATGCCTTCTCTCTCCTCAGACAGCTGGGATGTTTATGGTGAAATGGCCTGTACAAGTTTAACTAAGACAAC TTAACTTGCATTGTTAATCAAAAATTCTTTTCTCAAAGGGTTAACTGGTTGCCATTTTGAATAGTATGTTCAAGGGTGTAGCTTCCTGTT TCTTTCCAAATTATAAGTAGCTACCTAAATATAGTATAATTATATATTAATAATATGGCTTGCTGGCACAGTAGTTTACCCTGTTATCTG TGTTTCATAATGGGGGCTGTATGAATATTATTTAAAACTAATAAAATGTTGCCAGAATTATACTAAACTGTTGGATGAGATTAGGAGATC AGAGGCTGGACCTTCTCTTGATAATGCTTGTTTTGTTAAAGGTATAATGAAATAATTTGTATATGATTTGATGAAGATTAAAGACCCTTA TTTTCCACAGCTTTAAAAAAAAACCTTTATTTATGATCAAGTAATAAAGATAATATTCTACTTGTGGGATCTTACATTACGGAAATAGTT TGACGTTTTTGACCTCAAGAGTATGTATAATTTGAAGAGATACTTTGTAACTATGCTTGGGTGATATTGAGCAGTTCCTAAAGAATAATT CATTTAAAAAAAAAGAAGAAAAAAAAAGAAGAATTCATTTAAATAACCTGATCCTTTCATTTGCCCTTTTCGAATTTACAGATACTACTT GTACATTTGGCATAACTAGTTGAAATTGGCCATTCGTACCATGAATAAATCTGATAGTTTCCTTGTTAGGAAGAGATTGTAAGTAAATAC AGTCATTGCAGTCAGAACAGTATTAGTGAACCTTGTGTGGTGTTTTCAAGCTCTTTAAAATGGTACAATGTAGCACATTTGCTTTCATTT CTTTTTTTATTTTTGGCATTTGACCTTGTATTCTTTCTGAAGCTCTATATGTGTTTTTATTAGTCAATAATCTGGCAAGTAGCACTTTGC CTGTGCAGTTTGCTGGAGTGTAGATGTACATATGAGGATTTCCCGGGAGGTGCACTTCTTTGAAGAACTTCCTAAAGTACCTGTATAGTA GTTTTCATCTTAATATTCAGTATTTAATCTTCAGTTTGTGCTTTGTAAACTCATGACTTAATTGGTCAGAAACTTTTTAGTGTCTTTATA AAATTTTGTATACATATTTATACTAAACACATTGTGATACTGTATTTGAATGAATGGTGAAAAAATATTTGCTATTGGAATTATGTGCAC >72423_72423_2_RASAL2-KIF14_RASAL2_chr1_178252826_ENST00000448150_KIF14_chr1_200544819_ENST00000367350_length(amino acids)=721AA_BP=228 MKKAAPRRWEGEPPLPGSSASSPGRDPHTPEPGPTLGRGHAGSSSRPGSRPPTAGGLHRPSLGQWATGKEVRGVRAGCRSGPCPRCALSS SPYRRQGAEPRASPPRSRRLVPLPPRGTMELSPSSGGAAEALSWPEMFPALESDSPLPPEDLDAVVPVSGAVAGGMLDRILLESVCQQQS WVRVYDVKGPPTHRLSCGQSPYTETTTWERKYCILTDSQLVLLNKEKESNGSNRGEDAFCDPEDEWEPDITDAPVSSLSRRRSRSLMKNR RISGCLHDIQVHPIKNLHSSHSSGLMDKSSTIYSNSAESFLPGICKELIGSSLDFFGQSYDEERTIADSLINSFLKIYNGLFAISKAHEE QDEESQDNLFSSDRAIQSLTIQTACAFEQLVVLMKHWLSDLLPCTNIARLEDELRQEVKKLGGYLQLFLQGCCLDISSMIKEAQKNAIQI VQQAVKYVGQLAVLKGSKLHFLENGNNKAASVQEEFMDAVCDGVGLGMKILLDSGLEKAKELQHELFRQCTKNEVTKEMKTNAMGLIRSL ENIFAESKIKSFRRQVQEENFEYQDFKRMVNRAPEFLKLKHCLEKAIEIIISALKGCHSDINLLQTCVESIRNLASDFYSDFSVPSTSVG SYESRVTHIVHQELESLAKSLLFCFESEESPDLLKPWETYNQNTKEEHQQSKSSGIDGSKNKGVPKRVYELHGSSPAVSSEECTPSRIQW -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RASAL2-KIF14 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RASAL2-KIF14 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RASAL2-KIF14 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
