
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RBM10-TFE3 (FusionGDB2 ID:72752) |
Fusion Gene Summary for RBM10-TFE3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RBM10-TFE3 | Fusion gene ID: 72752 | Hgene | Tgene | Gene symbol | RBM10 | TFE3 | Gene ID | 8241 | 7030 |
| Gene name | RNA binding motif protein 10 | transcription factor binding to IGHM enhancer 3 | |
| Synonyms | DXS8237E|GPATC9|GPATCH9|S1-1|TARPS|ZRANB5 | RCCP2|RCCX1|TFEA|bHLHe33 | |
| Cytomap | Xp11.3 | Xp11.23 | |
| Type of gene | protein-coding | protein-coding | |
| Description | RNA-binding protein 10RNA-binding protein S1-1epididymis secretory sperm binding proteing patch domain-containing protein 9 | transcription factor E3class E basic helix-loop-helix protein 33transcription factor E family, member Atranscription factor for IgH enhancertranscription factor for immunoglobulin heavy-chain enhancer 3 | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | P19532 | |
| Ensembl transtripts involved in fusion gene | ENST00000329236, ENST00000345781, ENST00000377604, ENST00000478410, | ENST00000487451, ENST00000315869, | |
| Fusion gene scores | * DoF score | 17 X 8 X 9=1224 | 14 X 15 X 6=1260 |
| # samples | 18 | 15 | |
| ** MAII score | log2(18/1224*10)=-2.76553474636298 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/1260*10)=-3.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: RBM10 [Title/Abstract] AND TFE3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RBM10(47041725)-TFE3(48895639), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across RBM10 (5'-gene) Fusion gene breakpoints across RBM10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
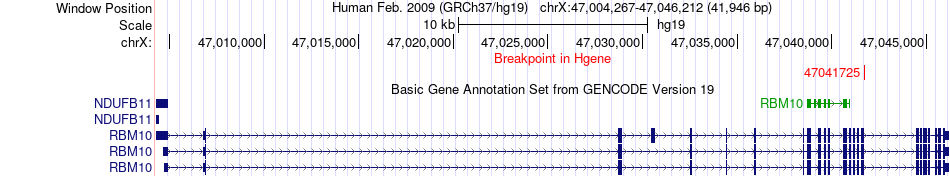 |
 Fusion gene breakpoints across TFE3 (3'-gene) Fusion gene breakpoints across TFE3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
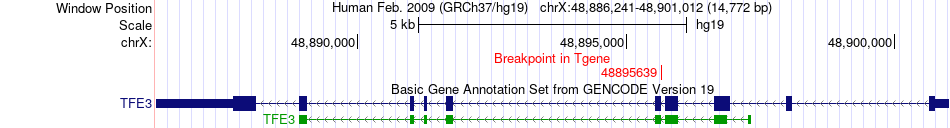 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KIRP | TCGA-DZ-6131-01A | RBM10 | chrX | 47041725 | - | TFE3 | chrX | 48895639 | - |
| ChimerDB4 | KIRP | TCGA-DZ-6131-01A | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| ChimerDB4 | KIRP | TCGA-DZ-6131 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| ChimerKB4 | . | . | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
Top |
Fusion Gene ORF analysis for RBM10-TFE3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000329236 | ENST00000487451 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| 5CDS-5UTR | ENST00000345781 | ENST00000487451 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| 5CDS-5UTR | ENST00000377604 | ENST00000487451 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| In-frame | ENST00000329236 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| In-frame | ENST00000345781 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| In-frame | ENST00000377604 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| intron-3CDS | ENST00000478410 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| intron-5UTR | ENST00000478410 | ENST00000487451 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - |
| intron-intron | ENST00000329236 | ENST00000315869 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000329236 | ENST00000487451 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000345781 | ENST00000315869 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000345781 | ENST00000487451 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000377604 | ENST00000315869 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000377604 | ENST00000487451 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000478410 | ENST00000315869 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
| intron-intron | ENST00000478410 | ENST00000487451 | RBM10 | chrX | 47041560 | + | TFE3 | chrX | 47041560 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000377604 | RBM10 | chrX | 47041725 | + | ENST00000315869 | TFE3 | chrX | 48895639 | - | 5067 | 2692 | 424 | 3639 | 1071 |
| ENST00000329236 | RBM10 | chrX | 47041725 | + | ENST00000315869 | TFE3 | chrX | 48895639 | - | 4470 | 2095 | 61 | 3042 | 993 |
| ENST00000345781 | RBM10 | chrX | 47041725 | + | ENST00000315869 | TFE3 | chrX | 48895639 | - | 4426 | 2051 | 14 | 2998 | 994 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000377604 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - | 0.004157387 | 0.99584264 |
| ENST00000329236 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - | 0.006056023 | 0.993944 |
| ENST00000345781 | ENST00000315869 | RBM10 | chrX | 47041725 | + | TFE3 | chrX | 48895639 | - | 0.005462841 | 0.9945372 |
Top |
Fusion Genomic Features for RBM10-TFE3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
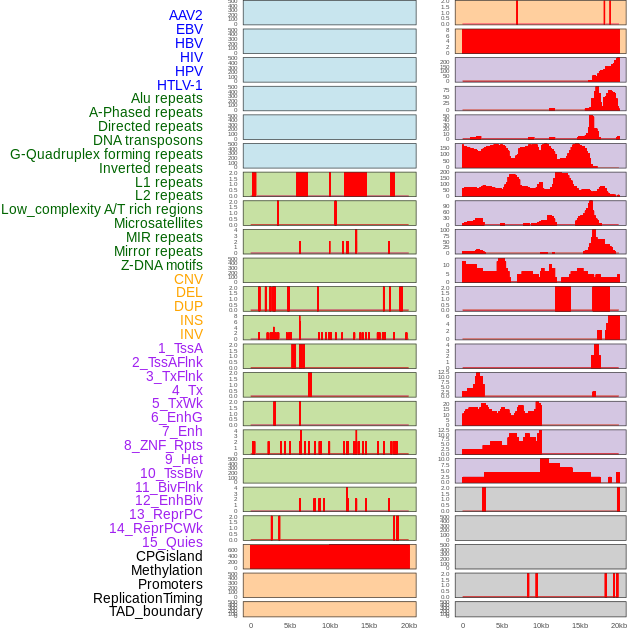 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for RBM10-TFE3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chrX:47041725/chrX:48895639) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | TFE3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcription factor that acts as a master regulator of lysosomal biogenesis and immune response (PubMed:2338243, PubMed:29146937, PubMed:30733432, PubMed:31672913). Specifically recognizes and binds E-box sequences (5'-CANNTG-3'); efficient DNA-binding requires dimerization with itself or with another MiT/TFE family member such as TFEB or MITF (By similarity). Involved in the cellular response to amino acid availability by acting downstream of MTOR: in the presence of nutrients, TFE3 phosphorylation by MTOR promotes its cytosolic retention and subsequent inactivation (PubMed:31672913). Upon starvation or lysosomal stress, inhibition of MTOR induces TFE3 dephosphorylation, resulting in nuclear localization and transcription factor activity (PubMed:31672913). In association with TFEB, activates the expression of CD40L in T-cells, thereby playing a role in T-cell-dependent antibody responses in activated CD4(+) T-cells and thymus-dependent humoral immunity (By similarity). Specifically recognizes the MUE3 box, a subset of E-boxes, present in the immunoglobulin enhancer (PubMed:2338243). It also binds very well to a USF/MLTF site (PubMed:2338243). May regulate lysosomal positioning in response to nutrient deprivation by promoting the expression of PIP4P1 (PubMed:29146937). Acts as a positive regulator of browning of adipose tissue by promoting expression of target genes; mTOR-dependent phosphorylation promotes cytoplasmic retention of TFE3 and inhibits browning of adipose tissue (By similarity). Maintains the pluripotent state of embryonic stem cells by promoting the expression of genes such as ESRRB; mTOR-dependent nuclear exclusion promotes exit from pluripotency (By similarity). Required to maintain the naive pluripotent state of hematopoietic stem cell; mTOR-dependent cytoplasmic retention of TFE3 promotes the exit of hematopoietic stem cell from pluripotency (PubMed:30733432). {ECO:0000250|UniProtKB:Q64092, ECO:0000269|PubMed:2338243, ECO:0000269|PubMed:29146937, ECO:0000269|PubMed:30733432, ECO:0000269|PubMed:31672913}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 113_125 | 572 | 853.0 | Compositional bias | Note=Poly-Glu |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 80_87 | 572 | 853.0 | Compositional bias | Note=Poly-Arg |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 113_125 | 573 | 854.0 | Compositional bias | Note=Poly-Glu |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 80_87 | 573 | 854.0 | Compositional bias | Note=Poly-Arg |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 113_125 | 650 | 931.0 | Compositional bias | Note=Poly-Glu |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 561_607 | 650 | 931.0 | Compositional bias | Note=Tyr-rich |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 80_87 | 650 | 931.0 | Compositional bias | Note=Poly-Arg |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 129_209 | 572 | 853.0 | Domain | RRM 1 |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 300_384 | 572 | 853.0 | Domain | RRM 2 |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 129_209 | 573 | 854.0 | Domain | RRM 1 |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 300_384 | 573 | 854.0 | Domain | RRM 2 |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 129_209 | 650 | 931.0 | Domain | RRM 1 |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 300_384 | 650 | 931.0 | Domain | RRM 2 |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 212_242 | 572 | 853.0 | Zinc finger | RanBP2-type |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 212_242 | 573 | 854.0 | Zinc finger | RanBP2-type |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 212_242 | 650 | 931.0 | Zinc finger | RanBP2-type |
| Tgene | TFE3 | chrX:47041725 | chrX:48895639 | ENST00000315869 | 3 | 10 | 346_399 | 260 | 576.0 | Domain | bHLH | |
| Tgene | TFE3 | chrX:47041725 | chrX:48895639 | ENST00000315869 | 3 | 10 | 260_271 | 260 | 576.0 | Region | Strong transcription activation domain | |
| Tgene | TFE3 | chrX:47041725 | chrX:48895639 | ENST00000315869 | 3 | 10 | 409_430 | 260 | 576.0 | Region | Note=Leucine-zipper |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 561_607 | 572 | 853.0 | Compositional bias | Note=Tyr-rich |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 561_607 | 573 | 854.0 | Compositional bias | Note=Tyr-rich |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 858_904 | 572 | 853.0 | Domain | G-patch |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 858_904 | 573 | 854.0 | Domain | G-patch |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 858_904 | 650 | 931.0 | Domain | G-patch |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000329236 | + | 16 | 23 | 759_784 | 572 | 853.0 | Zinc finger | C2H2-type%3B atypical |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000345781 | + | 16 | 23 | 759_784 | 573 | 854.0 | Zinc finger | C2H2-type%3B atypical |
| Hgene | RBM10 | chrX:47041725 | chrX:48895639 | ENST00000377604 | + | 17 | 24 | 759_784 | 650 | 931.0 | Zinc finger | C2H2-type%3B atypical |
Top |
Fusion Gene Sequence for RBM10-TFE3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >72752_72752_1_RBM10-TFE3_RBM10_chrX_47041725_ENST00000329236_TFE3_chrX_48895639_ENST00000315869_length(transcript)=4470nt_BP=2095nt GGTGGTCGGAGCGCCGACTCCCTTCTCGTCGTCGCCATTTTGAGCTGGTGACTGTGGCCGGCTGGGAGTAGGCGGCAGTGAGTTTCCCTG GGAGGGCAGCGCGCTTGGCGCTTCTCCCCTCCCCCCGATCTGCCTCCAGTCTCGGACTTGGTTGTTGCGCGCTCCGGCTCCGGCTGAGCT GGGAGAGTTGGAGGAGGTGGCGGCGGGCAGAGGTGATGTCTGGGAGCCCTTCCTTGACAGCCCGGGCCGAGAAGAGTCCCTGCAGGAAGC ATCACCCAGGCTGGCAGATCATGGTAGCAGCAGCGGGGGTGGCTGGGAAGTGAAACGGAGCCAGCGGCTGAGGAGGGGCCCCAGCAGCCC CCGAAGGCCCTATCAGGACATGGAGTATGAAAGACGTGGTGGTCGTGGTGACAGGACTGGCCGCTATGGAGCCACTGACCGCTCGCAGGA TGATGGTGGGGAGAACCGCAGCCGAGACCACGACTACCGGGACATGGACTACCGTTCATATCCTCGCGAGTATGGCAGCCAGGAGGGCAA GCATGACTATGACGACTCATCTGAGGAGCAGAGTGCGGAGATCCGTGGCCAGCTGCAGTCGCACGGCGTGCAAGCACGGGAGGTTCGGCT GATGCGGAACAAATCTTCAGGTCAGAGCCGGGGCTTCGCCTTCGTCGAGTTTAGTCACTTGCAGGACGCTACACGATGGATGGAAGCCAA TCAGCACTCCCTCAACATCCTGGGCCAGAAGGTGTCGATGCACTACAGTGACCCCAAGCCCAAGATCAATGAGGACTGGCTGTGCAATAA GTGTGGCGTCCAGAACTTCAAACGCCGAGAGAAGTGCTTCAAATGTGGCGTGCCCAAGTCAGAGGCAGAGCAGAAGCTGCCCCTCGGCAC GAGGCTGGATCAGCAGACACTGCCACTGGGTGGCCGGGAGCTGAGCCAGGGCCTGCTTCCCCTGCCGCAGCCCTACCAGGCCCAGGGAGT CCTGGCCTCCCAAGCCCTGTCACAGGGCTCGGAGCCAAGCTCAGAGAACGCCAATGACACCATCATTTTGCGCAACCTGAACCCACACAG CACCATGGATTCCATCCTGGGGGCCCTGGCACCCTACGCGGTGCTGTCCTCCTCCAACGTGCGCGTCATAAAGGACAAGCAGACCCAACT GAACCGCGGCTTTGCCTTCATCCAGCTCTCCACCATCGAGGCAGCCCAGCTGCTGCAGATCCTGCAGGCCCTGCACCCACCACTCACTAT CGACGGCAAGACCATCAATGTTGAGTTTGCCAAGGGTTCTAAGAGGGACATGGCCTCCAATGAAGGCAGTCGCATCAGTGCTGCCTCTGT GGCCAGCACTGCCATTGCTGCGGCCCAGTGGGCCATCTCACAGGCCTCCCAAGGTGGGGAGGGTACCTGGGCCACCTCCGAGGAGCCGCC GGTCGACTACAGCTACTACCAACAGGATGAGGGCTATGGCAACAGCCAGGGCACAGAGTCTTCCCTCTATGCCCATGGCTACCTCAAGGG CACCAAGGGCCCTGGCATCACTGGAACCAAAGGGGATCCCACTGGAGCAGGTCCCGAGGCCTCCCTAGAGCCTGGGGCCGACTCTGTGTC GATGCAGGCTTTCTCTCGCGCCCAGCCTGGTGCTGCTCCTGGCATCTACCAACAATCAGCCGAGGCGAGCAGTAGCCAGGGCACTGCTGC CAACAGCCAGTCGTATACCATCATGTCACCCGCTGTGCTCAAATCTGAGCTCCAGAGCCCTACCCATCCTAGTTCTGCTCTCCCACCGGC TACCAGCCCCACTGCCCAGGAATCCTACAGCCAGTACCCTGTTCCCGACGTCTCTACCTACCAGTACGATGAGACCTCCGGCTACTACTA TGACCCCCAGACCGGCCTCTACTATGACCCCAACTCCCAGTATTACTACAATGCTCAGAGCCAGCAGTACCTGTACTGGGATGGGGAGAG GCGGACCTATGTTCCCGCCCTGGAGCAGTCGGCCGACGGACATAAGGAGACAGGGGCACCCTCGAAGGAGGGCAAAGAGAAGAAGGAGAA GCACAAGACCAAGACAGCTCAACAGATTGATGATGTCATTGATGAGATCATCAGCCTGGAGTCCAGTTACAATGATGAAATGCTCAGCTA TCTGCCCGGAGGCACCACAGGACTGCAGCTCCCCAGCACGCTGCCTGTGTCAGGGAATCTGCTTGATGTGTACAGTAGTCAAGGCGTGGC CACACCAGCCATCACTGTCAGCAACTCCTGCCCAGCTGAGCTGCCCAACATCAAACGGGAGATCTCTGAGACCGAGGCAAAGGCCCTTTT GAAGGAACGGCAGAAGAAAGACAATCACAACCTAATTGAGCGTCGCAGGCGATTCAACATTAACGACAGGATCAAGGAACTGGGCACTCT CATCCCTAAGTCCAGTGACCCGGAGATGCGCTGGAACAAGGGCACCATCCTGAAGGCCTCTGTGGATTATATCCGCAAGCTGCAGAAGGA GCAGCAGCGCTCCAAAGACCTGGAGAGCCGGCAGCGATCCCTGGAGCAGGCCAACCGCAGCCTGCAGCTCCGAATTCAGGAACTAGAACT GCAGGCCCAGATCCATGGCCTGCCAGTACCTCCCACTCCAGGGCTGCTTTCCTTGGCCACGACTTCGGCTTCTGACAGCCTCAAGCCAGA GCAGCTGGACATTGAGGAGGAGGGCAGGCCAGGCGCAGCAACGTTCCATGTAGGGGGGGGACCTGCCCAGAATGCTCCCCATCAGCAGCC CCCTGCACCGCCCTCAGATGCCCTTCTGGACCTGCACTTTCCCAGCGACCACCTGGGGGACCTGGGAGACCCCTTCCACCTGGGGCTGGA GGACATTCTGATGGAGGAGGAGGAGGGGGTGGTGGGAGGACTGTCGGGGGGTGCCCTGTCCCCACTGCGGGCTGCCTCCGATCCCCTGCT CTCTTCAGTGTCCCCTGCTGTCTCCAAGGCCAGCAGCCGCCGCAGCAGCTTCAGCATGGAAGAGGAGTCCTGATCAGGCCTCACCCCTCC CCTGGGACTTTCCCACCCAGGAAAGGAGGACCAGTCAGGATGAGGCCCCGCCTTTTCCCCCACCCTCCCATGAGACTGCCCTGCCCAGGT ATCCTGGGGGAAGAGGAGATGTGATCAGGCCCCACCCCTGTAATCAGGCAAGGAGGAGGAGTCAGATGAGGCCCTGCACCTTCCCCAAAG GAACCGCCCAGTGCAGGTATTTCAGAAGGAGAAGGCTGGAGAAGGACATGAGATCAGGGCCTGCCCCCTGGGGATCACAGCCTCACCCCT GCCCCTGTGGGACTCATCCTTGCCCAGGTGAGGGAAGGAGACAGGATGAGGTCTCGACCCTGTCCCCTAGGGACTGTCCTAGCCAGGTCT CCTGGGAAAGGGAGATGTCAGGATGTTGCTCCATCCTTTGTCTTGGAACCACCAGTCTAGTCCGTCCTGGCACAGAAGAGGAGTCAAGTA ATGGAGGTCCCAGCCCTGGGGGTTTAAGCTCTGCCCCTTCCCCATGAACCCTGCCCTGCTCTGCCCAGGCAAGGAACAGAAGTGAGGATG AGACCCAGCCCCTTCCCCTGGGAACTCTCCTGGCCTTCTAGGAATGGAGGAGCCAGGCCCCACCCCTTCCCTATAGGAACAGCCCAGCAC AGGTATTTCAGGTGTGAAAGAATCAGTAGGACCAGGCCACCGCTAGTGCTTGTGGAGATCACAGCCCCACCCTTGTCCCTCAGCAACATC CCATCTAAGCATTCCACACTGCAGGGAGGAGTGGTACTTAAGCTCCCCTGCCTTAACCTGGGACCAACCTGACCTAACCTAGGAGGGCTC TGAGCCAACCTTGCTCTTGGGGAAGGGGACAGATTATGAAATTTCATGGATGAATTTTCCAGACCTATATCTGGAGTGAGAGGCCCCCAC CCTTGGGCAGAGTCCTGCCTTCTTCCTTGAGGGGCAGTTTGGGAAGGTGATGGGTATTAGTGGGGGACTGAGTTCAGGTTACCAGAACCA GTACCTCAGTATTCTTTTTCAACATGTAGGGCAAGAGGATGAAGGAAGGGGCTATCCTGGGACCTCCCCAGCCCAGGAAAAACTGGAAGC CTTCCCCCAGCAAGGCAGAAGCTTGGAGGAGGGTTGTAAAAGCATATTGTACCCCCTCATTTGTTTATCTGATTTTTTTATTGCTCCGCA TACTGAGAATCTAGGCCACCCCAACCTCTGTTCCCCACCCAGTTCTTCATTTGGAGGAATCACCCCATTTCAGAGTTATCAAGAGACACT CCCCCCTCCATTCCCACCCCTCATACCTACACCCAAGGTTGTCAGCTTTGGATTGCTGGGGCCAGGCCCCATGGAGGGTATACTGAGGGG >72752_72752_1_RBM10-TFE3_RBM10_chrX_47041725_ENST00000329236_TFE3_chrX_48895639_ENST00000315869_length(amino acids)=993AA_BP=661 MGVGGSEFPWEGSALGASPLPPICLQSRTWLLRAPAPAELGELEEVAAGRGDVWEPFLDSPGREESLQEASPRLADHGSSSGGGWEVKRS QRLRRGPSSPRRPYQDMEYERRGGRGDRTGRYGATDRSQDDGGENRSRDHDYRDMDYRSYPREYGSQEGKHDYDDSSEEQSAEIRGQLQS HGVQAREVRLMRNKSSGQSRGFAFVEFSHLQDATRWMEANQHSLNILGQKVSMHYSDPKPKINEDWLCNKCGVQNFKRREKCFKCGVPKS EAEQKLPLGTRLDQQTLPLGGRELSQGLLPLPQPYQAQGVLASQALSQGSEPSSENANDTIILRNLNPHSTMDSILGALAPYAVLSSSNV RVIKDKQTQLNRGFAFIQLSTIEAAQLLQILQALHPPLTIDGKTINVEFAKGSKRDMASNEGSRISAASVASTAIAAAQWAISQASQGGE GTWATSEEPPVDYSYYQQDEGYGNSQGTESSLYAHGYLKGTKGPGITGTKGDPTGAGPEASLEPGADSVSMQAFSRAQPGAAPGIYQQSA EASSSQGTAANSQSYTIMSPAVLKSELQSPTHPSSALPPATSPTAQESYSQYPVPDVSTYQYDETSGYYYDPQTGLYYDPNSQYYYNAQS QQYLYWDGERRTYVPALEQSADGHKETGAPSKEGKEKKEKHKTKTAQQIDDVIDEIISLESSYNDEMLSYLPGGTTGLQLPSTLPVSGNL LDVYSSQGVATPAITVSNSCPAELPNIKREISETEAKALLKERQKKDNHNLIERRRRFNINDRIKELGTLIPKSSDPEMRWNKGTILKAS VDYIRKLQKEQQRSKDLESRQRSLEQANRSLQLRIQELELQAQIHGLPVPPTPGLLSLATTSASDSLKPEQLDIEEEGRPGAATFHVGGG PAQNAPHQQPPAPPSDALLDLHFPSDHLGDLGDPFHLGLEDILMEEEEGVVGGLSGGALSPLRAASDPLLSSVSPAVSKASSRRSSFSME -------------------------------------------------------------- >72752_72752_2_RBM10-TFE3_RBM10_chrX_47041725_ENST00000345781_TFE3_chrX_48895639_ENST00000315869_length(transcript)=4426nt_BP=2051nt GTGACTGTGGCCGGCTGGGAGTAGGCGGCAGTGAGTTTCCCTGGGAGGGCAGCGCGCTTGGCGCTTCTCCCCTCCCCCCGATCTGCCTCC AGTCTCGGACTTGGTTGTTGCGCGCTCCGGCTCCGGCTGAGCTGGGAGAGTTGGAGGAGGTGGCGGCGGGCAGAGGTGATGTCTGGGAGC CCTTCCTTGACAGCCCGGGCCGAGAAGAGTCCCTGCAGGAAGCATCACCCAGGCTGGCAGATCATGGTAGCAGCAGCGGGGGTGGCTGGG AAGTGAAACGGAGCCAGCGGCTGAGGAGGGGCCCCAGCAGCCCCCGAAGGCCCTATCAGGACATGGAGTATGAAAGACGTGGTGGTCGTG GTGACAGGACTGGCCGCTATGGAGCCACTGACCGCTCGCAGGATGATGGTGGGGAGAACCGCAGCCGAGACCACGACTACCGGGACATGG ACTACCGTTCATATCCTCGCGAGTATGGCAGCCAGGAGGGCAAGCATGACTATGACGACTCATCTGAGGAGCAGAGTGCGGAGATCCGTG GCCAGCTGCAGTCGCACGGCGTGCAAGCACGGGAGGTTCGGCTGATGCGGAACAAATCTTCAGGTCAGAGCCGGGGCTTCGCCTTCGTCG AGTTTAGTCACTTGCAGGACGCTACACGATGGATGGAAGCCAATCAGCACTCCCTCAACATCCTGGGCCAGAAGGTGTCGATGCACTACA GTGACCCCAAGCCCAAGATCAATGAGGACTGGCTGTGCAATAAGTGTGGCGTCCAGAACTTCAAACGCCGAGAGAAGTGCTTCAAATGTG GCGTGCCCAAGTCAGAGGCAGAGCAGAAGCTGCCCCTCGGCACGAGGCTGGATCAGCAGACACTGCCACTGGGTGGCCGGGAGCTGAGCC AGGGCCTGCTTCCCCTGCCGCAGCCCTACCAGGCCCAGGGAGTCCTGGCCTCCCAAGCCCTGTCACAGGGCTCGGAGCCAAGCTCAGAGA ACGCCAATGACACCATCATTTTGCGCAACCTGAACCCACACAGCACCATGGATTCCATCCTGGGGGCCCTGGCACCCTACGCGGTGCTGT CCTCCTCCAACGTGCGCGTCATAAAGGACAAGCAGACCCAACTGAACCGCGGCTTTGCCTTCATCCAGCTCTCCACCATCGTGGAGGCAG CCCAGCTGCTGCAGATCCTGCAGGCCCTGCACCCACCACTCACTATCGACGGCAAGACCATCAATGTTGAGTTTGCCAAGGGTTCTAAGA GGGACATGGCCTCCAATGAAGGCAGTCGCATCAGTGCTGCCTCTGTGGCCAGCACTGCCATTGCTGCGGCCCAGTGGGCCATCTCACAGG CCTCCCAAGGTGGGGAGGGTACCTGGGCCACCTCCGAGGAGCCGCCGGTCGACTACAGCTACTACCAACAGGATGAGGGCTATGGCAACA GCCAGGGCACAGAGTCTTCCCTCTATGCCCATGGCTACCTCAAGGGCACCAAGGGCCCTGGCATCACTGGAACCAAAGGGGATCCCACTG GAGCAGGTCCCGAGGCCTCCCTAGAGCCTGGGGCCGACTCTGTGTCGATGCAGGCTTTCTCTCGCGCCCAGCCTGGTGCTGCTCCTGGCA TCTACCAACAATCAGCCGAGGCGAGCAGTAGCCAGGGCACTGCTGCCAACAGCCAGTCGTATACCATCATGTCACCCGCTGTGCTCAAAT CTGAGCTCCAGAGCCCTACCCATCCTAGTTCTGCTCTCCCACCGGCTACCAGCCCCACTGCCCAGGAATCCTACAGCCAGTACCCTGTTC CCGACGTCTCTACCTACCAGTACGATGAGACCTCCGGCTACTACTATGACCCCCAGACCGGCCTCTACTATGACCCCAACTCCCAGTATT ACTACAATGCTCAGAGCCAGCAGTACCTGTACTGGGATGGGGAGAGGCGGACCTATGTTCCCGCCCTGGAGCAGTCGGCCGACGGACATA AGGAGACAGGGGCACCCTCGAAGGAGGGCAAAGAGAAGAAGGAGAAGCACAAGACCAAGACAGCTCAACAGATTGATGATGTCATTGATG AGATCATCAGCCTGGAGTCCAGTTACAATGATGAAATGCTCAGCTATCTGCCCGGAGGCACCACAGGACTGCAGCTCCCCAGCACGCTGC CTGTGTCAGGGAATCTGCTTGATGTGTACAGTAGTCAAGGCGTGGCCACACCAGCCATCACTGTCAGCAACTCCTGCCCAGCTGAGCTGC CCAACATCAAACGGGAGATCTCTGAGACCGAGGCAAAGGCCCTTTTGAAGGAACGGCAGAAGAAAGACAATCACAACCTAATTGAGCGTC GCAGGCGATTCAACATTAACGACAGGATCAAGGAACTGGGCACTCTCATCCCTAAGTCCAGTGACCCGGAGATGCGCTGGAACAAGGGCA CCATCCTGAAGGCCTCTGTGGATTATATCCGCAAGCTGCAGAAGGAGCAGCAGCGCTCCAAAGACCTGGAGAGCCGGCAGCGATCCCTGG AGCAGGCCAACCGCAGCCTGCAGCTCCGAATTCAGGAACTAGAACTGCAGGCCCAGATCCATGGCCTGCCAGTACCTCCCACTCCAGGGC TGCTTTCCTTGGCCACGACTTCGGCTTCTGACAGCCTCAAGCCAGAGCAGCTGGACATTGAGGAGGAGGGCAGGCCAGGCGCAGCAACGT TCCATGTAGGGGGGGGACCTGCCCAGAATGCTCCCCATCAGCAGCCCCCTGCACCGCCCTCAGATGCCCTTCTGGACCTGCACTTTCCCA GCGACCACCTGGGGGACCTGGGAGACCCCTTCCACCTGGGGCTGGAGGACATTCTGATGGAGGAGGAGGAGGGGGTGGTGGGAGGACTGT CGGGGGGTGCCCTGTCCCCACTGCGGGCTGCCTCCGATCCCCTGCTCTCTTCAGTGTCCCCTGCTGTCTCCAAGGCCAGCAGCCGCCGCA GCAGCTTCAGCATGGAAGAGGAGTCCTGATCAGGCCTCACCCCTCCCCTGGGACTTTCCCACCCAGGAAAGGAGGACCAGTCAGGATGAG GCCCCGCCTTTTCCCCCACCCTCCCATGAGACTGCCCTGCCCAGGTATCCTGGGGGAAGAGGAGATGTGATCAGGCCCCACCCCTGTAAT CAGGCAAGGAGGAGGAGTCAGATGAGGCCCTGCACCTTCCCCAAAGGAACCGCCCAGTGCAGGTATTTCAGAAGGAGAAGGCTGGAGAAG GACATGAGATCAGGGCCTGCCCCCTGGGGATCACAGCCTCACCCCTGCCCCTGTGGGACTCATCCTTGCCCAGGTGAGGGAAGGAGACAG GATGAGGTCTCGACCCTGTCCCCTAGGGACTGTCCTAGCCAGGTCTCCTGGGAAAGGGAGATGTCAGGATGTTGCTCCATCCTTTGTCTT GGAACCACCAGTCTAGTCCGTCCTGGCACAGAAGAGGAGTCAAGTAATGGAGGTCCCAGCCCTGGGGGTTTAAGCTCTGCCCCTTCCCCA TGAACCCTGCCCTGCTCTGCCCAGGCAAGGAACAGAAGTGAGGATGAGACCCAGCCCCTTCCCCTGGGAACTCTCCTGGCCTTCTAGGAA TGGAGGAGCCAGGCCCCACCCCTTCCCTATAGGAACAGCCCAGCACAGGTATTTCAGGTGTGAAAGAATCAGTAGGACCAGGCCACCGCT AGTGCTTGTGGAGATCACAGCCCCACCCTTGTCCCTCAGCAACATCCCATCTAAGCATTCCACACTGCAGGGAGGAGTGGTACTTAAGCT CCCCTGCCTTAACCTGGGACCAACCTGACCTAACCTAGGAGGGCTCTGAGCCAACCTTGCTCTTGGGGAAGGGGACAGATTATGAAATTT CATGGATGAATTTTCCAGACCTATATCTGGAGTGAGAGGCCCCCACCCTTGGGCAGAGTCCTGCCTTCTTCCTTGAGGGGCAGTTTGGGA AGGTGATGGGTATTAGTGGGGGACTGAGTTCAGGTTACCAGAACCAGTACCTCAGTATTCTTTTTCAACATGTAGGGCAAGAGGATGAAG GAAGGGGCTATCCTGGGACCTCCCCAGCCCAGGAAAAACTGGAAGCCTTCCCCCAGCAAGGCAGAAGCTTGGAGGAGGGTTGTAAAAGCA TATTGTACCCCCTCATTTGTTTATCTGATTTTTTTATTGCTCCGCATACTGAGAATCTAGGCCACCCCAACCTCTGTTCCCCACCCAGTT CTTCATTTGGAGGAATCACCCCATTTCAGAGTTATCAAGAGACACTCCCCCCTCCATTCCCACCCCTCATACCTACACCCAAGGTTGTCA GCTTTGGATTGCTGGGGCCAGGCCCCATGGAGGGTATACTGAGGGGTCTATAGGTTTGTGATTAAAATAATAAAAGCTAGGCGTGTTTGA >72752_72752_2_RBM10-TFE3_RBM10_chrX_47041725_ENST00000345781_TFE3_chrX_48895639_ENST00000315869_length(amino acids)=994AA_BP=662 MGVGGSEFPWEGSALGASPLPPICLQSRTWLLRAPAPAELGELEEVAAGRGDVWEPFLDSPGREESLQEASPRLADHGSSSGGGWEVKRS QRLRRGPSSPRRPYQDMEYERRGGRGDRTGRYGATDRSQDDGGENRSRDHDYRDMDYRSYPREYGSQEGKHDYDDSSEEQSAEIRGQLQS HGVQAREVRLMRNKSSGQSRGFAFVEFSHLQDATRWMEANQHSLNILGQKVSMHYSDPKPKINEDWLCNKCGVQNFKRREKCFKCGVPKS EAEQKLPLGTRLDQQTLPLGGRELSQGLLPLPQPYQAQGVLASQALSQGSEPSSENANDTIILRNLNPHSTMDSILGALAPYAVLSSSNV RVIKDKQTQLNRGFAFIQLSTIVEAAQLLQILQALHPPLTIDGKTINVEFAKGSKRDMASNEGSRISAASVASTAIAAAQWAISQASQGG EGTWATSEEPPVDYSYYQQDEGYGNSQGTESSLYAHGYLKGTKGPGITGTKGDPTGAGPEASLEPGADSVSMQAFSRAQPGAAPGIYQQS AEASSSQGTAANSQSYTIMSPAVLKSELQSPTHPSSALPPATSPTAQESYSQYPVPDVSTYQYDETSGYYYDPQTGLYYDPNSQYYYNAQ SQQYLYWDGERRTYVPALEQSADGHKETGAPSKEGKEKKEKHKTKTAQQIDDVIDEIISLESSYNDEMLSYLPGGTTGLQLPSTLPVSGN LLDVYSSQGVATPAITVSNSCPAELPNIKREISETEAKALLKERQKKDNHNLIERRRRFNINDRIKELGTLIPKSSDPEMRWNKGTILKA SVDYIRKLQKEQQRSKDLESRQRSLEQANRSLQLRIQELELQAQIHGLPVPPTPGLLSLATTSASDSLKPEQLDIEEEGRPGAATFHVGG GPAQNAPHQQPPAPPSDALLDLHFPSDHLGDLGDPFHLGLEDILMEEEEGVVGGLSGGALSPLRAASDPLLSSVSPAVSKASSRRSSFSM -------------------------------------------------------------- >72752_72752_3_RBM10-TFE3_RBM10_chrX_47041725_ENST00000377604_TFE3_chrX_48895639_ENST00000315869_length(transcript)=5067nt_BP=2692nt GCCAACCATTTTCGGCCAGGCTGAGAAATTTCTTATTGATGGGTCTGCTCATTTCCTGCATAGGAAGAAATCGGTGACATTTGGGTGGAT ACAGTTCCCAGCCAATCGCCCCGAGTTCCCTCGCCCACTGGGGAAAGAGGCAGAGCTTCCGCTAGTCTTTCATCTTCAAAGGAAATCAAG CTTCTAAAAAGCGGGCCCTGCCGAATTGGCTAATAGCTACTCCTTTTCTCCACTAGGGGAGCGTTCGCAAGGGGTGTGGGGAGGTGGCCA GCGCTTGCCAATCAGAGGAGGCCAGGGCTGTCCCCTAGCGGGCGGTGACCAATCGCCGGGGCCGGCCAGAGCGCGCTTCCTTAGTAGGTG GATGGTGGTCGGAGCGCCGACTCCCTTCTCGTCGTCGCCATTTTGAGCTGGTGACTGTGGCCGGCTGGGAGTAGGCGGCAGTGAGTTTCC CTGGGAGGGCAGCGCGCTTGGCGCTTCTCCCCTCCCCCCGATCTGCCTCCAGTCTCGGACTTGGTTGTTGCGCGCTCCGGCTCCGGCTGA GCTGGGAGAGTTGGAGGAGGTGGCGGCGGGCAGAGGTGATGTCTGGGAGCCCTTCCTTGACAGCCCGGGCCGAGAAGAGTCCCTGCAGGA AGCATCACCCAGGCTGGCAGATCATGGTAGCAGCAGCGGGGGTGGCTGGGAAGTGAAACGGAGCCAGCGGCTGAGGAGGGGCCCCAGCAG CCCCCGAAGGCCCTATCAGGACATGGAGTATGAAAGACGTGGTGGTCGTGGTGACAGGACTGGCCGCTATGGAGCCACTGACCGCTCGCA GGATGATGGTGGGGAGAACCGCAGCCGAGACCACGACTACCGGGACATGGACTACCGTTCATATCCTCGCGAGTATGGCAGCCAGGAGGG CAAGCATGACTATGACGACTCATCTGAGGAGCAGAGTGCGGAGGATTCCTACGAGGCCTCCCCGGGCTCCGAGACTCAGCGTAGGCGGCG GCGGCGGCACAGGCACAGCCCCACCGGCCCGCCAGGCTTCCCCCGAGACGGCGACTATCGGGACCAGGACTATCGGACCGAGCAAGGGGA GGAGGAGGAGGAGGAGGAGGATGAGGAGGAGGAGGAGAAGGCCAGTAACATCGTCATGCTGAGGATGCTGCCACAGGCAGCCACTGAGGA TGACATCCGTGGCCAGCTGCAGTCGCACGGCGTGCAAGCACGGGAGGTTCGGCTGATGCGGAACAAATCTTCAGGTCAGAGCCGGGGCTT CGCCTTCGTCGAGTTTAGTCACTTGCAGGACGCTACACGATGGATGGAAGCCAATCAGCACTCCCTCAACATCCTGGGCCAGAAGGTGTC GATGCACTACAGTGACCCCAAGCCCAAGATCAATGAGGACTGGCTGTGCAATAAGTGTGGCGTCCAGAACTTCAAACGCCGAGAGAAGTG CTTCAAATGTGGCGTGCCCAAGTCAGAGGCAGAGCAGAAGCTGCCCCTCGGCACGAGGCTGGATCAGCAGACACTGCCACTGGGTGGCCG GGAGCTGAGCCAGGGCCTGCTTCCCCTGCCGCAGCCCTACCAGGCCCAGGGAGTCCTGGCCTCCCAAGCCCTGTCACAGGGCTCGGAGCC AAGCTCAGAGAACGCCAATGACACCATCATTTTGCGCAACCTGAACCCACACAGCACCATGGATTCCATCCTGGGGGCCCTGGCACCCTA CGCGGTGCTGTCCTCCTCCAACGTGCGCGTCATAAAGGACAAGCAGACCCAACTGAACCGCGGCTTTGCCTTCATCCAGCTCTCCACCAT CGTGGAGGCAGCCCAGCTGCTGCAGATCCTGCAGGCCCTGCACCCACCACTCACTATCGACGGCAAGACCATCAATGTTGAGTTTGCCAA GGGTTCTAAGAGGGACATGGCCTCCAATGAAGGCAGTCGCATCAGTGCTGCCTCTGTGGCCAGCACTGCCATTGCTGCGGCCCAGTGGGC CATCTCACAGGCCTCCCAAGGTGGGGAGGGTACCTGGGCCACCTCCGAGGAGCCGCCGGTCGACTACAGCTACTACCAACAGGATGAGGG CTATGGCAACAGCCAGGGCACAGAGTCTTCCCTCTATGCCCATGGCTACCTCAAGGGCACCAAGGGCCCTGGCATCACTGGAACCAAAGG GGATCCCACTGGAGCAGGTCCCGAGGCCTCCCTAGAGCCTGGGGCCGACTCTGTGTCGATGCAGGCTTTCTCTCGCGCCCAGCCTGGTGC TGCTCCTGGCATCTACCAACAATCAGCCGAGGCGAGCAGTAGCCAGGGCACTGCTGCCAACAGCCAGTCGTATACCATCATGTCACCCGC TGTGCTCAAATCTGAGCTCCAGAGCCCTACCCATCCTAGTTCTGCTCTCCCACCGGCTACCAGCCCCACTGCCCAGGAATCCTACAGCCA GTACCCTGTTCCCGACGTCTCTACCTACCAGTACGATGAGACCTCCGGCTACTACTATGACCCCCAGACCGGCCTCTACTATGACCCCAA CTCCCAGTATTACTACAATGCTCAGAGCCAGCAGTACCTGTACTGGGATGGGGAGAGGCGGACCTATGTTCCCGCCCTGGAGCAGTCGGC CGACGGACATAAGGAGACAGGGGCACCCTCGAAGGAGGGCAAAGAGAAGAAGGAGAAGCACAAGACCAAGACAGCTCAACAGATTGATGA TGTCATTGATGAGATCATCAGCCTGGAGTCCAGTTACAATGATGAAATGCTCAGCTATCTGCCCGGAGGCACCACAGGACTGCAGCTCCC CAGCACGCTGCCTGTGTCAGGGAATCTGCTTGATGTGTACAGTAGTCAAGGCGTGGCCACACCAGCCATCACTGTCAGCAACTCCTGCCC AGCTGAGCTGCCCAACATCAAACGGGAGATCTCTGAGACCGAGGCAAAGGCCCTTTTGAAGGAACGGCAGAAGAAAGACAATCACAACCT AATTGAGCGTCGCAGGCGATTCAACATTAACGACAGGATCAAGGAACTGGGCACTCTCATCCCTAAGTCCAGTGACCCGGAGATGCGCTG GAACAAGGGCACCATCCTGAAGGCCTCTGTGGATTATATCCGCAAGCTGCAGAAGGAGCAGCAGCGCTCCAAAGACCTGGAGAGCCGGCA GCGATCCCTGGAGCAGGCCAACCGCAGCCTGCAGCTCCGAATTCAGGAACTAGAACTGCAGGCCCAGATCCATGGCCTGCCAGTACCTCC CACTCCAGGGCTGCTTTCCTTGGCCACGACTTCGGCTTCTGACAGCCTCAAGCCAGAGCAGCTGGACATTGAGGAGGAGGGCAGGCCAGG CGCAGCAACGTTCCATGTAGGGGGGGGACCTGCCCAGAATGCTCCCCATCAGCAGCCCCCTGCACCGCCCTCAGATGCCCTTCTGGACCT GCACTTTCCCAGCGACCACCTGGGGGACCTGGGAGACCCCTTCCACCTGGGGCTGGAGGACATTCTGATGGAGGAGGAGGAGGGGGTGGT GGGAGGACTGTCGGGGGGTGCCCTGTCCCCACTGCGGGCTGCCTCCGATCCCCTGCTCTCTTCAGTGTCCCCTGCTGTCTCCAAGGCCAG CAGCCGCCGCAGCAGCTTCAGCATGGAAGAGGAGTCCTGATCAGGCCTCACCCCTCCCCTGGGACTTTCCCACCCAGGAAAGGAGGACCA GTCAGGATGAGGCCCCGCCTTTTCCCCCACCCTCCCATGAGACTGCCCTGCCCAGGTATCCTGGGGGAAGAGGAGATGTGATCAGGCCCC ACCCCTGTAATCAGGCAAGGAGGAGGAGTCAGATGAGGCCCTGCACCTTCCCCAAAGGAACCGCCCAGTGCAGGTATTTCAGAAGGAGAA GGCTGGAGAAGGACATGAGATCAGGGCCTGCCCCCTGGGGATCACAGCCTCACCCCTGCCCCTGTGGGACTCATCCTTGCCCAGGTGAGG GAAGGAGACAGGATGAGGTCTCGACCCTGTCCCCTAGGGACTGTCCTAGCCAGGTCTCCTGGGAAAGGGAGATGTCAGGATGTTGCTCCA TCCTTTGTCTTGGAACCACCAGTCTAGTCCGTCCTGGCACAGAAGAGGAGTCAAGTAATGGAGGTCCCAGCCCTGGGGGTTTAAGCTCTG CCCCTTCCCCATGAACCCTGCCCTGCTCTGCCCAGGCAAGGAACAGAAGTGAGGATGAGACCCAGCCCCTTCCCCTGGGAACTCTCCTGG CCTTCTAGGAATGGAGGAGCCAGGCCCCACCCCTTCCCTATAGGAACAGCCCAGCACAGGTATTTCAGGTGTGAAAGAATCAGTAGGACC AGGCCACCGCTAGTGCTTGTGGAGATCACAGCCCCACCCTTGTCCCTCAGCAACATCCCATCTAAGCATTCCACACTGCAGGGAGGAGTG GTACTTAAGCTCCCCTGCCTTAACCTGGGACCAACCTGACCTAACCTAGGAGGGCTCTGAGCCAACCTTGCTCTTGGGGAAGGGGACAGA TTATGAAATTTCATGGATGAATTTTCCAGACCTATATCTGGAGTGAGAGGCCCCCACCCTTGGGCAGAGTCCTGCCTTCTTCCTTGAGGG GCAGTTTGGGAAGGTGATGGGTATTAGTGGGGGACTGAGTTCAGGTTACCAGAACCAGTACCTCAGTATTCTTTTTCAACATGTAGGGCA AGAGGATGAAGGAAGGGGCTATCCTGGGACCTCCCCAGCCCAGGAAAAACTGGAAGCCTTCCCCCAGCAAGGCAGAAGCTTGGAGGAGGG TTGTAAAAGCATATTGTACCCCCTCATTTGTTTATCTGATTTTTTTATTGCTCCGCATACTGAGAATCTAGGCCACCCCAACCTCTGTTC CCCACCCAGTTCTTCATTTGGAGGAATCACCCCATTTCAGAGTTATCAAGAGACACTCCCCCCTCCATTCCCACCCCTCATACCTACACC CAAGGTTGTCAGCTTTGGATTGCTGGGGCCAGGCCCCATGGAGGGTATACTGAGGGGTCTATAGGTTTGTGATTAAAATAATAAAAGCTA >72752_72752_3_RBM10-TFE3_RBM10_chrX_47041725_ENST00000377604_TFE3_chrX_48895639_ENST00000315869_length(amino acids)=1071AA_BP=215 MGVGGSEFPWEGSALGASPLPPICLQSRTWLLRAPAPAELGELEEVAAGRGDVWEPFLDSPGREESLQEASPRLADHGSSSGGGWEVKRS QRLRRGPSSPRRPYQDMEYERRGGRGDRTGRYGATDRSQDDGGENRSRDHDYRDMDYRSYPREYGSQEGKHDYDDSSEEQSAEDSYEASP GSETQRRRRRRHRHSPTGPPGFPRDGDYRDQDYRTEQGEEEEEEEDEEEEEKASNIVMLRMLPQAATEDDIRGQLQSHGVQAREVRLMRN KSSGQSRGFAFVEFSHLQDATRWMEANQHSLNILGQKVSMHYSDPKPKINEDWLCNKCGVQNFKRREKCFKCGVPKSEAEQKLPLGTRLD QQTLPLGGRELSQGLLPLPQPYQAQGVLASQALSQGSEPSSENANDTIILRNLNPHSTMDSILGALAPYAVLSSSNVRVIKDKQTQLNRG FAFIQLSTIVEAAQLLQILQALHPPLTIDGKTINVEFAKGSKRDMASNEGSRISAASVASTAIAAAQWAISQASQGGEGTWATSEEPPVD YSYYQQDEGYGNSQGTESSLYAHGYLKGTKGPGITGTKGDPTGAGPEASLEPGADSVSMQAFSRAQPGAAPGIYQQSAEASSSQGTAANS QSYTIMSPAVLKSELQSPTHPSSALPPATSPTAQESYSQYPVPDVSTYQYDETSGYYYDPQTGLYYDPNSQYYYNAQSQQYLYWDGERRT YVPALEQSADGHKETGAPSKEGKEKKEKHKTKTAQQIDDVIDEIISLESSYNDEMLSYLPGGTTGLQLPSTLPVSGNLLDVYSSQGVATP AITVSNSCPAELPNIKREISETEAKALLKERQKKDNHNLIERRRRFNINDRIKELGTLIPKSSDPEMRWNKGTILKASVDYIRKLQKEQQ RSKDLESRQRSLEQANRSLQLRIQELELQAQIHGLPVPPTPGLLSLATTSASDSLKPEQLDIEEEGRPGAATFHVGGGPAQNAPHQQPPA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RBM10-TFE3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RBM10-TFE3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RBM10-TFE3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
