
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RERE-MFSD2A (FusionGDB2 ID:73503) |
Fusion Gene Summary for RERE-MFSD2A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RERE-MFSD2A | Fusion gene ID: 73503 | Hgene | Tgene | Gene symbol | RERE | MFSD2A | Gene ID | 473 | 84879 |
| Gene name | arginine-glutamic acid dipeptide repeats | major facilitator superfamily domain containing 2A | |
| Synonyms | ARG|ARP|ATN1L|DNB1|NEDBEH | MCPH15|MFSD2|NLS1 | |
| Cytomap | 1p36.23 | 1p34.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | arginine-glutamic acid dipeptide repeats proteinarginine-glutamic acid dipeptide (RE) repeatsatrophin 2atrophin-1 like proteinatrophin-1 related protein | sodium-dependent lysophosphatidylcholine symporter 1major facilitator superfamily domain-containing protein 2Asodium-dependent LPC symporter 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q8NA29 | |
| Ensembl transtripts involved in fusion gene | ENST00000337907, ENST00000377464, ENST00000400907, ENST00000400908, ENST00000460659, ENST00000476556, | ENST00000372809, ENST00000420632, ENST00000480630, ENST00000372811, | |
| Fusion gene scores | * DoF score | 27 X 28 X 11=8316 | 8 X 8 X 3=192 |
| # samples | 43 | 8 | |
| ** MAII score | log2(43/8316*10)=-4.27348119326891 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/192*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: RERE [Title/Abstract] AND MFSD2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RERE(8482786)-MFSD2A(40422758), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | RERE-MFSD2A seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. RERE-MFSD2A seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. RERE-MFSD2A seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MFSD2A | GO:0051977 | lysophospholipid transport | 24828044 |
 Fusion gene breakpoints across RERE (5'-gene) Fusion gene breakpoints across RERE (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
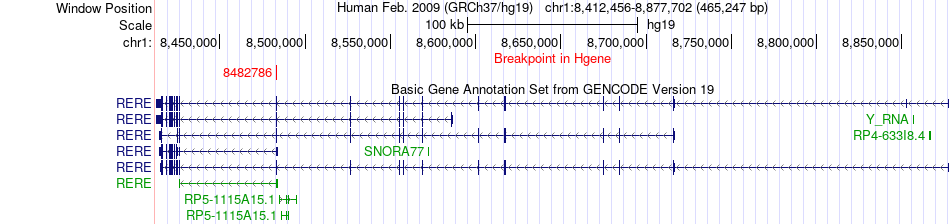 |
 Fusion gene breakpoints across MFSD2A (3'-gene) Fusion gene breakpoints across MFSD2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
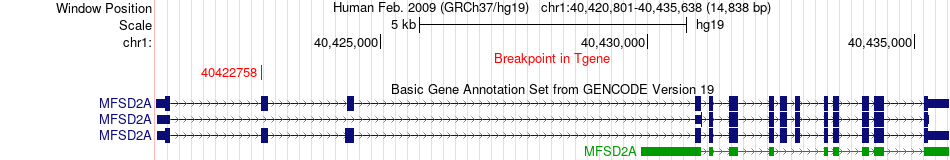 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-29-1778 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
Top |
Fusion Gene ORF analysis for RERE-MFSD2A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000337907 | ENST00000372809 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000337907 | ENST00000420632 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000337907 | ENST00000480630 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000377464 | ENST00000372809 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000377464 | ENST00000420632 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000377464 | ENST00000480630 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000400907 | ENST00000372809 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000400907 | ENST00000420632 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000400907 | ENST00000480630 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000400908 | ENST00000372809 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000400908 | ENST00000420632 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5CDS-intron | ENST00000400908 | ENST00000480630 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-3CDS | ENST00000460659 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-3CDS | ENST00000476556 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-intron | ENST00000460659 | ENST00000372809 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-intron | ENST00000460659 | ENST00000420632 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-intron | ENST00000460659 | ENST00000480630 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-intron | ENST00000476556 | ENST00000372809 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-intron | ENST00000476556 | ENST00000420632 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| 5UTR-intron | ENST00000476556 | ENST00000480630 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| Frame-shift | ENST00000400908 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| In-frame | ENST00000337907 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| In-frame | ENST00000377464 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
| In-frame | ENST00000400907 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000337907 | RERE | chr1 | 8482786 | - | ENST00000372811 | MFSD2A | chr1 | 40422758 | + | 3817 | 1919 | 533 | 3418 | 961 |
| ENST00000377464 | RERE | chr1 | 8482786 | - | ENST00000372811 | MFSD2A | chr1 | 40422758 | + | 2533 | 635 | 155 | 2134 | 659 |
| ENST00000400907 | RERE | chr1 | 8482786 | - | ENST00000372811 | MFSD2A | chr1 | 40422758 | + | 3256 | 1358 | 74 | 2857 | 927 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000337907 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + | 0.001124207 | 0.9988758 |
| ENST00000377464 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + | 0.003675117 | 0.99632484 |
| ENST00000400907 | ENST00000372811 | RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + | 0.001248496 | 0.9987515 |
Top |
Fusion Genomic Features for RERE-MFSD2A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + | 3.35E-08 | 1 |
| RERE | chr1 | 8482786 | - | MFSD2A | chr1 | 40422758 | + | 3.35E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
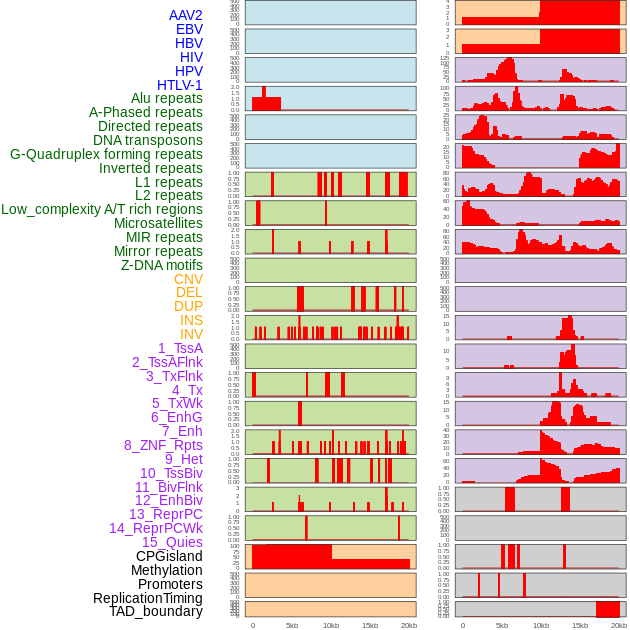 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
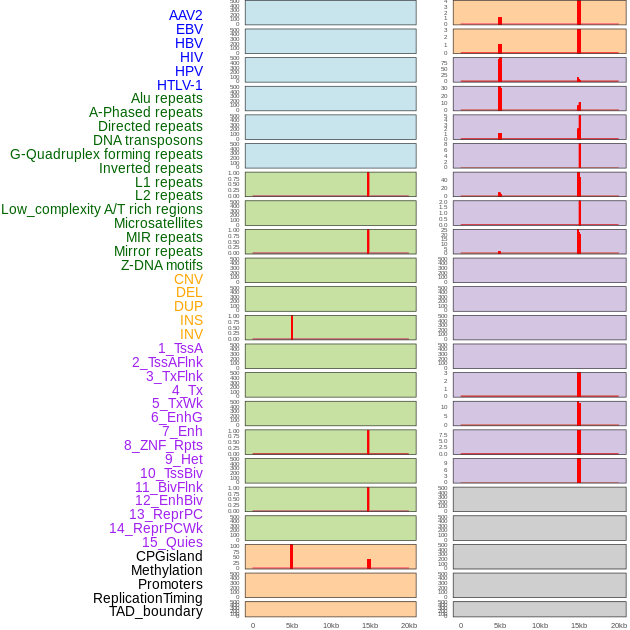 |
Top |
Fusion Protein Features for RERE-MFSD2A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:8482786/chr1:40422758) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MFSD2A |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Sodium-dependent lysophosphatidylcholine (LPC) symporter, which plays an essential role for blood-brain barrier formation and function (PubMed:24828040). Specifically expressed in endothelium of the blood-brain barrier of micro-vessels and transports LPC into the brain (By similarity). Transport of LPC is essential because it constitutes the major mechanism by which docosahexaenoic acid (DHA), an omega-3 fatty acid that is essential for normal brain growth and cognitive function, enters the brain (PubMed:26005868). Transports LPC carrying long-chain fatty acids such LPC oleate and LPC palmitate with a minimum acyl chain length of 14 carbons (By similarity). Does not transport docosahexaenoic acid in unesterified fatty acid (By similarity). Specifically required for blood-brain barrier formation and function, probably by mediating lipid transport (By similarity). Not required for central nervous system vascular morphogenesis (By similarity). Acts as a transporter for tunicamycin, an inhibitor of asparagine-linked glycosylation (PubMed:21677192). In placenta, acts as a receptor for ERVFRD-1/syncytin-2 and is required for trophoblast fusion (PubMed:18988732, PubMed:23177091). {ECO:0000250|UniProtKB:Q9DA75, ECO:0000269|PubMed:18988732, ECO:0000269|PubMed:21677192, ECO:0000269|PubMed:23177091, ECO:0000269|PubMed:24828040, ECO:0000269|PubMed:26005868}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 103_283 | 428 | 1567.0 | Domain | BAH |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 284_387 | 428 | 1567.0 | Domain | ELM2 |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 103_283 | 428 | 1567.0 | Domain | BAH |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 284_387 | 428 | 1567.0 | Domain | ELM2 |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 35_38 | 31 | 544.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 35_38 | 31 | 531.0 | Compositional bias | Note=Poly-Lys | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 102_128 | 31 | 544.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 150_160 | 31 | 544.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 182_256 | 31 | 544.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 278_310 | 31 | 544.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 332_344 | 31 | 544.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 366_370 | 31 | 544.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 392_393 | 31 | 544.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 415_437 | 31 | 544.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 459_480 | 31 | 544.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 502_543 | 31 | 544.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 68_80 | 31 | 544.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 102_128 | 31 | 531.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 150_160 | 31 | 531.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 182_256 | 31 | 531.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 278_310 | 31 | 531.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 332_344 | 31 | 531.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 366_370 | 31 | 531.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 392_393 | 31 | 531.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 415_437 | 31 | 531.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 459_480 | 31 | 531.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 502_543 | 31 | 531.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 68_80 | 31 | 531.0 | Topological domain | Cytoplasmic | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 129_149 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 161_181 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 257_277 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 311_331 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 345_365 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 371_391 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 394_414 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 438_458 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 47_67 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 481_501 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 81_101 | 31 | 544.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 129_149 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 161_181 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 257_277 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 311_331 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 345_365 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 371_391 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 394_414 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 438_458 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 47_67 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 481_501 | 31 | 531.0 | Transmembrane | Helical | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 81_101 | 31 | 531.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 1156_1211 | 428 | 1567.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 1156_1211 | 428 | 1567.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 1156_1211 | 0 | 1013.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 1179_1204 | 428 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 1300_1322 | 428 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 1425_1445 | 428 | 1567.0 | Compositional bias | Note=His-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 1451_1510 | 428 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 738_1118 | 428 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 1179_1204 | 428 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 1300_1322 | 428 | 1567.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 1425_1445 | 428 | 1567.0 | Compositional bias | Note=His-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 1451_1510 | 428 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 738_1118 | 428 | 1567.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 1179_1204 | 0 | 1013.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 1300_1322 | 0 | 1013.0 | Compositional bias | Note=Arg/Glu-rich (mixed charge) |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 1425_1445 | 0 | 1013.0 | Compositional bias | Note=His-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 1451_1510 | 0 | 1013.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 738_1118 | 0 | 1013.0 | Compositional bias | Note=Pro-rich |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 391_443 | 428 | 1567.0 | Domain | SANT |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 391_443 | 428 | 1567.0 | Domain | SANT |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 103_283 | 0 | 1013.0 | Domain | BAH |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 284_387 | 0 | 1013.0 | Domain | ELM2 |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 391_443 | 0 | 1013.0 | Domain | SANT |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000337907 | - | 13 | 24 | 507_532 | 428 | 1567.0 | Zinc finger | Note=GATA-type |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000400908 | - | 12 | 23 | 507_532 | 428 | 1567.0 | Zinc finger | Note=GATA-type |
| Hgene | RERE | chr1:8482786 | chr1:40422758 | ENST00000476556 | - | 2 | 13 | 507_532 | 0 | 1013.0 | Zinc finger | Note=GATA-type |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372809 | 0 | 14 | 1_46 | 31 | 544.0 | Topological domain | Extracellular | |
| Tgene | MFSD2A | chr1:8482786 | chr1:40422758 | ENST00000372811 | 0 | 14 | 1_46 | 31 | 531.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for RERE-MFSD2A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >73503_73503_1_RERE-MFSD2A_RERE_chr1_8482786_ENST00000337907_MFSD2A_chr1_40422758_ENST00000372811_length(transcript)=3817nt_BP=1919nt AAAATCACCATCATCCTCAGGGAGCCCCCTCGCCCTCGGAAAACAAAACCAGACATGATCCTGGGCTCCTGCTTCTGAAAGATCTGCCCG ACCGAGGAGGAGGGAGCCTAGGTTCAAGTCGCAGGAAAGGTCACGGCAGGGAAGCCCCCTCAGCCCCTCCGCGTCGCCTCCCCGCGGGCC GCCCCTCTCGGCCCTGCGCCCCGCGGCCCCGCAGCCTCCGCGCGGCCTCCCGCCCCATCCCCGCCACTTTCCCCGGAGCTGGGCCGCGAA CACCTCACTATTGATGGTGTAGCGCTTTAGGGGAAGGAGGTGATTGTCTAGGAGTAACTGTATGTGCGCTCAGCACCGGGTCACAACCAC CGTGGGATCCACTTACAGAGAAGATTTGTGGAGAGCTTCAGCCTGTTTATGGTGTTACTGAGAAAGGAATTGTGGCCATTTGATATAATT CTGGATGGAAGACCTCTTATTTGTATTACATTGAATTAGAGCATTTTTGAAACGTTGGGGTTGTTGGAGTGGTTGGATTTTCCCTGGAAT TGAGTGAGAAATTCAGAAGACTGAAGCCCAGGCTTACTGTCTACCTTTCACGGAGGCCTAGCCGTGAGAGGACAGAAGAAGGCACGTGGC GAATCATGACAGCGGACAAAGACAAAGACAAAGACAAAGAGAAGGACCGGGACCGAGACCGGGACCGAGAGAGAGAGAAAAGAGACAAAG CAAGAGAGAGTGAGAATTCAAGGCCACGCCGGAGCTGTACCTTGGAAGGAGGAGCCAAAAATTATGCTGAGAGTGATCACAGTGAAGACG AGGACAATGACAACAATAGTGCCACCGCAGAGGAGTCCACGAAGAAGAATAAGAAGAAACCACCGAAAAAAAAGTCTCGTTATGAAAGGA CAGATACCGGTGAGATAACATCCTACATCACTGAAGATGATGTGGTCTACAGACCAGGAGACTGTGTGTATATCGAGAGTCGGAGGCCAA ACACACCGTATTTCATCTGTAGCATTCAAGACTTCAAACTGGTCCACAACTCCCAGGCCTGTTGCAGATCTCCAACTCCTGCTTTGTGTG ACCCCCCAGCATGCTCTCTGCCGGTGGCATCACAGCCACCACAGCATCTTTCTGAAGCCGGGAGAGGGCCTGTAGGGAGTAAGAGGGACC ATCTCCTCATGAACGTCAAATGGTACTACCGTCAATCTGAGGTTCCAGATTCTGTGTATCAGCATTTGGTTCAGGATCGACATAATGAAA ATGACTCTGGAAGAGAACTTGTCATTACAGACCCAGTTATCAAGAACCGAGAGCTCTTCATTTCTGATTACGTTGACACTTACCATGCTG CTGCCCTTAGAGGGAAGTGTAACATCTCCCATTTTTCTGACATATTTGCTGCTAGAGAGTTTAAAGCCCGAGTGGATTCATTTTTCTACA TATTAGGATATAACCCTGAGACAAGGAGGCTGAACAGTACCCAGGGGGAGATTCGTGTCGGTCCTAGTCATCAGGCCAAACTTCCAGATC TGCAACCATTTCCTTCTCCAGATGGTGATACAGTGACCCAACATGAGGAACTGGTCTGGATGCCTGGAGTTAACGACTGTGACCTCCTTA TGTACTTGAGGGCAGCAAGGAGCATGGCGGCATTTGCAGGAATGTGTGATGGAGGCTCTACAGAGGACGGCTGTGTCGCAGCCTCTCGGG ATGACACCACTCTGAATGCACTGAACACACTGCATGAAAGCGGTTACGATGCTGGCAAAGCCCTGCAGCGCCTGGTGAAGAAGCCTGTGC CCAAGCTCATCGAGAAGTGCTGGACCGAGGACGAAGTGAAACGCTTCGTTAAGGGACTCAGGCAGTACGGGAAGAACTTCTTCAGAATTA GAAAGGAGCTGCTTCCCAATAAGGAAACAAAAGAACCGAAAAAGAAGAAACAACAGTTGTCTGTTTGCAACAAGCTTTGCTATGCACTTG GGGGAGCCCCCTACCAGGTGACGGGCTGTGCCCTGGGTTTCTTCCTTCAGATCTACCTATTGGATGTGGCTCAGGTGGGCCCTTTCTCTG CCTCCATCATCCTGTTTGTGGGCCGAGCCTGGGATGCCATCACAGACCCCCTGGTGGGCCTCTGCATCAGCAAATCCCCCTGGACCTGCC TGGGTCGCCTTATGCCCTGGATCATCTTCTCCACGCCCCTGGCCGTCATTGCCTACTTCCTCATCTGGTTCGTGCCCGACTTCCCACACG GCCAGACCTATTGGTACCTGCTTTTCTATTGCCTCTTTGAAACAATGGTCACGTGTTTCCATGTTCCCTACTCGGCTCTCACCATGTTCA TCAGCACCGAGCAGACTGAGCGGGATTCTGCCACCGCCTATCGGATGACTGTGGAAGTGCTGGGCACAGTGCTGGGCACGGCGATCCAGG GACAAATCGTGGGCCAAGCAGACACGCCTTGTTTCCAGGACCTCAATAGCTCTACAGTAGCTTCACAAAGTGCCAACCATACACATGGCA CCACCTCACACAGGGAAACGCAAAAGGCATACCTGCTGGCAGCGGGGGTCATTGTCTGTATCTATATAATCTGTGCTGTCATCCTGATCC TGGGCGTGCGGGAGCAGAGAGAACCCTATGAAGCCCAGCAGTCTGAGCCAATCGCCTACTTCCGGGGCCTACGGCTGGTCATGAGCCACG GCCCATACATCAAACTTATTACTGGCTTCCTCTTCACCTCCTTGGCTTTCATGCTGGTGGAGGGGAACTTTGTCTTGTTTTGCACCTACA CCTTGGGCTTCCGCAATGAATTCCAGAATCTACTCCTGGCCATCATGCTCTCGGCCACTTTAACCATTCCCATCTGGCAGTGGTTCTTGA CCCGGTTTGGCAAGAAGACAGCTGTATATGTTGGGATCTCATCAGCAGTGCCATTTCTCATCTTGGTGGCCCTCATGGAGAGTAACCTCA TCATTACATATGCGGTAGCTGTGGCAGCTGGCATCAGTGTGGCAGCTGCCTTCTTACTACCCTGGTCCATGCTGCCTGATGTCATTGACG ACTTCCATCTGAAGCAGCCCCACTTCCATGGAACCGAGCCCATCTTCTTCTCCTTCTATGTCTTCTTCACCAAGTTTGCCTCTGGAGTGT CACTGGGCATTTCTACCCTCAGTCTGGACTTTGCAGGGTACCAGACCCGTGGCTGCTCGCAGCCGGAACGTGTCAAGTTTACACTGAACA TGCTCGTGACCATGGCTCCCATAGTTCTCATCCTGCTGGGCCTGCTGCTCTTCAAAATGTACCCCATTGATGAGGAGAGGCGGCGGCAGA ATAAGAAGGCCCTGCAGGCACTGAGGGACGAGGCCAGCAGCTCTGGCTGCTCAGAAACAGACTCCACAGAGCTGGCTAGCATCCTCTAGG GCCCGCCACGTTGCCCGAAGCCACCATGCAGAAGGCCACAGAAGGGATCAGGACCTGTCTGCCGGCTTGCTGAGCAGCTGGACTGCAGGT GCTAGGAAGGGAACTGAAGACTCAAGGAGGTGGCCCAGGACACTTGCTGTGCTCACTGTGGGGCCGGCTGCTCTGTGGCCTCCTGCCTCC CCTCTGCCTGCCTGTGGGGCCAAGCCCTGGGGCTGCCACTGTGAATATGCCAAGGACTGATCGGGCCTAGCCCGGAACACTAATGTAGAA ACCTTTTTTTTTACAGAGCCTAATTAATAACTTAATGACTGTGTACATAGCAATGTGTGTGTATGTATATGTCTGTGAGCTATTAATGTT >73503_73503_1_RERE-MFSD2A_RERE_chr1_8482786_ENST00000337907_MFSD2A_chr1_40422758_ENST00000372811_length(amino acids)=961AA_BP=462 MELSEKFRRLKPRLTVYLSRRPSRERTEEGTWRIMTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHS EDEDNDNNSATAEESTKKNKKKPPKKKSRYERTDTGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPA LCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHLLMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTY HAAALRGKCNISHFSDIFAAREFKARVDSFFYILGYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCD LLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVKRFVKGLRQYGKNFF RIRKELLPNKETKEPKKKKQQLSVCNKLCYALGGAPYQVTGCALGFFLQIYLLDVAQVGPFSASIILFVGRAWDAITDPLVGLCISKSPW TCLGRLMPWIIFSTPLAVIAYFLIWFVPDFPHGQTYWYLLFYCLFETMVTCFHVPYSALTMFISTEQTERDSATAYRMTVEVLGTVLGTA IQGQIVGQADTPCFQDLNSSTVASQSANHTHGTTSHRETQKAYLLAAGVIVCIYIICAVILILGVREQREPYEAQQSEPIAYFRGLRLVM SHGPYIKLITGFLFTSLAFMLVEGNFVLFCTYTLGFRNEFQNLLLAIMLSATLTIPIWQWFLTRFGKKTAVYVGISSAVPFLILVALMES NLIITYAVAVAAGISVAAAFLLPWSMLPDVIDDFHLKQPHFHGTEPIFFSFYVFFTKFASGVSLGISTLSLDFAGYQTRGCSQPERVKFT -------------------------------------------------------------- >73503_73503_2_RERE-MFSD2A_RERE_chr1_8482786_ENST00000377464_MFSD2A_chr1_40422758_ENST00000372811_length(transcript)=2533nt_BP=635nt GGCTTAGGTCATAGCACAAGCTCAGTGAAAGCTCTATGAGGTCTCTGAGACTGCATGTGCCTCCATTTTGAGTTGTTAGTGTAGAAAAGG GCAGAGTTTTAGCAGAAAGTAAAGGTTCAGGAATCCAGCTAGACTCACCAGAGCCCGTGTTAGTCATGGATGATCCATTCAGCCCCTGCA GGAGGCTGAACAGTACCCAGGGGGAGATTCGTGTCGGTCCTAGTCATCAGGCCAAACTTCCAGATCTGCAACCATTTCCTTCTCCAGATG GTGATACAGTGACCCAACATGAGGAACTGGTCTGGATGCCTGGAGTTAACGACTGTGACCTCCTTATGTACTTGAGGGCAGCAAGGAGCA TGGCGGCATTTGCAGGAATGTGTGATGGAGGCTCTACAGAGGACGGCTGTGTCGCAGCCTCTCGGGATGACACCACTCTGAATGCACTGA ACACACTGCATGAAAGCGGTTACGATGCTGGCAAAGCCCTGCAGCGCCTGGTGAAGAAGCCTGTGCCCAAGCTCATCGAGAAGTGCTGGA CCGAGGACGAAGTGAAACGCTTCGTTAAGGGACTCAGGCAGTACGGGAAGAACTTCTTCAGAATTAGAAAGGAGCTGCTTCCCAATAAGG AAACAAAAGAACCGAAAAAGAAGAAACAACAGTTGTCTGTTTGCAACAAGCTTTGCTATGCACTTGGGGGAGCCCCCTACCAGGTGACGG GCTGTGCCCTGGGTTTCTTCCTTCAGATCTACCTATTGGATGTGGCTCAGGTGGGCCCTTTCTCTGCCTCCATCATCCTGTTTGTGGGCC GAGCCTGGGATGCCATCACAGACCCCCTGGTGGGCCTCTGCATCAGCAAATCCCCCTGGACCTGCCTGGGTCGCCTTATGCCCTGGATCA TCTTCTCCACGCCCCTGGCCGTCATTGCCTACTTCCTCATCTGGTTCGTGCCCGACTTCCCACACGGCCAGACCTATTGGTACCTGCTTT TCTATTGCCTCTTTGAAACAATGGTCACGTGTTTCCATGTTCCCTACTCGGCTCTCACCATGTTCATCAGCACCGAGCAGACTGAGCGGG ATTCTGCCACCGCCTATCGGATGACTGTGGAAGTGCTGGGCACAGTGCTGGGCACGGCGATCCAGGGACAAATCGTGGGCCAAGCAGACA CGCCTTGTTTCCAGGACCTCAATAGCTCTACAGTAGCTTCACAAAGTGCCAACCATACACATGGCACCACCTCACACAGGGAAACGCAAA AGGCATACCTGCTGGCAGCGGGGGTCATTGTCTGTATCTATATAATCTGTGCTGTCATCCTGATCCTGGGCGTGCGGGAGCAGAGAGAAC CCTATGAAGCCCAGCAGTCTGAGCCAATCGCCTACTTCCGGGGCCTACGGCTGGTCATGAGCCACGGCCCATACATCAAACTTATTACTG GCTTCCTCTTCACCTCCTTGGCTTTCATGCTGGTGGAGGGGAACTTTGTCTTGTTTTGCACCTACACCTTGGGCTTCCGCAATGAATTCC AGAATCTACTCCTGGCCATCATGCTCTCGGCCACTTTAACCATTCCCATCTGGCAGTGGTTCTTGACCCGGTTTGGCAAGAAGACAGCTG TATATGTTGGGATCTCATCAGCAGTGCCATTTCTCATCTTGGTGGCCCTCATGGAGAGTAACCTCATCATTACATATGCGGTAGCTGTGG CAGCTGGCATCAGTGTGGCAGCTGCCTTCTTACTACCCTGGTCCATGCTGCCTGATGTCATTGACGACTTCCATCTGAAGCAGCCCCACT TCCATGGAACCGAGCCCATCTTCTTCTCCTTCTATGTCTTCTTCACCAAGTTTGCCTCTGGAGTGTCACTGGGCATTTCTACCCTCAGTC TGGACTTTGCAGGGTACCAGACCCGTGGCTGCTCGCAGCCGGAACGTGTCAAGTTTACACTGAACATGCTCGTGACCATGGCTCCCATAG TTCTCATCCTGCTGGGCCTGCTGCTCTTCAAAATGTACCCCATTGATGAGGAGAGGCGGCGGCAGAATAAGAAGGCCCTGCAGGCACTGA GGGACGAGGCCAGCAGCTCTGGCTGCTCAGAAACAGACTCCACAGAGCTGGCTAGCATCCTCTAGGGCCCGCCACGTTGCCCGAAGCCAC CATGCAGAAGGCCACAGAAGGGATCAGGACCTGTCTGCCGGCTTGCTGAGCAGCTGGACTGCAGGTGCTAGGAAGGGAACTGAAGACTCA AGGAGGTGGCCCAGGACACTTGCTGTGCTCACTGTGGGGCCGGCTGCTCTGTGGCCTCCTGCCTCCCCTCTGCCTGCCTGTGGGGCCAAG CCCTGGGGCTGCCACTGTGAATATGCCAAGGACTGATCGGGCCTAGCCCGGAACACTAATGTAGAAACCTTTTTTTTTACAGAGCCTAAT TAATAACTTAATGACTGTGTACATAGCAATGTGTGTGTATGTATATGTCTGTGAGCTATTAATGTTATTAATTTTCATAAAAGCTGGAAA >73503_73503_2_RERE-MFSD2A_RERE_chr1_8482786_ENST00000377464_MFSD2A_chr1_40422758_ENST00000372811_length(amino acids)=659AA_BP=160 MDDPFSPCRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASR DDTTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVKRFVKGLRQYGKNFFRIRKELLPNKETKEPKKKKQQLSVCNKLCYAL GGAPYQVTGCALGFFLQIYLLDVAQVGPFSASIILFVGRAWDAITDPLVGLCISKSPWTCLGRLMPWIIFSTPLAVIAYFLIWFVPDFPH GQTYWYLLFYCLFETMVTCFHVPYSALTMFISTEQTERDSATAYRMTVEVLGTVLGTAIQGQIVGQADTPCFQDLNSSTVASQSANHTHG TTSHRETQKAYLLAAGVIVCIYIICAVILILGVREQREPYEAQQSEPIAYFRGLRLVMSHGPYIKLITGFLFTSLAFMLVEGNFVLFCTY TLGFRNEFQNLLLAIMLSATLTIPIWQWFLTRFGKKTAVYVGISSAVPFLILVALMESNLIITYAVAVAAGISVAAAFLLPWSMLPDVID DFHLKQPHFHGTEPIFFSFYVFFTKFASGVSLGISTLSLDFAGYQTRGCSQPERVKFTLNMLVTMAPIVLILLGLLLFKMYPIDEERRRQ -------------------------------------------------------------- >73503_73503_3_RERE-MFSD2A_RERE_chr1_8482786_ENST00000400907_MFSD2A_chr1_40422758_ENST00000372811_length(transcript)=3256nt_BP=1358nt TGAAGCCCAGGCTTACTGTCTACCTTTCACGGAGGCCTAGCCGTGAGAGGACAGAAGAAGGCACGTGGCGAATCATGACAGCGGACAAAG ACAAAGACAAAGACAAAGAGAAGGACCGGGACCGAGACCGGGACCGAGAGAGAGAGAAAAGAGACAAAGCAAGAGAGAGTGAGAATTCAA GGCCACGCCGGAGCTGTACCTTGGAAGGAGGAGCCAAAAATTATGCTGAGAGTGATCACAGTGAAGACGAGGACAATGACAACAATAGTG CCACCGCAGAGGAGTCCACGAAGAAGAATAAGAAGAAACCACCGAAAAAAAAGTCTCGTTATGAAAGGACAGATACCGGTGAGATAACAT CCTACATCACTGAAGATGATGTGGTCTACAGACCAGGAGACTGTGTGTATATCGAGAGTCGGAGGCCAAACACACCGTATTTCATCTGTA GCATTCAAGACTTCAAACTGGTCCACAACTCCCAGGCCTGTTGCAGATCTCCAACTCCTGCTTTGTGTGACCCCCCAGCATGCTCTCTGC CGGTGGCATCACAGCCACCACAGCATCTTTCTGAAGCCGGGAGAGGGCCTGTAGGGAGTAAGAGGGACCATCTCCTCATGAACGTCAAAT GGTACTACCGTCAATCTGAGGTTCCAGATTCTGTGTATCAGCATTTGGTTCAGGATCGACATAATGAAAATGACTCTGGAAGAGAACTTG TCATTACAGACCCAGTTATCAAGAACCGAGAGCTCTTCATTTCTGATTACGTTGACACTTACCATGCTGCTGCCCTTAGAGGGAAGTGTA ACATCTCCCATTTTTCTGACATATTTGCTGCTAGAGAGTTTAAAGCCCGAGTGGATTCATTTTTCTACATATTAGGATATAACCCTGAGA CAAGGAGGCTGAACAGTACCCAGGGGGAGATTCGTGTCGGTCCTAGTCATCAGGCCAAACTTCCAGATCTGCAACCATTTCCTTCTCCAG ATGGTGATACAGTGACCCAACATGAGGAACTGGTCTGGATGCCTGGAGTTAACGACTGTGACCTCCTTATGTACTTGAGGGCAGCAAGGA GCATGGCGGCATTTGCAGGAATGTGTGATGGAGGCTCTACAGAGGACGGCTGTGTCGCAGCCTCTCGGGATGACACCACTCTGAATGCAC TGAACACACTGCATGAAAGCGGTTACGATGCTGGCAAAGCCCTGCAGCGCCTGGTGAAGAAGCCTGTGCCCAAGCTCATCGAGAAGTGCT GGACCGAGGACGAAGTGAAACGCTTCGTTAAGGGACTCAGGCAGTACGGGAAGAACTTCTTCAGAATTAGAAAGGAGCTGCTTCCCAATA AGGAAACAAAAGAACCGAAAAAGAAGAAACAACAGTTGTCTGTTTGCAACAAGCTTTGCTATGCACTTGGGGGAGCCCCCTACCAGGTGA CGGGCTGTGCCCTGGGTTTCTTCCTTCAGATCTACCTATTGGATGTGGCTCAGGTGGGCCCTTTCTCTGCCTCCATCATCCTGTTTGTGG GCCGAGCCTGGGATGCCATCACAGACCCCCTGGTGGGCCTCTGCATCAGCAAATCCCCCTGGACCTGCCTGGGTCGCCTTATGCCCTGGA TCATCTTCTCCACGCCCCTGGCCGTCATTGCCTACTTCCTCATCTGGTTCGTGCCCGACTTCCCACACGGCCAGACCTATTGGTACCTGC TTTTCTATTGCCTCTTTGAAACAATGGTCACGTGTTTCCATGTTCCCTACTCGGCTCTCACCATGTTCATCAGCACCGAGCAGACTGAGC GGGATTCTGCCACCGCCTATCGGATGACTGTGGAAGTGCTGGGCACAGTGCTGGGCACGGCGATCCAGGGACAAATCGTGGGCCAAGCAG ACACGCCTTGTTTCCAGGACCTCAATAGCTCTACAGTAGCTTCACAAAGTGCCAACCATACACATGGCACCACCTCACACAGGGAAACGC AAAAGGCATACCTGCTGGCAGCGGGGGTCATTGTCTGTATCTATATAATCTGTGCTGTCATCCTGATCCTGGGCGTGCGGGAGCAGAGAG AACCCTATGAAGCCCAGCAGTCTGAGCCAATCGCCTACTTCCGGGGCCTACGGCTGGTCATGAGCCACGGCCCATACATCAAACTTATTA CTGGCTTCCTCTTCACCTCCTTGGCTTTCATGCTGGTGGAGGGGAACTTTGTCTTGTTTTGCACCTACACCTTGGGCTTCCGCAATGAAT TCCAGAATCTACTCCTGGCCATCATGCTCTCGGCCACTTTAACCATTCCCATCTGGCAGTGGTTCTTGACCCGGTTTGGCAAGAAGACAG CTGTATATGTTGGGATCTCATCAGCAGTGCCATTTCTCATCTTGGTGGCCCTCATGGAGAGTAACCTCATCATTACATATGCGGTAGCTG TGGCAGCTGGCATCAGTGTGGCAGCTGCCTTCTTACTACCCTGGTCCATGCTGCCTGATGTCATTGACGACTTCCATCTGAAGCAGCCCC ACTTCCATGGAACCGAGCCCATCTTCTTCTCCTTCTATGTCTTCTTCACCAAGTTTGCCTCTGGAGTGTCACTGGGCATTTCTACCCTCA GTCTGGACTTTGCAGGGTACCAGACCCGTGGCTGCTCGCAGCCGGAACGTGTCAAGTTTACACTGAACATGCTCGTGACCATGGCTCCCA TAGTTCTCATCCTGCTGGGCCTGCTGCTCTTCAAAATGTACCCCATTGATGAGGAGAGGCGGCGGCAGAATAAGAAGGCCCTGCAGGCAC TGAGGGACGAGGCCAGCAGCTCTGGCTGCTCAGAAACAGACTCCACAGAGCTGGCTAGCATCCTCTAGGGCCCGCCACGTTGCCCGAAGC CACCATGCAGAAGGCCACAGAAGGGATCAGGACCTGTCTGCCGGCTTGCTGAGCAGCTGGACTGCAGGTGCTAGGAAGGGAACTGAAGAC TCAAGGAGGTGGCCCAGGACACTTGCTGTGCTCACTGTGGGGCCGGCTGCTCTGTGGCCTCCTGCCTCCCCTCTGCCTGCCTGTGGGGCC AAGCCCTGGGGCTGCCACTGTGAATATGCCAAGGACTGATCGGGCCTAGCCCGGAACACTAATGTAGAAACCTTTTTTTTTACAGAGCCT AATTAATAACTTAATGACTGTGTACATAGCAATGTGTGTGTATGTATATGTCTGTGAGCTATTAATGTTATTAATTTTCATAAAAGCTGG >73503_73503_3_RERE-MFSD2A_RERE_chr1_8482786_ENST00000400907_MFSD2A_chr1_40422758_ENST00000372811_length(amino acids)=927AA_BP=428 MTADKDKDKDKEKDRDRDRDREREKRDKARESENSRPRRSCTLEGGAKNYAESDHSEDEDNDNNSATAEESTKKNKKKPPKKKSRYERTD TGEITSYITEDDVVYRPGDCVYIESRRPNTPYFICSIQDFKLVHNSQACCRSPTPALCDPPACSLPVASQPPQHLSEAGRGPVGSKRDHL LMNVKWYYRQSEVPDSVYQHLVQDRHNENDSGRELVITDPVIKNRELFISDYVDTYHAAALRGKCNISHFSDIFAAREFKARVDSFFYIL GYNPETRRLNSTQGEIRVGPSHQAKLPDLQPFPSPDGDTVTQHEELVWMPGVNDCDLLMYLRAARSMAAFAGMCDGGSTEDGCVAASRDD TTLNALNTLHESGYDAGKALQRLVKKPVPKLIEKCWTEDEVKRFVKGLRQYGKNFFRIRKELLPNKETKEPKKKKQQLSVCNKLCYALGG APYQVTGCALGFFLQIYLLDVAQVGPFSASIILFVGRAWDAITDPLVGLCISKSPWTCLGRLMPWIIFSTPLAVIAYFLIWFVPDFPHGQ TYWYLLFYCLFETMVTCFHVPYSALTMFISTEQTERDSATAYRMTVEVLGTVLGTAIQGQIVGQADTPCFQDLNSSTVASQSANHTHGTT SHRETQKAYLLAAGVIVCIYIICAVILILGVREQREPYEAQQSEPIAYFRGLRLVMSHGPYIKLITGFLFTSLAFMLVEGNFVLFCTYTL GFRNEFQNLLLAIMLSATLTIPIWQWFLTRFGKKTAVYVGISSAVPFLILVALMESNLIITYAVAVAAGISVAAAFLLPWSMLPDVIDDF HLKQPHFHGTEPIFFSFYVFFTKFASGVSLGISTLSLDFAGYQTRGCSQPERVKFTLNMLVTMAPIVLILLGLLLFKMYPIDEERRRQNK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RERE-MFSD2A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RERE-MFSD2A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RERE-MFSD2A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
