
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RNF139-EIF2A (FusionGDB2 ID:74788) |
Fusion Gene Summary for RNF139-EIF2A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RNF139-EIF2A | Fusion gene ID: 74788 | Hgene | Tgene | Gene symbol | RNF139 | EIF2A | Gene ID | 11236 | 83939 |
| Gene name | ring finger protein 139 | eukaryotic translation initiation factor 2A | |
| Synonyms | HRCA1|RCA1|TRC8 | CDA02|EIF-2A|MST089|MSTP004|MSTP089 | |
| Cytomap | 8q24.13 | 3q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase RNF139RING-type E3 ubiquitin transferase RNF139multiple membrane spanning receptor TRC8patched related protein translocated in renal cancertranslocation in renal carcinoma on chromosome 8 proteintranslocation in renal carc | eukaryotic translation initiation factor 2A65 kDa eukaryotic translation initiation factor 2Aeukaryotic translation initiation factor 2A, 65kDa | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9P2K8 | |
| Ensembl transtripts involved in fusion gene | ENST00000303545, | ENST00000383043, ENST00000482471, ENST00000273435, ENST00000406576, ENST00000460851, ENST00000487799, | |
| Fusion gene scores | * DoF score | 6 X 3 X 5=90 | 3 X 3 X 3=27 |
| # samples | 6 | 4 | |
| ** MAII score | log2(6/90*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/27*10)=0.567040592723894 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: RNF139 [Title/Abstract] AND EIF2A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RNF139(125487531)-EIF2A(150270143), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | RNF139 | GO:0008285 | negative regulation of cell proliferation | 17016439 |
| Hgene | RNF139 | GO:0016567 | protein ubiquitination | 10500182|17016439 |
| Hgene | RNF139 | GO:0017148 | negative regulation of translation | 20068067 |
| Hgene | RNF139 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport | 19706601 |
| Hgene | RNF139 | GO:0070613 | regulation of protein processing | 19706601 |
 Fusion gene breakpoints across RNF139 (5'-gene) Fusion gene breakpoints across RNF139 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
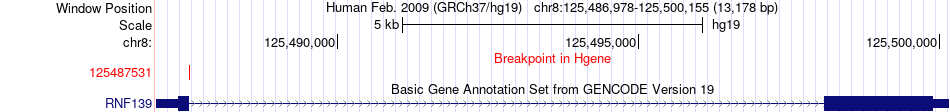 |
 Fusion gene breakpoints across EIF2A (3'-gene) Fusion gene breakpoints across EIF2A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
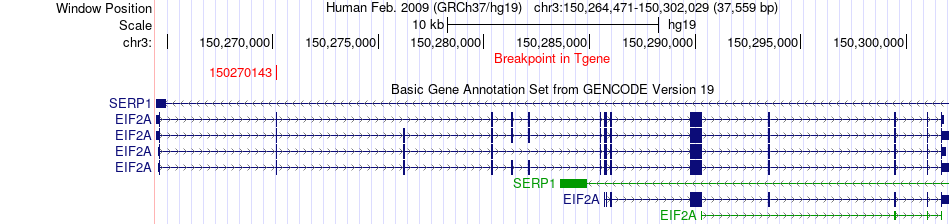 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-69-8254-01A | RNF139 | chr8 | 125487531 | - | EIF2A | chr3 | 150270143 | + |
| ChimerDB4 | LUAD | TCGA-69-8254-01A | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
Top |
Fusion Gene ORF analysis for RNF139-EIF2A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000303545 | ENST00000383043 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
| 5CDS-intron | ENST00000303545 | ENST00000482471 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
| In-frame | ENST00000303545 | ENST00000273435 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
| In-frame | ENST00000303545 | ENST00000406576 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
| In-frame | ENST00000303545 | ENST00000460851 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
| In-frame | ENST00000303545 | ENST00000487799 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000303545 | RNF139 | chr8 | 125487531 | + | ENST00000487799 | EIF2A | chr3 | 150270143 | + | 2320 | 553 | 186 | 2207 | 673 |
| ENST00000303545 | RNF139 | chr8 | 125487531 | + | ENST00000460851 | EIF2A | chr3 | 150270143 | + | 2614 | 553 | 186 | 2282 | 698 |
| ENST00000303545 | RNF139 | chr8 | 125487531 | + | ENST00000406576 | EIF2A | chr3 | 150270143 | + | 2304 | 553 | 186 | 2099 | 637 |
| ENST00000303545 | RNF139 | chr8 | 125487531 | + | ENST00000273435 | EIF2A | chr3 | 150270143 | + | 2599 | 553 | 186 | 2267 | 693 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000303545 | ENST00000487799 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + | 0.002323724 | 0.9976763 |
| ENST00000303545 | ENST00000460851 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + | 0.000957737 | 0.9990422 |
| ENST00000303545 | ENST00000406576 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + | 0.001438803 | 0.9985612 |
| ENST00000303545 | ENST00000273435 | RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270143 | + | 0.001010258 | 0.99898976 |
Top |
Fusion Genomic Features for RNF139-EIF2A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270142 | + | 5.52E-10 | 1 |
| RNF139 | chr8 | 125487531 | + | EIF2A | chr3 | 150270142 | + | 5.52E-10 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
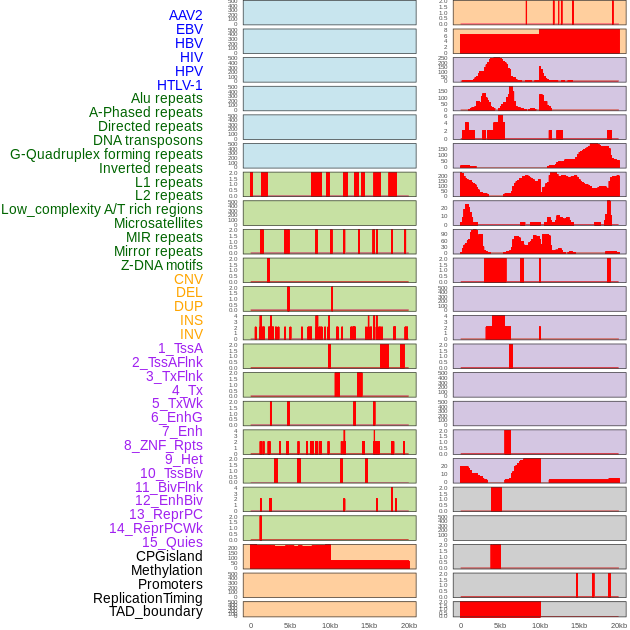 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
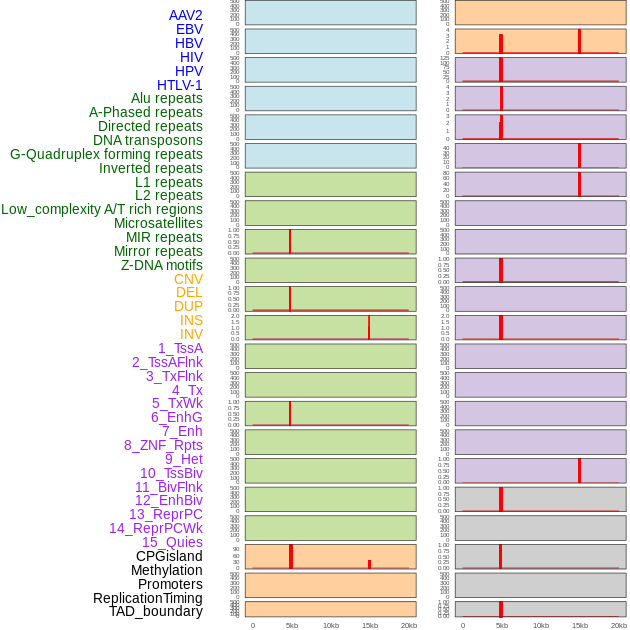 |
Top |
Fusion Protein Features for RNF139-EIF2A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:125487531/chr3:150270143) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | EIF2A |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Metabolic-stress sensing protein kinase that phosphorylates the alpha subunit of eukaryotic translation initiation factor 2 (EIF2S1/eIF-2-alpha) in response to low amino acid availability (PubMed:25329545). Plays a role as an activator of the integrated stress response (ISR) required for adaptation to amino acid starvation (By similarity). EIF2S1/eIF-2-alpha phosphorylation in response to stress converts EIF2S1/eIF-2-alpha in a global protein synthesis inhibitor, leading to a global attenuation of cap-dependent translation, and thus to a reduced overall utilization of amino acids, while concomitantly initiating the preferential translation of ISR-specific mRNAs, such as the transcriptional activator ATF4, and hence allowing ATF4-mediated reprogramming of amino acid biosynthetic gene expression to alleviate nutrient depletion (By similarity). Binds uncharged tRNAs (By similarity). Involved in cell cycle arrest by promoting cyclin D1 mRNA translation repression after the unfolded protein response pathway (UPR) activation or cell cycle inhibitor CDKN1A/p21 mRNA translation activation in response to amino acid deprivation (PubMed:26102367). Plays a role in the consolidation of synaptic plasticity, learning as well as formation of long-term memory (By similarity). Plays a role in neurite outgrowth inhibition (By similarity). Plays a proapoptotic role in response to glucose deprivation (By similarity). Promotes global cellular protein synthesis repression in response to UV irradiation independently of the stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) and p38 MAPK signaling pathways (By similarity). Plays a role in the antiviral response against alphavirus infection; impairs early viral mRNA translation of the incoming genomic virus RNA, thus preventing alphavirus replication (By similarity). {ECO:0000250|UniProtKB:P15442, ECO:0000250|UniProtKB:Q9QZ05, ECO:0000269|PubMed:25329545, ECO:0000269|PubMed:26102367}.; FUNCTION: (Microbial infection) Plays a role in modulating the adaptive immune response to yellow fever virus infection; promotes dendritic cells to initiate autophagy and antigene presentation to both CD4(+) and CD8(+) T-cells under amino acid starvation (PubMed:24310610). {ECO:0000269|PubMed:24310610}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 531_582 | 0 | 372.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 531_582 | 9 | 586.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 101_159 | 0 | 372.0 | Repeat | Note=WD 2 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 160_210 | 0 | 372.0 | Repeat | Note=WD 3 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 211_264 | 0 | 372.0 | Repeat | Note=WD 4 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 265_306 | 0 | 372.0 | Repeat | Note=WD 5 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 307_348 | 0 | 372.0 | Repeat | Note=WD 6 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 349_391 | 0 | 372.0 | Repeat | Note=WD 7 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000383043 | 0 | 8 | 56_100 | 0 | 372.0 | Repeat | Note=WD 1 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 101_159 | 9 | 586.0 | Repeat | Note=WD 2 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 160_210 | 9 | 586.0 | Repeat | Note=WD 3 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 211_264 | 9 | 586.0 | Repeat | Note=WD 4 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 265_306 | 9 | 586.0 | Repeat | Note=WD 5 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 307_348 | 9 | 586.0 | Repeat | Note=WD 6 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 349_391 | 9 | 586.0 | Repeat | Note=WD 7 | |
| Tgene | EIF2A | chr8:125487531 | chr3:150270143 | ENST00000460851 | 0 | 14 | 56_100 | 9 | 586.0 | Repeat | Note=WD 1 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 125_145 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 154_174 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 178_198 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 293_313 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 323_343 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 356_376 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 390_410 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 420_440 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 469_489 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 495_512 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 51_71 | 60 | 665.0 | Transmembrane | Helical |
| Hgene | RNF139 | chr8:125487531 | chr3:150270143 | ENST00000303545 | + | 1 | 2 | 85_105 | 60 | 665.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for RNF139-EIF2A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >74788_74788_1_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000273435_length(transcript)=2599nt_BP=553nt GACTGACGGCGCAGCCTCCGGGCCTAGCCACAGCAGCAACGGCAGAGGCCAGCGGGCGAGGTCAAGATGGTGGCTCCGCGGGCGGGGGAG GCAGTGGAGGGAGGAGGAGTCAGACCTTAGCCAGCCGGAAACACCGAAACCCAGAGACCTCCTGGGGAGCCGCCGCCGCCGCCCTCTCGG CCATCGCTGCCTCCGCCGCCTGCTCCACCTCGAGGGACGCGAGCGGGCGGCGGGGCTGGCCGTGAGAGAGACAGGAGAGGAAGGAGGGCA GGGGCGGAGTTGCCCGCCTTAGCCCCCGCCCCCGGCCGCGGCCCCGGGCCCTGCCCCGCGCGGCCCTGCCCGGCCCACCGAGCCCTGGTG TGGCAGCGGCTCATGGCGGCCGTGGGGCCCCCGCAGCAGCAGGTGCGGATGGCCCATCAGCAGGTCTGGGCGGCGCTCGAAGTGGCGCTC CGGGTGCCCTGCCTTTACATCATCGACGCCATCTTCAACTCCTACCCGGATTCCAGCCAAAGCCGGTTCTGCATCGTGCTCCAGATCTTC CTCCGGCTCTTTGTCCGAGGATCAGAAGGACTGTACATGGTGAATGGACCACCACATTTTACAGAAAGCACAGTGTTTCCAAGGGAATCT GGGAAGAATTGCAAAGTCTGTATCTTTAGTAAGGATGGGACCTTGTTTGCCTGGGGCAATGGAGAAAATGTCACTAACAAGGGACTACTG CACTCCTTCGACCTCCTGAAGGCAGTTTGCCTTGAATTCTCACCCAAAAATACTGTCCTGGCAACGTGGCAGCCTTACACTACTTCTAAA GATGGCACAGCTGGGATACCCAACCTACAACTTTATGATGTGAAAACTGGGACATGTTTGAAATCTTTCATCCAGAAAAAAATGCAAAAT TGGTGTCCATCCTGGTCAGAAGATGAAACTCTTTGTGCCCGCAATGTTAACAATGAAGTTCACTTCTTTGAAAACAACAATTTTAACACA ATTGCAAATAAATTGCATTTGCAAAAAATTAATGATTTTGTATTATCACCTGGACCCCAACCATACAAGGTGGCTGTCTATGTTCCAGGA AGTAAAGGTGCACCTTCATTTGTTAGATTATATCAGTACCCCAACTTTGCTGGACCTCATGCAGCTTTAGCTAATAAAAGTTTCTTTAAG GCAGATAAAGTTACAATGCTGTGGAATAAAAAAGCTACTGCTGTGTTGGTAATAGCTAGCACAGATGTTGACAAGACAGGAGCTTCCTAC TATGGAGAACAAACTCTACACTACATTGCAACAAATGGAGAAAGTGCTGTAGTGCAATTACCAAAAAATGGCCCCATTTATGATGTAGTT TGGAATTCTAGTTCTACTGAGTTTTGTGCTGTATATGGTTTTATGCCTGCCAAAGCGACAATTTTCAACTTGAAATGTGATCCTGTATTT GACTTTGGAACTGGTCCTCGTAATGCAGCCTACTATAGCCCTCATGGACATATATTAGTATTAGCTGGATTTGGAAATCTGAGGGGACAA ATGGAAGTGTGGGATGTGAAAAACTACAAACTTATTTCTAAACCGGTGGCTTCTGATTCTACATATTTTGCTTGGTGCCCGGATGGTGAG CATATTTTAACAGCTACATGTGCTCCCAGGTTACGGGTTAATAATGGATACAAAATTTGGCATTATACTGGCTCTATCTTGCACAAGTAT GATGTGCCATCAAATGCAGAATTATGGCAGGTTTCTTGGCAGCCATTTTTGGATGGAATATTTCCAGCAAAAACAATAACTTACCAAGCA GTTCCAAGTGAAGTACCCAATGAGGAACCTAAAGTTGCAACAGCTTATAGACCCCCAGCTTTAAGAAATAAACCAATCACCAATTCCAAA TTGCATGAAGAGGAACCACCTCAGAATATGAAACCACAATCAGGAAACGATAAGCCATTATCAAAAACAGCTCTTAAAAATCAAAGGAAG CATGAAGCTAAGAAAGCTGCAAAGCAGGAAGCAAGAAGTGACAAGAGTCCAGATTTGGCACCTACTCCTGCCCCACAGAGCACACCACGA AACACTGTCTCTCAGTCAATTTCTGGGGACCCTGAGATAGACAAAAAAATCAAGAACCTAAAGAAGAAACTGAAAGCAATCGAACAACTG AAAGAACAAGCAGCAACTGGAAAACAGCTAGAAAAAAATCAGTTGGAGAAAATTCAGAAAGAAACAGCCCTTCTCCAGGAGCTGGAAGAT TTGGAATTGGGTATTTAAAGATTCACGGAAAGCAAGTTGATGACCAGAAATCAGTGCAAACACATCTTCTGTTAAACCCATTGGTATACA CAGAATATTCCTGTGCCCACACTTAATGTCAATCTATAATTTTAACCATTTATCCAAGATTCTACTAAGTGTAAAATTATTTAATAATGT CTATTAAATTGATATTTATATCTTGCATCCTATATCATGTCAATATGTGATATAGAAAAGAGATACGTGAATTTTTTAGCTAAGCTTGAC >74788_74788_1_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000273435_length(amino acids)=693AA_BP=113 MPPPPAPPRGTRAGGGAGRERDRRGRRAGAELPALAPAPGRGPGPCPARPCPAHRALVWQRLMAAVGPPQQQVRMAHQQVWAALEVALRV PCLYIIDAIFNSYPDSSQSRFCIVLQIFLRLFVRGSEGLYMVNGPPHFTESTVFPRESGKNCKVCIFSKDGTLFAWGNGENVTNKGLLHS FDLLKAVCLEFSPKNTVLATWQPYTTSKDGTAGIPNLQLYDVKTGTCLKSFIQKKMQNWCPSWSEDETLCARNVNNEVHFFENNNFNTIA NKLHLQKINDFVLSPGPQPYKVAVYVPGSKGAPSFVRLYQYPNFAGPHAALANKSFFKADKVTMLWNKKATAVLVIASTDVDKTGASYYG EQTLHYIATNGESAVVQLPKNGPIYDVVWNSSSTEFCAVYGFMPAKATIFNLKCDPVFDFGTGPRNAAYYSPHGHILVLAGFGNLRGQME VWDVKNYKLISKPVASDSTYFAWCPDGEHILTATCAPRLRVNNGYKIWHYTGSILHKYDVPSNAELWQVSWQPFLDGIFPAKTITYQAVP SEVPNEEPKVATAYRPPALRNKPITNSKLHEEEPPQNMKPQSGNDKPLSKTALKNQRKHEAKKAAKQEARSDKSPDLAPTPAPQSTPRNT -------------------------------------------------------------- >74788_74788_2_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000406576_length(transcript)=2304nt_BP=553nt GACTGACGGCGCAGCCTCCGGGCCTAGCCACAGCAGCAACGGCAGAGGCCAGCGGGCGAGGTCAAGATGGTGGCTCCGCGGGCGGGGGAG GCAGTGGAGGGAGGAGGAGTCAGACCTTAGCCAGCCGGAAACACCGAAACCCAGAGACCTCCTGGGGAGCCGCCGCCGCCGCCCTCTCGG CCATCGCTGCCTCCGCCGCCTGCTCCACCTCGAGGGACGCGAGCGGGCGGCGGGGCTGGCCGTGAGAGAGACAGGAGAGGAAGGAGGGCA GGGGCGGAGTTGCCCGCCTTAGCCCCCGCCCCCGGCCGCGGCCCCGGGCCCTGCCCCGCGCGGCCCTGCCCGGCCCACCGAGCCCTGGTG TGGCAGCGGCTCATGGCGGCCGTGGGGCCCCCGCAGCAGCAGGTGCGGATGGCCCATCAGCAGGTCTGGGCGGCGCTCGAAGTGGCGCTC CGGGTGCCCTGCCTTTACATCATCGACGCCATCTTCAACTCCTACCCGGATTCCAGCCAAAGCCGGTTCTGCATCGTGCTCCAGATCTTC CTCCGGCTCTTTGTCCGAGGATCAGAAGGACTGTACATGGTGAATGGACCACCACATTTTACAGAAAGCACAGTGTTTCCAAGGGAATCT GGGAAGAATTGCAAAGTCTGTATCTTTAGTAAGGATGGGACCTTGTTTGCCTGGGGCAATGGAGAAAAAGTAAATATTATCAGTGTCACT AACAAGGGACTACTGCACTCCTTCGACCTCCTGAAGGCAGTTTGCCTTGAATTCTCACCCAAAAATACTGTCCTGGCAACGTGGCAGCCT TACACTAACACAATTGCAAATAAATTGCATTTGCAAAAAATTAATGATTTTGTATTATCACCTGGACCCCAACCATACAAGGTGGCTGTC TATGTTCCAGGAAGTAAAGGTGCACCTTCATTTGTTAGATTATATCAGTACCCCAACTTTGCTGGACCTCATGCAGCTTTAGCTAATAAA AGTTTCTTTAAGGCAGATAAAGTTACAATGCTGTGGAATAAAAAAGCTACTGCTGTGTTGGTAATAGCTAGCACAGATGTTGACAAGACA GGAGCTTCCTACTATGGAGAACAAACTCTACACTACATTGCAACAAATGGAGAAAGTGCTGTAGTGCAATTACCAAAAAATGGCCCCATT TATGATGTAGTTTGGAATTCTAGTTCTACTGAGTTTTGTGCTGTATATGGTTTTATGCCTGCCAAAGCGACAATTTTCAACTTGAAATGT GATCCTGTATTTGACTTTGGAACTGGTCCTCGTAATGCAGCCTACTATAGCCCTCATGGACATATATTAGTATTAGCTGGATTTGGAAAT CTGAGGGGACAAATGGAAGTGTGGGATGTGAAAAACTACAAACTTATTTCTAAACCGGTGGCTTCTGATTCTACATATTTTGCTTGGTGC CCGGATGGTGAGCATATTTTAACAGCTACATGTGCTCCCAGGTTACGGGTTAATAATGGATACAAAATTTGGCATTATACTGGCTCTATC TTGCACAAGTATGATGTGCCATCAAATGCAGAATTATGGCAGGTTTCTTGGCAGCCATTTTTGGATGGAATATTTCCAGCAAAAACAATA ACTTACCAAGCAGTTCCAAGTGAAGTACCCAATGAGGAACCTAAAGTTGCAACAGCTTATAGACCCCCAGCTTTAAGAAATAAACCAATC ACCAATTCCAAATTGCATGAAGAGGAACCACCTCAGAATATGAAACCACAATCAGGAAACGATAAGCCATTATCAAAAACAGCTCTTAAA AATCAAAGGAAGCATGAAGCTAAGAAAGCTGCAAAGCAGGAAGCAAGAAGTGACAAGAGTCCAGATTTGGCACCTACTCCTGCCCCACAG AGCACACCACGAAACACTGTCTCTCAGTCAATTTCTGGGGACCCTGAGATAGACAAAAAAATCAAGAACCTAAAGAAGAAACTGAAAGCA ATCGAACAACTGAAAGAACAAGCAGCAACTGGAAAACAGCTAGAAAAAAATCAGTTGGAGAAAATTCAGAAAGAAACAGCCCTTCTCCAG GAGCTGGAAGATTTGGAATTGGGTATTTAAAGATTCACGGAAAGCAAGTTGATGACCAGAAATCAGTGCAAACACATCTTCTGTTAAACC CATTGGTATACACAGAATATTCCTGTGCCCACACTTAATGTCAATCTATAATTTTAACCATTTATCCAAGATTCTACTAAGTGTAAAATT >74788_74788_2_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000406576_length(amino acids)=637AA_BP=113 MPPPPAPPRGTRAGGGAGRERDRRGRRAGAELPALAPAPGRGPGPCPARPCPAHRALVWQRLMAAVGPPQQQVRMAHQQVWAALEVALRV PCLYIIDAIFNSYPDSSQSRFCIVLQIFLRLFVRGSEGLYMVNGPPHFTESTVFPRESGKNCKVCIFSKDGTLFAWGNGEKVNIISVTNK GLLHSFDLLKAVCLEFSPKNTVLATWQPYTNTIANKLHLQKINDFVLSPGPQPYKVAVYVPGSKGAPSFVRLYQYPNFAGPHAALANKSF FKADKVTMLWNKKATAVLVIASTDVDKTGASYYGEQTLHYIATNGESAVVQLPKNGPIYDVVWNSSSTEFCAVYGFMPAKATIFNLKCDP VFDFGTGPRNAAYYSPHGHILVLAGFGNLRGQMEVWDVKNYKLISKPVASDSTYFAWCPDGEHILTATCAPRLRVNNGYKIWHYTGSILH KYDVPSNAELWQVSWQPFLDGIFPAKTITYQAVPSEVPNEEPKVATAYRPPALRNKPITNSKLHEEEPPQNMKPQSGNDKPLSKTALKNQ RKHEAKKAAKQEARSDKSPDLAPTPAPQSTPRNTVSQSISGDPEIDKKIKNLKKKLKAIEQLKEQAATGKQLEKNQLEKIQKETALLQEL -------------------------------------------------------------- >74788_74788_3_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000460851_length(transcript)=2614nt_BP=553nt GACTGACGGCGCAGCCTCCGGGCCTAGCCACAGCAGCAACGGCAGAGGCCAGCGGGCGAGGTCAAGATGGTGGCTCCGCGGGCGGGGGAG GCAGTGGAGGGAGGAGGAGTCAGACCTTAGCCAGCCGGAAACACCGAAACCCAGAGACCTCCTGGGGAGCCGCCGCCGCCGCCCTCTCGG CCATCGCTGCCTCCGCCGCCTGCTCCACCTCGAGGGACGCGAGCGGGCGGCGGGGCTGGCCGTGAGAGAGACAGGAGAGGAAGGAGGGCA GGGGCGGAGTTGCCCGCCTTAGCCCCCGCCCCCGGCCGCGGCCCCGGGCCCTGCCCCGCGCGGCCCTGCCCGGCCCACCGAGCCCTGGTG TGGCAGCGGCTCATGGCGGCCGTGGGGCCCCCGCAGCAGCAGGTGCGGATGGCCCATCAGCAGGTCTGGGCGGCGCTCGAAGTGGCGCTC CGGGTGCCCTGCCTTTACATCATCGACGCCATCTTCAACTCCTACCCGGATTCCAGCCAAAGCCGGTTCTGCATCGTGCTCCAGATCTTC CTCCGGCTCTTTGTCCGAGGATCAGAAGGACTGTACATGGTGAATGGACCACCACATTTTACAGAAAGCACAGTGTTTCCAAGGGAATCT GGGAAGAATTGCAAAGTCTGTATCTTTAGTAAGGATGGGACCTTGTTTGCCTGGGGCAATGGAGAAAAAGTAAATATTATCAGTGTCACT AACAAGGGACTACTGCACTCCTTCGACCTCCTGAAGGCAGTTTGCCTTGAATTCTCACCCAAAAATACTGTCCTGGCAACGTGGCAGCCT TACACTACTTCTAAAGATGGCACAGCTGGGATACCCAACCTACAACTTTATGATGTGAAAACTGGGACATGTTTGAAATCTTTCATCCAG AAAAAAATGCAAAATTGGTGTCCATCCTGGTCAGAAGATGAAACTCTTTGTGCCCGCAATGTTAACAATGAAGTTCACTTCTTTGAAAAC AACAATTTTAACACAATTGCAAATAAATTGCATTTGCAAAAAATTAATGATTTTGTATTATCACCTGGACCCCAACCATACAAGGTGGCT GTCTATGTTCCAGGAAGTAAAGGTGCACCTTCATTTGTTAGATTATATCAGTACCCCAACTTTGCTGGACCTCATGCAGCTTTAGCTAAT AAAAGTTTCTTTAAGGCAGATAAAGTTACAATGCTGTGGAATAAAAAAGCTACTGCTGTGTTGGTAATAGCTAGCACAGATGTTGACAAG ACAGGAGCTTCCTACTATGGAGAACAAACTCTACACTACATTGCAACAAATGGAGAAAGTGCTGTAGTGCAATTACCAAAAAATGGCCCC ATTTATGATGTAGTTTGGAATTCTAGTTCTACTGAGTTTTGTGCTGTATATGGTTTTATGCCTGCCAAAGCGACAATTTTCAACTTGAAA TGTGATCCTGTATTTGACTTTGGAACTGGTCCTCGTAATGCAGCCTACTATAGCCCTCATGGACATATATTAGTATTAGCTGGATTTGGA AATCTGAGGGGACAAATGGAAGTGTGGGATGTGAAAAACTACAAACTTATTTCTAAACCGGTGGCTTCTGATTCTACATATTTTGCTTGG TGCCCGGATGGTGAGCATATTTTAACAGCTACATGTGCTCCCAGGTTACGGGTTAATAATGGATACAAAATTTGGCATTATACTGGCTCT ATCTTGCACAAGTATGATGTGCCATCAAATGCAGAATTATGGCAGGTTTCTTGGCAGCCATTTTTGGATGGAATATTTCCAGCAAAAACA ATAACTTACCAAGCAGTTCCAAGTGAAGTACCCAATGAGGAACCTAAAGTTGCAACAGCTTATAGACCCCCAGCTTTAAGAAATAAACCA ATCACCAATTCCAAATTGCATGAAGAGGAACCACCTCAGAATATGAAACCACAATCAGGAAACGATAAGCCATTATCAAAAACAGCTCTT AAAAATCAAAGGAAGCATGAAGCTAAGAAAGCTGCAAAGCAGGAAGCAAGAAGTGACAAGAGTCCAGATTTGGCACCTACTCCTGCCCCA CAGAGCACACCACGAAACACTGTCTCTCAGTCAATTTCTGGGGACCCTGAGATAGACAAAAAAATCAAGAACCTAAAGAAGAAACTGAAA GCAATCGAACAACTGAAAGAACAAGCAGCAACTGGAAAACAGCTAGAAAAAAATCAGTTGGAGAAAATTCAGAAAGAAACAGCCCTTCTC CAGGAGCTGGAAGATTTGGAATTGGGTATTTAAAGATTCACGGAAAGCAAGTTGATGACCAGAAATCAGTGCAAACACATCTTCTGTTAA ACCCATTGGTATACACAGAATATTCCTGTGCCCACACTTAATGTCAATCTATAATTTTAACCATTTATCCAAGATTCTACTAAGTGTAAA ATTATTTAATAATGTCTATTAAATTGATATTTATATCTTGCATCCTATATCATGTCAATATGTGATATAGAAAAGAGATACGTGAATTTT TTAGCTAAGCTTGACAGATTGAAAGACAAGTGTCATTTTTTTTTGTAGAGGGTGATATATACCATGTAAATGAACAAAGACATTTTAAAT >74788_74788_3_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000460851_length(amino acids)=698AA_BP=113 MPPPPAPPRGTRAGGGAGRERDRRGRRAGAELPALAPAPGRGPGPCPARPCPAHRALVWQRLMAAVGPPQQQVRMAHQQVWAALEVALRV PCLYIIDAIFNSYPDSSQSRFCIVLQIFLRLFVRGSEGLYMVNGPPHFTESTVFPRESGKNCKVCIFSKDGTLFAWGNGEKVNIISVTNK GLLHSFDLLKAVCLEFSPKNTVLATWQPYTTSKDGTAGIPNLQLYDVKTGTCLKSFIQKKMQNWCPSWSEDETLCARNVNNEVHFFENNN FNTIANKLHLQKINDFVLSPGPQPYKVAVYVPGSKGAPSFVRLYQYPNFAGPHAALANKSFFKADKVTMLWNKKATAVLVIASTDVDKTG ASYYGEQTLHYIATNGESAVVQLPKNGPIYDVVWNSSSTEFCAVYGFMPAKATIFNLKCDPVFDFGTGPRNAAYYSPHGHILVLAGFGNL RGQMEVWDVKNYKLISKPVASDSTYFAWCPDGEHILTATCAPRLRVNNGYKIWHYTGSILHKYDVPSNAELWQVSWQPFLDGIFPAKTIT YQAVPSEVPNEEPKVATAYRPPALRNKPITNSKLHEEEPPQNMKPQSGNDKPLSKTALKNQRKHEAKKAAKQEARSDKSPDLAPTPAPQS -------------------------------------------------------------- >74788_74788_4_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000487799_length(transcript)=2320nt_BP=553nt GACTGACGGCGCAGCCTCCGGGCCTAGCCACAGCAGCAACGGCAGAGGCCAGCGGGCGAGGTCAAGATGGTGGCTCCGCGGGCGGGGGAG GCAGTGGAGGGAGGAGGAGTCAGACCTTAGCCAGCCGGAAACACCGAAACCCAGAGACCTCCTGGGGAGCCGCCGCCGCCGCCCTCTCGG CCATCGCTGCCTCCGCCGCCTGCTCCACCTCGAGGGACGCGAGCGGGCGGCGGGGCTGGCCGTGAGAGAGACAGGAGAGGAAGGAGGGCA GGGGCGGAGTTGCCCGCCTTAGCCCCCGCCCCCGGCCGCGGCCCCGGGCCCTGCCCCGCGCGGCCCTGCCCGGCCCACCGAGCCCTGGTG TGGCAGCGGCTCATGGCGGCCGTGGGGCCCCCGCAGCAGCAGGTGCGGATGGCCCATCAGCAGGTCTGGGCGGCGCTCGAAGTGGCGCTC CGGGTGCCCTGCCTTTACATCATCGACGCCATCTTCAACTCCTACCCGGATTCCAGCCAAAGCCGGTTCTGCATCGTGCTCCAGATCTTC CTCCGGCTCTTTGTCCGAGGATCAGAAGGACTGTACATGGTGAATGGACCACCACATTTTACAGAAAGCACAGTGTTTCCAAGAGTAAAT ATTATCAGTGTCACTAACAAGGGACTACTGCACTCCTTCGACCTCCTGAAGGCAGTTTGCCTTGAATTCTCACCCAAAAATACTGTCCTG GCAACGTGGCAGCCTTACACTACTTCTAAAGATGGCACAGCTGGGATACCCAACCTACAACTTTATGATGTGAAAACTGGGACATGTTTG AAATCTTTCATCCAGAAAAAAATGCAAAATTGGTGTCCATCCTGGTCAGAAGATGAAACTCTTTGTGCCCGCAATGTTAACAATGAAGTT CACTTCTTTGAAAACAACAATTTTAACACAATTGCAAATAAATTGCATTTGCAAAAAATTAATGATTTTGTATTATCACCTGGACCCCAA CCATACAAGGTGGCTGTCTATGTTCCAGGAAGTAAAGGTGCACCTTCATTTGTTAGATTATATCAGTACCCCAACTTTGCTGGACCTCAT GCAGCTTTAGCTAATAAAAGTTTCTTTAAGGCAGATAAAGTTACAATGCTGTGGAATAAAAAAGCTACTGCTGTGTTGGTAATAGCTAGC ACAGATGTTGACAAGACAGGAGCTTCCTACTATGGAGAACAAACTCTACACTACATTGCAACAAATGGAGAAAGTGCTGTAGTGCAATTA CCAAAAAATGGCCCCATTTATGATGTAGTTTGGAATTCTAGTTCTACTGAGTTTTGTGCTGTATATGGTTTTATGCCTGCCAAAGCGACA ATTTTCAACTTGAAATGTGATCCTGTATTTGACTTTGGAACTGGTCCTCGTAATGCAGCCTACTATAGCCCTCATGGACATATATTAGTA TTAGCTGGATTTGGAAATCTGAGGGGACAAATGGAAGTGTGGGATGTGAAAAACTACAAACTTATTTCTAAACCGGTGGCTTCTGATTCT ACATATTTTGCTTGGTGCCCGGATGGTGAGCATATTTTAACAGCTACATGTGCTCCCAGGTTACGGGTTAATAATGGATACAAAATTTGG CATTATACTGGCTCTATCTTGCACAAGTATGATGTGCCATCAAATGCAGAATTATGGCAGGTTTCTTGGCAGCCATTTTTGGATGGAATA TTTCCAGCAAAAACAATAACTTACCAAGCAGTTCCAAGTGAAGTACCCAATGAGGAACCTAAAGTTGCAACAGCTTATAGACCCCCAGCT TTAAGAAATAAACCAATCACCAATTCCAAATTGCATGAAGAGGAACCACCTCAGAATATGAAACCACAATCAGGAAACGATAAGCCATTA TCAAAAACAGCTCTTAAAAATCAAAGGAAGCATGAAGCTAAGAAAGCTGCAAAGCAGGAAGCAAGAAGTGACAAGAGTCCAGATTTGGCA CCTACTCCTGCCCCACAGAGCACACCACGAAACACTGTCTCTCAGTCAATTTCTGGGGACCCTGAGATAGACAAAAAAATCAAGAACCTA AAGAAGAAACTGAAAGCAATCGAACAACTGAAAGAACAAGCAGCAACTGGAAAACAGCTAGAAAAAAATCAGTTGGAGAAAATTCAGAAA GAAACAGCCCTTCTCCAGGAGCTGGAAGATTTGGAATTGGGTATTTAAAGATTCACGGAAAGCAAGTTGATGACCAGAAATCAGTGCAAA >74788_74788_4_RNF139-EIF2A_RNF139_chr8_125487531_ENST00000303545_EIF2A_chr3_150270143_ENST00000487799_length(amino acids)=673AA_BP=113 MPPPPAPPRGTRAGGGAGRERDRRGRRAGAELPALAPAPGRGPGPCPARPCPAHRALVWQRLMAAVGPPQQQVRMAHQQVWAALEVALRV PCLYIIDAIFNSYPDSSQSRFCIVLQIFLRLFVRGSEGLYMVNGPPHFTESTVFPRVNIISVTNKGLLHSFDLLKAVCLEFSPKNTVLAT WQPYTTSKDGTAGIPNLQLYDVKTGTCLKSFIQKKMQNWCPSWSEDETLCARNVNNEVHFFENNNFNTIANKLHLQKINDFVLSPGPQPY KVAVYVPGSKGAPSFVRLYQYPNFAGPHAALANKSFFKADKVTMLWNKKATAVLVIASTDVDKTGASYYGEQTLHYIATNGESAVVQLPK NGPIYDVVWNSSSTEFCAVYGFMPAKATIFNLKCDPVFDFGTGPRNAAYYSPHGHILVLAGFGNLRGQMEVWDVKNYKLISKPVASDSTY FAWCPDGEHILTATCAPRLRVNNGYKIWHYTGSILHKYDVPSNAELWQVSWQPFLDGIFPAKTITYQAVPSEVPNEEPKVATAYRPPALR NKPITNSKLHEEEPPQNMKPQSGNDKPLSKTALKNQRKHEAKKAAKQEARSDKSPDLAPTPAPQSTPRNTVSQSISGDPEIDKKIKNLKK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RNF139-EIF2A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RNF139-EIF2A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RNF139-EIF2A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
