
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RNF165-ADAM10 (FusionGDB2 ID:74858) |
Fusion Gene Summary for RNF165-ADAM10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RNF165-ADAM10 | Fusion gene ID: 74858 | Hgene | Tgene | Gene symbol | RNF165 | ADAM10 | Gene ID | 494470 | 102 |
| Gene name | ring finger protein 165 | ADAM metallopeptidase domain 10 | |
| Synonyms | ARKL2|Ark2C|RNF111L2 | AD10|AD18|CD156c|CDw156|HsT18717|MADM|RAK|kuz | |
| Cytomap | 18q21.1 | 15q21.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | E3 ubiquitin-protein ligase RNF165 | disintegrin and metalloproteinase domain-containing protein 10a disintegrin and metalloprotease domain 10kuzbanian protein homologmammalian disintegrin-metalloprotease | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | O14672 | |
| Ensembl transtripts involved in fusion gene | ENST00000269439, ENST00000587853, ENST00000588679, ENST00000590330, ENST00000543885, | ENST00000558733, ENST00000396140, ENST00000402627, ENST00000561288, ENST00000260408, | |
| Fusion gene scores | * DoF score | 5 X 3 X 5=75 | 12 X 10 X 8=960 |
| # samples | 5 | 13 | |
| ** MAII score | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/960*10)=-2.88452278258006 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: RNF165 [Title/Abstract] AND ADAM10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RNF165(43914298)-ADAM10(58957396), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | RNF165-ADAM10 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. RNF165-ADAM10 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | ADAM10 | GO:0006509 | membrane protein ectodomain proteolysis | 12714508|17557115|18355449|18419754|18676862|19114711 |
| Tgene | ADAM10 | GO:0007162 | negative regulation of cell adhesion | 12714508 |
| Tgene | ADAM10 | GO:0034612 | response to tumor necrosis factor | 11831872 |
| Tgene | ADAM10 | GO:0051089 | constitutive protein ectodomain proteolysis | 12714508 |
 Fusion gene breakpoints across RNF165 (5'-gene) Fusion gene breakpoints across RNF165 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
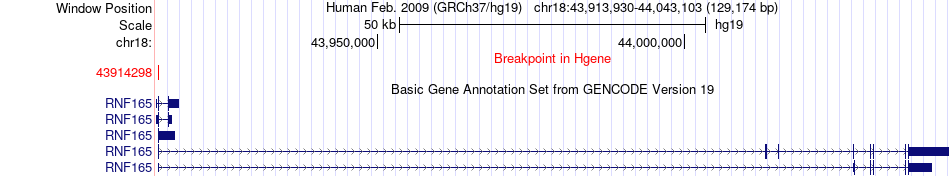 |
 Fusion gene breakpoints across ADAM10 (3'-gene) Fusion gene breakpoints across ADAM10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
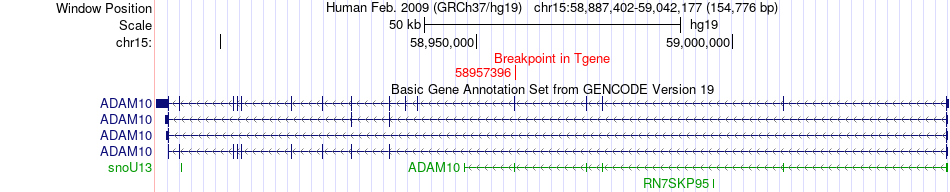 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-19-4065-02A | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
Top |
Fusion Gene ORF analysis for RNF165-ADAM10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000269439 | ENST00000558733 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-5UTR | ENST00000587853 | ENST00000558733 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-5UTR | ENST00000588679 | ENST00000558733 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-5UTR | ENST00000590330 | ENST00000558733 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000269439 | ENST00000396140 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000269439 | ENST00000402627 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000269439 | ENST00000561288 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000587853 | ENST00000396140 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000587853 | ENST00000402627 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000587853 | ENST00000561288 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000588679 | ENST00000396140 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000588679 | ENST00000402627 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000588679 | ENST00000561288 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000590330 | ENST00000396140 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000590330 | ENST00000402627 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5CDS-intron | ENST00000590330 | ENST00000561288 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5UTR-3CDS | ENST00000543885 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5UTR-5UTR | ENST00000543885 | ENST00000558733 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5UTR-intron | ENST00000543885 | ENST00000396140 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5UTR-intron | ENST00000543885 | ENST00000402627 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| 5UTR-intron | ENST00000543885 | ENST00000561288 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| Frame-shift | ENST00000588679 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| In-frame | ENST00000269439 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| In-frame | ENST00000587853 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
| In-frame | ENST00000590330 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000269439 | RNF165 | chr18 | 43914298 | + | ENST00000260408 | ADAM10 | chr15 | 58957396 | - | 4218 | 112 | 51 | 1874 | 607 |
| ENST00000587853 | RNF165 | chr18 | 43914298 | + | ENST00000260408 | ADAM10 | chr15 | 58957396 | - | 4357 | 251 | 1 | 2013 | 670 |
| ENST00000590330 | RNF165 | chr18 | 43914298 | + | ENST00000260408 | ADAM10 | chr15 | 58957396 | - | 4414 | 308 | 7 | 2070 | 687 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000269439 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - | 0.000102676 | 0.99989736 |
| ENST00000587853 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - | 0.000103464 | 0.9998965 |
| ENST00000590330 | ENST00000260408 | RNF165 | chr18 | 43914298 | + | ADAM10 | chr15 | 58957396 | - | 0.000107897 | 0.9998921 |
Top |
Fusion Genomic Features for RNF165-ADAM10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
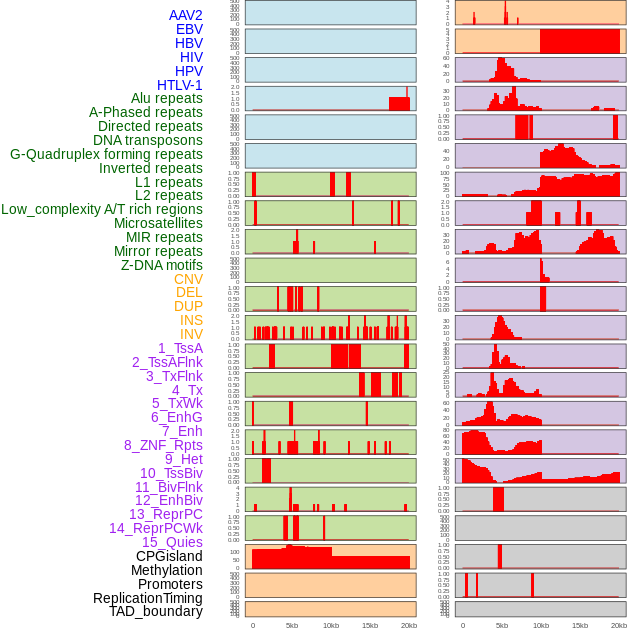 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for RNF165-ADAM10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:43914298/chr15:58957396) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ADAM10 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Cleaves the membrane-bound precursor of TNF-alpha at '76-Ala-|-Val-77' to its mature soluble form. Responsible for the proteolytical release of soluble JAM3 from endothelial cells surface (PubMed:20592283). Responsible for the proteolytic release of several other cell-surface proteins, including heparin-binding epidermal growth-like factor, ephrin-A2, CD44, CDH2 and for constitutive and regulated alpha-secretase cleavage of amyloid precursor protein (APP) (PubMed:26686862, PubMed:11786905, PubMed:29224781). Contributes to the normal cleavage of the cellular prion protein (PubMed:11477090). Involved in the cleavage of the adhesion molecule L1 at the cell surface and in released membrane vesicles, suggesting a vesicle-based protease activity (PubMed:12475894). Controls also the proteolytic processing of Notch and mediates lateral inhibition during neurogenesis (By similarity). Responsible for the FasL ectodomain shedding and for the generation of the remnant ADAM10-processed FasL (FasL APL) transmembrane form (PubMed:17557115). Also cleaves the ectodomain of the integral membrane proteins CORIN and ITM2B (PubMed:19114711, PubMed:21288900). Mediates the proteolytic cleavage of LAG3, leading to release the secreted form of LAG3 (By similarity). Mediates the proteolytic cleavage of IL6R and IL11RA, leading to the release of secreted forms of IL6R and IL11RA (PubMed:26876177). Enhances the cleavage of CHL1 by BACE1 (By similarity). Cleaves NRCAM (By similarity). Cleaves TREM2, resulting in shedding of the TREM2 ectodomain (PubMed:24990881). Involved in the development and maturation of glomerular and coronary vasculature (By similarity). During development of the cochlear organ of Corti, promotes pillar cell separation by forming a ternary complex with CADH1 and EPHA4 and cleaving CADH1 at adherens junctions (By similarity). May regulate the EFNA5-EPHA3 signaling (PubMed:16239146). {ECO:0000250|UniProtKB:O35598, ECO:0000269|PubMed:11477090, ECO:0000269|PubMed:11786905, ECO:0000269|PubMed:12475894, ECO:0000269|PubMed:16239146, ECO:0000269|PubMed:17557115, ECO:0000269|PubMed:19114711, ECO:0000269|PubMed:20592283, ECO:0000269|PubMed:21288900, ECO:0000269|PubMed:24990881, ECO:0000269|PubMed:26686862, ECO:0000269|PubMed:26876177, ECO:0000269|PubMed:29224781}.; FUNCTION: (Microbial infection) Promotes the cytotoxic activity of S.aureus hly by binding to the toxin at zonula adherens and promoting formation of toxin pores. {ECO:0000269|PubMed:20624979, ECO:0000269|PubMed:30463011}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 555_673 | 161 | 749.0 | Compositional bias | Cys-rich | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 220_456 | 161 | 749.0 | Domain | Peptidase M12B | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 457_551 | 161 | 749.0 | Domain | Disintegrin | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 171_178 | 161 | 749.0 | Motif | Cysteine switch | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 708_715 | 161 | 749.0 | Motif | SH3-binding | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 722_728 | 161 | 749.0 | Motif | SH3-binding | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 694_748 | 161 | 749.0 | Topological domain | Cytoplasmic | |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 673_693 | 161 | 749.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RNF165 | chr18:43914298 | chr15:58957396 | ENST00000269439 | + | 1 | 8 | 266_268 | 20 | 347.0 | Region | Ubiquitin binding |
| Hgene | RNF165 | chr18:43914298 | chr15:58957396 | ENST00000269439 | + | 1 | 8 | 309_313 | 20 | 347.0 | Region | Ubiquitin binding |
| Hgene | RNF165 | chr18:43914298 | chr15:58957396 | ENST00000543885 | + | 1 | 6 | 266_268 | 0 | 155.0 | Region | Ubiquitin binding |
| Hgene | RNF165 | chr18:43914298 | chr15:58957396 | ENST00000543885 | + | 1 | 6 | 309_313 | 0 | 155.0 | Region | Ubiquitin binding |
| Hgene | RNF165 | chr18:43914298 | chr15:58957396 | ENST00000269439 | + | 1 | 8 | 294_335 | 20 | 347.0 | Zinc finger | RING-type%3B atypical |
| Hgene | RNF165 | chr18:43914298 | chr15:58957396 | ENST00000543885 | + | 1 | 6 | 294_335 | 0 | 155.0 | Zinc finger | RING-type%3B atypical |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 20_672 | 161 | 749.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for RNF165-ADAM10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >74858_74858_1_RNF165-ADAM10_RNF165_chr18_43914298_ENST00000269439_ADAM10_chr15_58957396_ENST00000260408_length(transcript)=4218nt_BP=112nt ACAAAGGGCCCGCGCGCAGCCGCCGCCGCCGCCGCCGCGCGAGGAGCCAGGATGGTCCTGGTCCACGTCGGCTATCTCGTGCTTCCAGTG TTTGGCTCTGTGCGAAACAGAGACTATCCCCATAAATACGGTCCTCAGGGGGGCTGTGCAGATCATTCAGTATTTGAAAGAATGAGGAAA TACCAGATGACTGGTGTAGAGGAAGTAACACAGATACCTCAAGAAGAACATGCTGCTAATGGTCCAGAACTTCTGAGGAAAAAACGTACA ACTTCAGCTGAAAAAAATACTTGTCAGCTTTATATTCAGACTGATCATTTGTTCTTTAAATATTACGGAACACGAGAAGCTGTGATTGCC CAGATATCCAGTCATGTTAAAGCGATTGATACAATTTACCAGACCACAGACTTCTCCGGAATCCGTAACATCAGTTTCATGGTGAAACGC ATAAGAATCAATACAACTGCTGATGAGAAGGACCCTACAAATCCTTTCCGTTTCCCAAATATTGGTGTGGAGAAGTTTCTGGAATTGAAT TCTGAGCAGAATCATGATGACTACTGTTTGGCCTATGTCTTCACAGACCGAGATTTTGATGATGGCGTACTTGGTCTGGCTTGGGTTGGA GCACCTTCAGGAAGCTCTGGAGGAATATGTGAAAAAAGTAAACTCTATTCAGATGGTAAGAAGAAGTCCTTAAACACTGGAATTATTACT GTTCAGAACTATGGGTCTCATGTACCTCCCAAAGTCTCTCACATTACTTTTGCTCACGAAGTTGGACATAACTTTGGATCCCCACATGAT TCTGGAACAGAGTGCACACCAGGAGAATCTAAGAATTTGGGTCAAAAAGAAAATGGCAATTACATCATGTATGCAAGAGCAACATCTGGG GACAAACTTAACAACAATAAATTCTCACTCTGTAGTATTAGAAATATAAGCCAAGTTCTTGAGAAGAAGAGAAACAACTGTTTTGTTGAA TCTGGCCAACCTATTTGTGGAAATGGAATGGTAGAACAAGGTGAAGAATGTGATTGTGGCTATAGTGACCAGTGTAAAGATGAATGCTGC TTCGATGCAAATCAACCAGAGGGAAGAAAATGCAAACTGAAACCTGGGAAACAGTGCAGTCCAAGTCAAGGTCCTTGTTGTACAGCACAG TGTGCATTCAAGTCAAAGTCTGAGAAGTGTCGGGATGATTCAGACTGTGCAAGGGAAGGAATATGTAATGGCTTCACAGCTCTCTGCCCA GCATCTGACCCTAAACCAAACTTCACAGACTGTAATAGGCATACACAAGTGTGCATTAATGGGCAATGTGCAGGTTCTATCTGTGAGAAA TATGGCTTAGAGGAGTGTACGTGTGCCAGTTCTGATGGCAAAGATGATAAAGAATTATGCCATGTATGCTGTATGAAGAAAATGGACCCA TCAACTTGTGCCAGTACAGGGTCTGTGCAGTGGAGTAGGCACTTCAGTGGTCGAACCATCACCCTGCAACCTGGATCCCCTTGCAACGAT TTTAGAGGTTACTGTGATGTTTTCATGCGGTGCAGATTAGTAGATGCTGATGGTCCTCTAGCTAGGCTTAAAAAAGCAATTTTTAGTCCA GAGCTCTATGAAAACATTGCTGAATGGATTGTGGCTCATTGGTGGGCAGTATTACTTATGGGAATTGCTCTGATCATGCTAATGGCTGGA TTTATTAAGATATGCAGTGTTCATACTCCAAGTAGTAATCCAAAGTTGCCTCCTCCTAAACCACTTCCAGGCACTTTAAAGAGGAGGAGA CCTCCACAGCCCATTCAGCAACCCCAGCGTCAGCGGCCCCGAGAGAGTTATCAAATGGGACACATGAGACGCTAACTGCAGCTTTTGCCT TGGTTCTTCCTAGTGCCTACAATGGGAAAACTTCACTCCAAAGAGAAACCTATTAAGTCATCATCTCCAAACTAAACCCTCACAAGTAAC AGTTGAAGAAAAAATGGCAAGAGATCATATCCTCAGACCAGGTGGAATTACTTAAATTTTAAAGCCTGAAAATTCCAATTTGGGGGTGGG AGGTGGAAAAGGAACCCAATTTTCTTATGAACAGATATTTTTAACTTAATGGCACAAAGTCTTAGAATATTATTATGTGCCCCGTGTTCC CTGTTCTTCGTTGCTGCATTTTCTTCACTTGCAGGCAAACTTGGCTCTCAATAAACTTTTACCACAAATTGAAATAAATATATTTTTTTC AACTGCCAATCAAGGCTAGGAGGCTCGACCACCTCAACATTGGAGACATCACTTGCCAATGTACATACCTTGTTATATGCAGACATGTAT TTCTTACGTACACTGTACTTCTGTGTGCAATTGTAAACAGAAATTGCAATATGGATGTTTCTTTGTATTATAAAATTTTTCCGCTCTTAA TTAAAAATTACTGTTTAATTGACATACTCAGGATAACAGAGAATGGTGGTATTCAGTGGTCCAGGATTCTGTAATGCTTTACACAGGCAG TTTTGAAATGAAAATCAATTTACCTTTCTGTTACGATGGAGTTGGTTTTGATACTCATTTTTTCTTTATCACATGGCTGCTACGGGCACA AGTGACTATACTGAAGAACACAGTTAAGTGTTGTGCAAACTGGACATAGCAGCACATACTACTTCAGAGTTCATGATGTAGATGTCTGGT TTCTGCTTACGTCTTTTAAACTTTCTAATTCAATTCCATTTTTCAATTAATAGGTGAAATTTTATTCATGCTTTGATAGAAATTATGTCA ATGAAATGATTCTTTTTATTTGTAGCCTACTTATTTGTGTTTTTCATATATCTGAAATATGCTAATTATGTTTTCTGTCTGATATGGAAA AGAAAAGCTGTGTCTTTATCAAAATATTTAAACGGTTTTTTCAGCATATCATCACTGATCATTGGTAACCACTAAAGATGAGTAATTTGC TTAAGTAGTAGTTAAAATTGTAGATAGGCCTTCTGACATTTTTTTTCCTAAAATTTTTAACAGCATTGAAGGTGAAACAGCACAATGTCC CATTCCAAATTTATTTTTGAAACAGATGTAAATAATTGGCATTTTAAAGAGAAAGCAAAAACATTTAATGTATTAACAGGCTTATTGCTA TGCAGGAAATAGAAGGGGCATTACAAAAATTGAAGCTTGTGACATATTTATTGCTTCTGTTTTCCAACTACATCACTTCAACTAGAAGTA AAGCTATGATTTTCCTGACTTCACATAGGAGGCAAATTTAGAGAAAGTTGTAAAGATTTCTATGTTTTGGGTTTTTTTTTTTCCTTTTTT TTTTTAAGAGTATAAGGTTTACACAATCATTCTCATAATGTGACGCAAGCCAGCAAGGCCAAAAATGCTAGAGAAAATAACGGGATCTCT TCCTTGTAAACTTGTACAGTATGTGGTGACTTTTTCAAAATACAGCTTTTTGTACATGATTTAGAGACAAATTTTGTACATGAAACCCCA GATAGACTATAAATAATTCTAAACAAACAAGTAGGTAGATATGTATGTAATTGCTTTTAAATCATTTAAATGCCTTTGTTTTTGGACTGT GCAAAGGTTGGAAGTGGGTTTGCATTTCTAAAATGGTGACTTTTATTCTGCAAGAGTTCTTAGTAACTTCTTGAGTGTGGTAGACTTTGG AACATGTAAATTTTTTGCTTGTAATGTTATCCTGTGGTAGGATTTTGGCAGGTACACACACTGCCCTATTTTATTTTGAGTCTAAGTTAA ATGTTTTCTGAAAAGAGATACATGCACTGAACTCTTTCCACTGCGAATCAAGATGTGGTAATATAAAAGGATCAAGACAAATGAGATCTA ATACTACTGTCAGTTTTAATGTCCACTGTGTTTTATACAGTATCTTTTTTTGTTCACTTTGGAAATTTTTACTAAAAATTGCAAAAAATA AAGTATTGTGCAAAGATGTAAGGTTTTTTGAAACTTGAAATGCATTAATAAATAGACGATTAAATCAACTTGAAGGTTCTATACTCTTTG AACTCTGAGAACTATCACAAGAAGCTTCCCACAAGGCAGTGTTTTCTTACAGTTGTCTCTTCCTACAAAAGTATAGATTATCTTTATTCT >74858_74858_1_RNF165-ADAM10_RNF165_chr18_43914298_ENST00000269439_ADAM10_chr15_58957396_ENST00000260408_length(amino acids)=607AA_BP=20 MVLVHVGYLVLPVFGSVRNRDYPHKYGPQGGCADHSVFERMRKYQMTGVEEVTQIPQEEHAANGPELLRKKRTTSAEKNTCQLYIQTDHL FFKYYGTREAVIAQISSHVKAIDTIYQTTDFSGIRNISFMVKRIRINTTADEKDPTNPFRFPNIGVEKFLELNSEQNHDDYCLAYVFTDR DFDDGVLGLAWVGAPSGSSGGICEKSKLYSDGKKKSLNTGIITVQNYGSHVPPKVSHITFAHEVGHNFGSPHDSGTECTPGESKNLGQKE NGNYIMYARATSGDKLNNNKFSLCSIRNISQVLEKKRNNCFVESGQPICGNGMVEQGEECDCGYSDQCKDECCFDANQPEGRKCKLKPGK QCSPSQGPCCTAQCAFKSKSEKCRDDSDCAREGICNGFTALCPASDPKPNFTDCNRHTQVCINGQCAGSICEKYGLEECTCASSDGKDDK ELCHVCCMKKMDPSTCASTGSVQWSRHFSGRTITLQPGSPCNDFRGYCDVFMRCRLVDADGPLARLKKAIFSPELYENIAEWIVAHWWAV -------------------------------------------------------------- >74858_74858_2_RNF165-ADAM10_RNF165_chr18_43914298_ENST00000587853_ADAM10_chr15_58957396_ENST00000260408_length(transcript)=4357nt_BP=251nt GCTGTCCCCCGCGGTTCCGGCGAGGCCGCGCCGCGCTGCGCTCCGCTCCCATTGGCTGCGCGCGCCTTTGTGTGGCGGCAGCTGCGGCCC CAGCGCCTTTGAATGTCGAGAGGGGCCGGGAAGGGAGCCTTTCCATGGGCCATCTGGCCCGCGCGCAGCCGCCGCCGCCGCCGCCGCGCG AGGAGCCAGGATGGTCCTGGTCCACGTCGGCTATCTCGTGCTTCCAGTGTTTGGCTCTGTGCGAAACAGAGACTATCCCCATAAATACGG TCCTCAGGGGGGCTGTGCAGATCATTCAGTATTTGAAAGAATGAGGAAATACCAGATGACTGGTGTAGAGGAAGTAACACAGATACCTCA AGAAGAACATGCTGCTAATGGTCCAGAACTTCTGAGGAAAAAACGTACAACTTCAGCTGAAAAAAATACTTGTCAGCTTTATATTCAGAC TGATCATTTGTTCTTTAAATATTACGGAACACGAGAAGCTGTGATTGCCCAGATATCCAGTCATGTTAAAGCGATTGATACAATTTACCA GACCACAGACTTCTCCGGAATCCGTAACATCAGTTTCATGGTGAAACGCATAAGAATCAATACAACTGCTGATGAGAAGGACCCTACAAA TCCTTTCCGTTTCCCAAATATTGGTGTGGAGAAGTTTCTGGAATTGAATTCTGAGCAGAATCATGATGACTACTGTTTGGCCTATGTCTT CACAGACCGAGATTTTGATGATGGCGTACTTGGTCTGGCTTGGGTTGGAGCACCTTCAGGAAGCTCTGGAGGAATATGTGAAAAAAGTAA ACTCTATTCAGATGGTAAGAAGAAGTCCTTAAACACTGGAATTATTACTGTTCAGAACTATGGGTCTCATGTACCTCCCAAAGTCTCTCA CATTACTTTTGCTCACGAAGTTGGACATAACTTTGGATCCCCACATGATTCTGGAACAGAGTGCACACCAGGAGAATCTAAGAATTTGGG TCAAAAAGAAAATGGCAATTACATCATGTATGCAAGAGCAACATCTGGGGACAAACTTAACAACAATAAATTCTCACTCTGTAGTATTAG AAATATAAGCCAAGTTCTTGAGAAGAAGAGAAACAACTGTTTTGTTGAATCTGGCCAACCTATTTGTGGAAATGGAATGGTAGAACAAGG TGAAGAATGTGATTGTGGCTATAGTGACCAGTGTAAAGATGAATGCTGCTTCGATGCAAATCAACCAGAGGGAAGAAAATGCAAACTGAA ACCTGGGAAACAGTGCAGTCCAAGTCAAGGTCCTTGTTGTACAGCACAGTGTGCATTCAAGTCAAAGTCTGAGAAGTGTCGGGATGATTC AGACTGTGCAAGGGAAGGAATATGTAATGGCTTCACAGCTCTCTGCCCAGCATCTGACCCTAAACCAAACTTCACAGACTGTAATAGGCA TACACAAGTGTGCATTAATGGGCAATGTGCAGGTTCTATCTGTGAGAAATATGGCTTAGAGGAGTGTACGTGTGCCAGTTCTGATGGCAA AGATGATAAAGAATTATGCCATGTATGCTGTATGAAGAAAATGGACCCATCAACTTGTGCCAGTACAGGGTCTGTGCAGTGGAGTAGGCA CTTCAGTGGTCGAACCATCACCCTGCAACCTGGATCCCCTTGCAACGATTTTAGAGGTTACTGTGATGTTTTCATGCGGTGCAGATTAGT AGATGCTGATGGTCCTCTAGCTAGGCTTAAAAAAGCAATTTTTAGTCCAGAGCTCTATGAAAACATTGCTGAATGGATTGTGGCTCATTG GTGGGCAGTATTACTTATGGGAATTGCTCTGATCATGCTAATGGCTGGATTTATTAAGATATGCAGTGTTCATACTCCAAGTAGTAATCC AAAGTTGCCTCCTCCTAAACCACTTCCAGGCACTTTAAAGAGGAGGAGACCTCCACAGCCCATTCAGCAACCCCAGCGTCAGCGGCCCCG AGAGAGTTATCAAATGGGACACATGAGACGCTAACTGCAGCTTTTGCCTTGGTTCTTCCTAGTGCCTACAATGGGAAAACTTCACTCCAA AGAGAAACCTATTAAGTCATCATCTCCAAACTAAACCCTCACAAGTAACAGTTGAAGAAAAAATGGCAAGAGATCATATCCTCAGACCAG GTGGAATTACTTAAATTTTAAAGCCTGAAAATTCCAATTTGGGGGTGGGAGGTGGAAAAGGAACCCAATTTTCTTATGAACAGATATTTT TAACTTAATGGCACAAAGTCTTAGAATATTATTATGTGCCCCGTGTTCCCTGTTCTTCGTTGCTGCATTTTCTTCACTTGCAGGCAAACT TGGCTCTCAATAAACTTTTACCACAAATTGAAATAAATATATTTTTTTCAACTGCCAATCAAGGCTAGGAGGCTCGACCACCTCAACATT GGAGACATCACTTGCCAATGTACATACCTTGTTATATGCAGACATGTATTTCTTACGTACACTGTACTTCTGTGTGCAATTGTAAACAGA AATTGCAATATGGATGTTTCTTTGTATTATAAAATTTTTCCGCTCTTAATTAAAAATTACTGTTTAATTGACATACTCAGGATAACAGAG AATGGTGGTATTCAGTGGTCCAGGATTCTGTAATGCTTTACACAGGCAGTTTTGAAATGAAAATCAATTTACCTTTCTGTTACGATGGAG TTGGTTTTGATACTCATTTTTTCTTTATCACATGGCTGCTACGGGCACAAGTGACTATACTGAAGAACACAGTTAAGTGTTGTGCAAACT GGACATAGCAGCACATACTACTTCAGAGTTCATGATGTAGATGTCTGGTTTCTGCTTACGTCTTTTAAACTTTCTAATTCAATTCCATTT TTCAATTAATAGGTGAAATTTTATTCATGCTTTGATAGAAATTATGTCAATGAAATGATTCTTTTTATTTGTAGCCTACTTATTTGTGTT TTTCATATATCTGAAATATGCTAATTATGTTTTCTGTCTGATATGGAAAAGAAAAGCTGTGTCTTTATCAAAATATTTAAACGGTTTTTT CAGCATATCATCACTGATCATTGGTAACCACTAAAGATGAGTAATTTGCTTAAGTAGTAGTTAAAATTGTAGATAGGCCTTCTGACATTT TTTTTCCTAAAATTTTTAACAGCATTGAAGGTGAAACAGCACAATGTCCCATTCCAAATTTATTTTTGAAACAGATGTAAATAATTGGCA TTTTAAAGAGAAAGCAAAAACATTTAATGTATTAACAGGCTTATTGCTATGCAGGAAATAGAAGGGGCATTACAAAAATTGAAGCTTGTG ACATATTTATTGCTTCTGTTTTCCAACTACATCACTTCAACTAGAAGTAAAGCTATGATTTTCCTGACTTCACATAGGAGGCAAATTTAG AGAAAGTTGTAAAGATTTCTATGTTTTGGGTTTTTTTTTTTCCTTTTTTTTTTTAAGAGTATAAGGTTTACACAATCATTCTCATAATGT GACGCAAGCCAGCAAGGCCAAAAATGCTAGAGAAAATAACGGGATCTCTTCCTTGTAAACTTGTACAGTATGTGGTGACTTTTTCAAAAT ACAGCTTTTTGTACATGATTTAGAGACAAATTTTGTACATGAAACCCCAGATAGACTATAAATAATTCTAAACAAACAAGTAGGTAGATA TGTATGTAATTGCTTTTAAATCATTTAAATGCCTTTGTTTTTGGACTGTGCAAAGGTTGGAAGTGGGTTTGCATTTCTAAAATGGTGACT TTTATTCTGCAAGAGTTCTTAGTAACTTCTTGAGTGTGGTAGACTTTGGAACATGTAAATTTTTTGCTTGTAATGTTATCCTGTGGTAGG ATTTTGGCAGGTACACACACTGCCCTATTTTATTTTGAGTCTAAGTTAAATGTTTTCTGAAAAGAGATACATGCACTGAACTCTTTCCAC TGCGAATCAAGATGTGGTAATATAAAAGGATCAAGACAAATGAGATCTAATACTACTGTCAGTTTTAATGTCCACTGTGTTTTATACAGT ATCTTTTTTTGTTCACTTTGGAAATTTTTACTAAAAATTGCAAAAAATAAAGTATTGTGCAAAGATGTAAGGTTTTTTGAAACTTGAAAT GCATTAATAAATAGACGATTAAATCAACTTGAAGGTTCTATACTCTTTGAACTCTGAGAACTATCACAAGAAGCTTCCCACAAGGCAGTG TTTTCTTACAGTTGTCTCTTCCTACAAAAGTATAGATTATCTTTATTCTTAATACTTTGGAATCCATGTAGAAAATTTCCAGTTAGATAC >74858_74858_2_RNF165-ADAM10_RNF165_chr18_43914298_ENST00000587853_ADAM10_chr15_58957396_ENST00000260408_length(amino acids)=670AA_BP=83 LSPAVPARPRRAALRSHWLRAPLCGGSCGPSAFECREGPGREPFHGPSGPRAAAAAAAARGARMVLVHVGYLVLPVFGSVRNRDYPHKYG PQGGCADHSVFERMRKYQMTGVEEVTQIPQEEHAANGPELLRKKRTTSAEKNTCQLYIQTDHLFFKYYGTREAVIAQISSHVKAIDTIYQ TTDFSGIRNISFMVKRIRINTTADEKDPTNPFRFPNIGVEKFLELNSEQNHDDYCLAYVFTDRDFDDGVLGLAWVGAPSGSSGGICEKSK LYSDGKKKSLNTGIITVQNYGSHVPPKVSHITFAHEVGHNFGSPHDSGTECTPGESKNLGQKENGNYIMYARATSGDKLNNNKFSLCSIR NISQVLEKKRNNCFVESGQPICGNGMVEQGEECDCGYSDQCKDECCFDANQPEGRKCKLKPGKQCSPSQGPCCTAQCAFKSKSEKCRDDS DCAREGICNGFTALCPASDPKPNFTDCNRHTQVCINGQCAGSICEKYGLEECTCASSDGKDDKELCHVCCMKKMDPSTCASTGSVQWSRH FSGRTITLQPGSPCNDFRGYCDVFMRCRLVDADGPLARLKKAIFSPELYENIAEWIVAHWWAVLLMGIALIMLMAGFIKICSVHTPSSNP -------------------------------------------------------------- >74858_74858_3_RNF165-ADAM10_RNF165_chr18_43914298_ENST00000590330_ADAM10_chr15_58957396_ENST00000260408_length(transcript)=4414nt_BP=308nt CGCGCCTTTGTGTGGCGGCAGCTGCGGCCCCAGCGCCTTTGAATGTCGAGAGGGGCCGGGAAGGGAGCCTTTCCATGGGCCATCTGTCTC CCCGCCAGCCGCCGCCCCACCGCCGCCCGCGCCCCGCGCTCCGCCTCCCGCCCCGGCGGCTCCCGCAGCCCCGCCGCCGCCCGCGCCCGC GCCCCGGAGCCGCGCCACAAAGGGCCCGCGCGCAGCCGCCGCCGCCGCCGCCGCGCGAGGAGCCAGGATGGTCCTGGTCCACGTCGGCTA TCTCGTGCTTCCAGTGTTTGGCTCTGTGCGAAACAGAGACTATCCCCATAAATACGGTCCTCAGGGGGGCTGTGCAGATCATTCAGTATT TGAAAGAATGAGGAAATACCAGATGACTGGTGTAGAGGAAGTAACACAGATACCTCAAGAAGAACATGCTGCTAATGGTCCAGAACTTCT GAGGAAAAAACGTACAACTTCAGCTGAAAAAAATACTTGTCAGCTTTATATTCAGACTGATCATTTGTTCTTTAAATATTACGGAACACG AGAAGCTGTGATTGCCCAGATATCCAGTCATGTTAAAGCGATTGATACAATTTACCAGACCACAGACTTCTCCGGAATCCGTAACATCAG TTTCATGGTGAAACGCATAAGAATCAATACAACTGCTGATGAGAAGGACCCTACAAATCCTTTCCGTTTCCCAAATATTGGTGTGGAGAA GTTTCTGGAATTGAATTCTGAGCAGAATCATGATGACTACTGTTTGGCCTATGTCTTCACAGACCGAGATTTTGATGATGGCGTACTTGG TCTGGCTTGGGTTGGAGCACCTTCAGGAAGCTCTGGAGGAATATGTGAAAAAAGTAAACTCTATTCAGATGGTAAGAAGAAGTCCTTAAA CACTGGAATTATTACTGTTCAGAACTATGGGTCTCATGTACCTCCCAAAGTCTCTCACATTACTTTTGCTCACGAAGTTGGACATAACTT TGGATCCCCACATGATTCTGGAACAGAGTGCACACCAGGAGAATCTAAGAATTTGGGTCAAAAAGAAAATGGCAATTACATCATGTATGC AAGAGCAACATCTGGGGACAAACTTAACAACAATAAATTCTCACTCTGTAGTATTAGAAATATAAGCCAAGTTCTTGAGAAGAAGAGAAA CAACTGTTTTGTTGAATCTGGCCAACCTATTTGTGGAAATGGAATGGTAGAACAAGGTGAAGAATGTGATTGTGGCTATAGTGACCAGTG TAAAGATGAATGCTGCTTCGATGCAAATCAACCAGAGGGAAGAAAATGCAAACTGAAACCTGGGAAACAGTGCAGTCCAAGTCAAGGTCC TTGTTGTACAGCACAGTGTGCATTCAAGTCAAAGTCTGAGAAGTGTCGGGATGATTCAGACTGTGCAAGGGAAGGAATATGTAATGGCTT CACAGCTCTCTGCCCAGCATCTGACCCTAAACCAAACTTCACAGACTGTAATAGGCATACACAAGTGTGCATTAATGGGCAATGTGCAGG TTCTATCTGTGAGAAATATGGCTTAGAGGAGTGTACGTGTGCCAGTTCTGATGGCAAAGATGATAAAGAATTATGCCATGTATGCTGTAT GAAGAAAATGGACCCATCAACTTGTGCCAGTACAGGGTCTGTGCAGTGGAGTAGGCACTTCAGTGGTCGAACCATCACCCTGCAACCTGG ATCCCCTTGCAACGATTTTAGAGGTTACTGTGATGTTTTCATGCGGTGCAGATTAGTAGATGCTGATGGTCCTCTAGCTAGGCTTAAAAA AGCAATTTTTAGTCCAGAGCTCTATGAAAACATTGCTGAATGGATTGTGGCTCATTGGTGGGCAGTATTACTTATGGGAATTGCTCTGAT CATGCTAATGGCTGGATTTATTAAGATATGCAGTGTTCATACTCCAAGTAGTAATCCAAAGTTGCCTCCTCCTAAACCACTTCCAGGCAC TTTAAAGAGGAGGAGACCTCCACAGCCCATTCAGCAACCCCAGCGTCAGCGGCCCCGAGAGAGTTATCAAATGGGACACATGAGACGCTA ACTGCAGCTTTTGCCTTGGTTCTTCCTAGTGCCTACAATGGGAAAACTTCACTCCAAAGAGAAACCTATTAAGTCATCATCTCCAAACTA AACCCTCACAAGTAACAGTTGAAGAAAAAATGGCAAGAGATCATATCCTCAGACCAGGTGGAATTACTTAAATTTTAAAGCCTGAAAATT CCAATTTGGGGGTGGGAGGTGGAAAAGGAACCCAATTTTCTTATGAACAGATATTTTTAACTTAATGGCACAAAGTCTTAGAATATTATT ATGTGCCCCGTGTTCCCTGTTCTTCGTTGCTGCATTTTCTTCACTTGCAGGCAAACTTGGCTCTCAATAAACTTTTACCACAAATTGAAA TAAATATATTTTTTTCAACTGCCAATCAAGGCTAGGAGGCTCGACCACCTCAACATTGGAGACATCACTTGCCAATGTACATACCTTGTT ATATGCAGACATGTATTTCTTACGTACACTGTACTTCTGTGTGCAATTGTAAACAGAAATTGCAATATGGATGTTTCTTTGTATTATAAA ATTTTTCCGCTCTTAATTAAAAATTACTGTTTAATTGACATACTCAGGATAACAGAGAATGGTGGTATTCAGTGGTCCAGGATTCTGTAA TGCTTTACACAGGCAGTTTTGAAATGAAAATCAATTTACCTTTCTGTTACGATGGAGTTGGTTTTGATACTCATTTTTTCTTTATCACAT GGCTGCTACGGGCACAAGTGACTATACTGAAGAACACAGTTAAGTGTTGTGCAAACTGGACATAGCAGCACATACTACTTCAGAGTTCAT GATGTAGATGTCTGGTTTCTGCTTACGTCTTTTAAACTTTCTAATTCAATTCCATTTTTCAATTAATAGGTGAAATTTTATTCATGCTTT GATAGAAATTATGTCAATGAAATGATTCTTTTTATTTGTAGCCTACTTATTTGTGTTTTTCATATATCTGAAATATGCTAATTATGTTTT CTGTCTGATATGGAAAAGAAAAGCTGTGTCTTTATCAAAATATTTAAACGGTTTTTTCAGCATATCATCACTGATCATTGGTAACCACTA AAGATGAGTAATTTGCTTAAGTAGTAGTTAAAATTGTAGATAGGCCTTCTGACATTTTTTTTCCTAAAATTTTTAACAGCATTGAAGGTG AAACAGCACAATGTCCCATTCCAAATTTATTTTTGAAACAGATGTAAATAATTGGCATTTTAAAGAGAAAGCAAAAACATTTAATGTATT AACAGGCTTATTGCTATGCAGGAAATAGAAGGGGCATTACAAAAATTGAAGCTTGTGACATATTTATTGCTTCTGTTTTCCAACTACATC ACTTCAACTAGAAGTAAAGCTATGATTTTCCTGACTTCACATAGGAGGCAAATTTAGAGAAAGTTGTAAAGATTTCTATGTTTTGGGTTT TTTTTTTTCCTTTTTTTTTTTAAGAGTATAAGGTTTACACAATCATTCTCATAATGTGACGCAAGCCAGCAAGGCCAAAAATGCTAGAGA AAATAACGGGATCTCTTCCTTGTAAACTTGTACAGTATGTGGTGACTTTTTCAAAATACAGCTTTTTGTACATGATTTAGAGACAAATTT TGTACATGAAACCCCAGATAGACTATAAATAATTCTAAACAAACAAGTAGGTAGATATGTATGTAATTGCTTTTAAATCATTTAAATGCC TTTGTTTTTGGACTGTGCAAAGGTTGGAAGTGGGTTTGCATTTCTAAAATGGTGACTTTTATTCTGCAAGAGTTCTTAGTAACTTCTTGA GTGTGGTAGACTTTGGAACATGTAAATTTTTTGCTTGTAATGTTATCCTGTGGTAGGATTTTGGCAGGTACACACACTGCCCTATTTTAT TTTGAGTCTAAGTTAAATGTTTTCTGAAAAGAGATACATGCACTGAACTCTTTCCACTGCGAATCAAGATGTGGTAATATAAAAGGATCA AGACAAATGAGATCTAATACTACTGTCAGTTTTAATGTCCACTGTGTTTTATACAGTATCTTTTTTTGTTCACTTTGGAAATTTTTACTA AAAATTGCAAAAAATAAAGTATTGTGCAAAGATGTAAGGTTTTTTGAAACTTGAAATGCATTAATAAATAGACGATTAAATCAACTTGAA GGTTCTATACTCTTTGAACTCTGAGAACTATCACAAGAAGCTTCCCACAAGGCAGTGTTTTCTTACAGTTGTCTCTTCCTACAAAAGTAT AGATTATCTTTATTCTTAATACTTTGGAATCCATGTAGAAAATTTCCAGTTAGATACTCTGCGTACACACAATAAACCTTTTTAAAACAC >74858_74858_3_RNF165-ADAM10_RNF165_chr18_43914298_ENST00000590330_ADAM10_chr15_58957396_ENST00000260408_length(amino acids)=687AA_BP=45 MCGGSCGPSAFECREGPGREPFHGPSVSPPAAAPPPPAPRAPPPAPAAPAAPPPPAPAPRSRATKGPRAAAAAAAARGARMVLVHVGYLV LPVFGSVRNRDYPHKYGPQGGCADHSVFERMRKYQMTGVEEVTQIPQEEHAANGPELLRKKRTTSAEKNTCQLYIQTDHLFFKYYGTREA VIAQISSHVKAIDTIYQTTDFSGIRNISFMVKRIRINTTADEKDPTNPFRFPNIGVEKFLELNSEQNHDDYCLAYVFTDRDFDDGVLGLA WVGAPSGSSGGICEKSKLYSDGKKKSLNTGIITVQNYGSHVPPKVSHITFAHEVGHNFGSPHDSGTECTPGESKNLGQKENGNYIMYARA TSGDKLNNNKFSLCSIRNISQVLEKKRNNCFVESGQPICGNGMVEQGEECDCGYSDQCKDECCFDANQPEGRKCKLKPGKQCSPSQGPCC TAQCAFKSKSEKCRDDSDCAREGICNGFTALCPASDPKPNFTDCNRHTQVCINGQCAGSICEKYGLEECTCASSDGKDDKELCHVCCMKK MDPSTCASTGSVQWSRHFSGRTITLQPGSPCNDFRGYCDVFMRCRLVDADGPLARLKKAIFSPELYENIAEWIVAHWWAVLLMGIALIML -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RNF165-ADAM10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | ADAM10 | chr18:43914298 | chr15:58957396 | ENST00000260408 | 3 | 16 | 734_748 | 161.33333333333334 | 749.0 | AP2A1%2C AP2A2 and AP2M1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RNF165-ADAM10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RNF165-ADAM10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
