
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ATG3-CLTC (FusionGDB2 ID:7558) |
Fusion Gene Summary for ATG3-CLTC |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ATG3-CLTC | Fusion gene ID: 7558 | Hgene | Tgene | Gene symbol | ATG3 | CLTC | Gene ID | 64422 | 1213 |
| Gene name | autophagy related 3 | clathrin heavy chain | |
| Synonyms | APG3|APG3-LIKE|APG3L|PC3-96 | CHC|CHC17|CLH-17|CLTCL2|Hc|MRD56 | |
| Cytomap | 3q13.2 | 17q23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ubiquitin-like-conjugating enzyme ATG32610016C12RikAPG3 autophagy 3-likeATG3 autophagy related 3 homologautophagy-related protein 3hApg3 | clathrin heavy chain 1clathrin heavy chain on chromosome 17clathrin, heavy polypeptide (Hc)clathrin, heavy polypeptide-like 2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9NT62 | P53675 | |
| Ensembl transtripts involved in fusion gene | ENST00000283290, ENST00000402314, ENST00000495756, | ENST00000579815, ENST00000269122, ENST00000393043, ENST00000579456, | |
| Fusion gene scores | * DoF score | 5 X 5 X 2=50 | 18 X 18 X 8=2592 |
| # samples | 5 | 22 | |
| ** MAII score | log2(5/50*10)=0 | log2(22/2592*10)=-3.55849028935997 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ATG3 [Title/Abstract] AND CLTC [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ATG3(112262904)-CLTC(57767997), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | ATG3-CLTC seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. ATG3-CLTC seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. ATG3-CLTC seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ATG3 | GO:0006464 | cellular protein modification process | 11825910 |
| Hgene | ATG3 | GO:0016567 | protein ubiquitination | 11825910 |
| Tgene | CLTC | GO:1900126 | negative regulation of hyaluronan biosynthetic process | 24251095 |
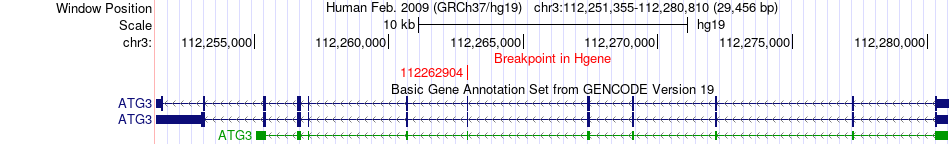
 Fusion gene breakpoints across ATG3 (5'-gene) Fusion gene breakpoints across ATG3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
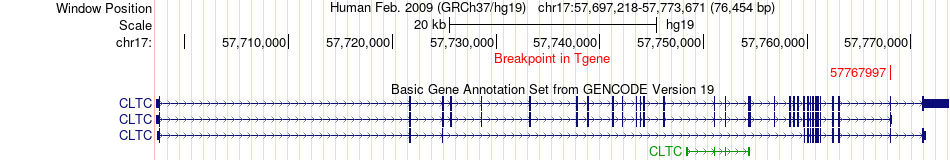
 Fusion gene breakpoints across CLTC (3'-gene) Fusion gene breakpoints across CLTC (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-BR-8080-01A | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
Top |
Fusion Gene ORF analysis for ATG3-CLTC |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000283290 | ENST00000579815 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| 5CDS-intron | ENST00000402314 | ENST00000579815 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| 5UTR-3CDS | ENST00000495756 | ENST00000269122 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| 5UTR-3CDS | ENST00000495756 | ENST00000393043 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| 5UTR-3CDS | ENST00000495756 | ENST00000579456 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| 5UTR-intron | ENST00000495756 | ENST00000579815 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| Frame-shift | ENST00000283290 | ENST00000393043 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| Frame-shift | ENST00000402314 | ENST00000393043 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| In-frame | ENST00000283290 | ENST00000269122 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| In-frame | ENST00000283290 | ENST00000579456 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| In-frame | ENST00000402314 | ENST00000269122 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
| In-frame | ENST00000402314 | ENST00000579456 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000283290 | ATG3 | chr3 | 112262904 | - | ENST00000269122 | CLTC | chr17 | 57767997 | + | 3487 | 828 | 363 | 1028 | 221 |
| ENST00000283290 | ATG3 | chr3 | 112262904 | - | ENST00000579456 | CLTC | chr17 | 57767997 | + | 1291 | 828 | 363 | 1028 | 221 |
| ENST00000402314 | ATG3 | chr3 | 112262904 | - | ENST00000269122 | CLTC | chr17 | 57767997 | + | 3440 | 781 | 316 | 981 | 221 |
| ENST00000402314 | ATG3 | chr3 | 112262904 | - | ENST00000579456 | CLTC | chr17 | 57767997 | + | 1244 | 781 | 316 | 981 | 221 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000283290 | ENST00000269122 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + | 0.00069011 | 0.9993099 |
| ENST00000283290 | ENST00000579456 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + | 0.002391371 | 0.99760866 |
| ENST00000402314 | ENST00000269122 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + | 0.000641282 | 0.9993587 |
| ENST00000402314 | ENST00000579456 | ATG3 | chr3 | 112262904 | - | CLTC | chr17 | 57767997 | + | 0.002027643 | 0.9979723 |
Top |
Fusion Genomic Features for ATG3-CLTC |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| ATG3 | chr3 | 112262903 | - | CLTC | chr17 | 57767996 | + | 5.95E-05 | 0.9999404 |
| ATG3 | chr3 | 112262903 | - | CLTC | chr17 | 57767996 | + | 5.95E-05 | 0.9999404 |
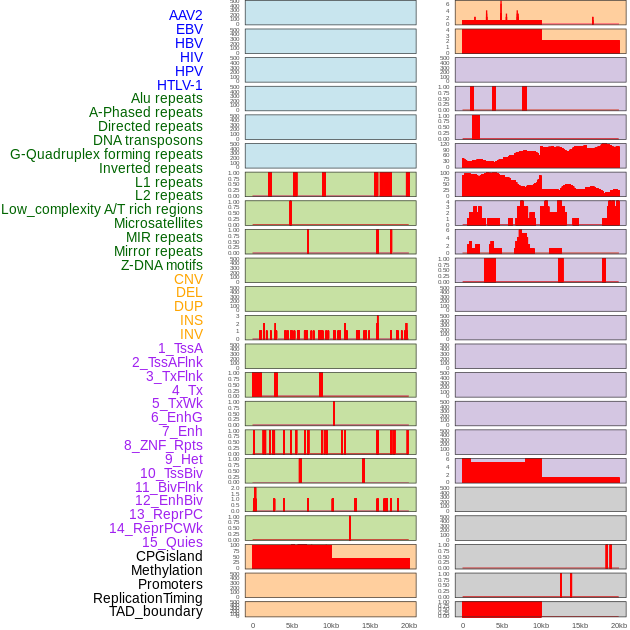
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
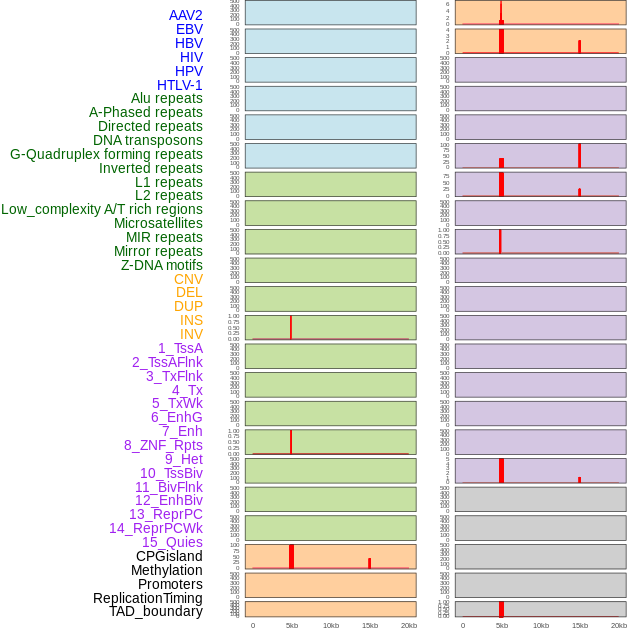
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for ATG3-CLTC |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:112262904/chr17:57767997) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ATG3 | CLTC |
| FUNCTION: E2 conjugating enzyme required for the cytoplasm to vacuole transport (Cvt), autophagy, and mitochondrial homeostasis. Responsible for the E2-like covalent binding of phosphatidylethanolamine to the C-terminal Gly of ATG8-like proteins (GABARAP, GABARAPL1, GABARAPL2 or MAP1LC3A). The ATG12-ATG5 conjugate plays a role of an E3 and promotes the transfer of ATG8-like proteins from ATG3 to phosphatidylethanolamine (PE). This step is required for the membrane association of ATG8-like proteins. The formation of the ATG8-phosphatidylethanolamine conjugates is essential for autophagy and for the cytoplasm to vacuole transport (Cvt). Preferred substrate is MAP1LC3A. Also acts as an autocatalytic E2-like enzyme, catalyzing the conjugation of ATG12 to itself, ATG12 conjugation to ATG3 playing a role in mitochondrial homeostasis but not in autophagy. ATG7 (E1-like enzyme) facilitates this reaction by forming an E1-E2 complex with ATG3. Promotes primary ciliogenesis by removing OFD1 from centriolar satellites via the autophagic pathway. {ECO:0000269|PubMed:11825910, ECO:0000269|PubMed:12207896, ECO:0000269|PubMed:12890687, ECO:0000269|PubMed:16704426, ECO:0000269|PubMed:20723759}. | FUNCTION: Clathrin is the major protein of the polyhedral coat of coated pits and vesicles. Two different adapter protein complexes link the clathrin lattice either to the plasma membrane or to the trans-Golgi network (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATG3 | chr3:112262904 | chr17:57767997 | ENST00000283290 | - | 6 | 12 | 166_169 | 131 | 315.0 | Motif | Note=Caspase cleavage motif LETD |
| Hgene | ATG3 | chr3:112262904 | chr17:57767997 | ENST00000402314 | - | 6 | 11 | 166_169 | 131 | 312.0 | Motif | Note=Caspase cleavage motif LETD |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 108_149 | 1609 | 1676.0 | Region | Note=WD40-like repeat 3 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 1213_1522 | 1609 | 1676.0 | Region | Involved in binding clathrin light chain | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 150_195 | 1609 | 1676.0 | Region | Note=WD40-like repeat 4 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 1550_1675 | 1609 | 1676.0 | Region | Trimerization | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 196_257 | 1609 | 1676.0 | Region | Note=WD40-like repeat 5 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 24_67 | 1609 | 1676.0 | Region | Note=WD40-like repeat 1 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 258_301 | 1609 | 1676.0 | Region | Note=WD40-like repeat 6 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 2_479 | 1609 | 1676.0 | Region | Note=Globular terminal domain | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 302_330 | 1609 | 1676.0 | Region | Note=WD40-like repeat 7 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 449_465 | 1609 | 1676.0 | Region | Binding site for the uncoating ATPase%2C involved in lattice disassembly | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 480_523 | 1609 | 1676.0 | Region | Note=Flexible linker | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 524_1675 | 1609 | 1676.0 | Region | Note=Heavy chain arm | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 524_634 | 1609 | 1676.0 | Region | Note=Distal segment | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 639_1675 | 1609 | 1676.0 | Region | Note=Proximal segment | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 68_107 | 1609 | 1676.0 | Region | Note=WD40-like repeat 2 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 108_149 | 1609 | 1640.0 | Region | Note=WD40-like repeat 3 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 1213_1522 | 1609 | 1640.0 | Region | Involved in binding clathrin light chain | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 150_195 | 1609 | 1640.0 | Region | Note=WD40-like repeat 4 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 1550_1675 | 1609 | 1640.0 | Region | Trimerization | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 196_257 | 1609 | 1640.0 | Region | Note=WD40-like repeat 5 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 24_67 | 1609 | 1640.0 | Region | Note=WD40-like repeat 1 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 258_301 | 1609 | 1640.0 | Region | Note=WD40-like repeat 6 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 2_479 | 1609 | 1640.0 | Region | Note=Globular terminal domain | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 302_330 | 1609 | 1640.0 | Region | Note=WD40-like repeat 7 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 449_465 | 1609 | 1640.0 | Region | Binding site for the uncoating ATPase%2C involved in lattice disassembly | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 480_523 | 1609 | 1640.0 | Region | Note=Flexible linker | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 524_1675 | 1609 | 1640.0 | Region | Note=Heavy chain arm | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 524_634 | 1609 | 1640.0 | Region | Note=Distal segment | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 639_1675 | 1609 | 1640.0 | Region | Note=Proximal segment | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 68_107 | 1609 | 1640.0 | Region | Note=WD40-like repeat 2 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 1128_1269 | 1609 | 1676.0 | Repeat | CHCR 5 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 1274_1420 | 1609 | 1676.0 | Repeat | CHCR 6 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 1423_1566 | 1609 | 1676.0 | Repeat | CHCR 7 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 537_683 | 1609 | 1676.0 | Repeat | CHCR 1 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 686_828 | 1609 | 1676.0 | Repeat | CHCR 2 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 833_972 | 1609 | 1676.0 | Repeat | CHCR 3 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000269122 | 29 | 32 | 979_1124 | 1609 | 1676.0 | Repeat | CHCR 4 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 1128_1269 | 1609 | 1640.0 | Repeat | CHCR 5 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 1274_1420 | 1609 | 1640.0 | Repeat | CHCR 6 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 1423_1566 | 1609 | 1640.0 | Repeat | CHCR 7 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 537_683 | 1609 | 1640.0 | Repeat | CHCR 1 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 686_828 | 1609 | 1640.0 | Repeat | CHCR 2 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 833_972 | 1609 | 1640.0 | Repeat | CHCR 3 | |
| Tgene | CLTC | chr3:112262904 | chr17:57767997 | ENST00000393043 | 29 | 31 | 979_1124 | 1609 | 1640.0 | Repeat | CHCR 4 |
Top |
Fusion Gene Sequence for ATG3-CLTC |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >7558_7558_1_ATG3-CLTC_ATG3_chr3_112262904_ENST00000283290_CLTC_chr17_57767997_ENST00000269122_length(transcript)=3487nt_BP=828nt TAGTGTCACGTGAGGCCCCGGTGGCGGCGCAGCTACGGCAAGAGAGTGAGAAGGAAGGGAAGCCGGAAGGGGCGCGAGTGAAGCAAAGCG AGGACAGACAGCTCGCAGAGGGCGAGGGGTGCGTGTGCGTCCGCTTCTCACCTCAGGTCTCCCTTCGGCCCCGCTGCCCTCCCTCGCGGC TGGGTGACAGCTGGGTCCGGTCCGTCGCGGGCTGCCTGGGGTGCGAGGATCGCGCACCCCGTCTTCGCGCGCTGTGCCTGCCGCCCCGCC CCCTCGTCCCGCCCGTCCCGTCGCGTCGCGTCCCGTCCCCTCGGGTGCTGCCAGCCGGGTGCTGATGCGAGTCGGTGGCAGCGAGGACAT TTTCTGACTCCCTGGCCCCTGACACGGCTGCACTTTCCATCCCGTCGCGGGGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATT AATACTGTGAAGGGAAAGGCACTGGAAGTGGCTGAGTACCTGACCCCGGTCCTCAAGGAATCAAAGTTTAAGGAAACAGGTGTAATTACC CCAGAAGAGTTTGTGGCAGCTGGAGATCACCTAGTCCACCACTGTCCAACATGGCAATGGGCTACAGGGGAAGAATTGAAAGTGAAGGCA TACCTACCAACAGGCAAACAATTTTTGGTAACCAAAAATGTGCCGTGCTATAAGCGGTGCAAACAGATGGAATATTCAGATGAATTGGAA GCTATCATTGAAGAAGATGATGGTGATGGCGGATGGGTAGATACATATCACAACACAGGTATTACAGGAATAACGGAAGCCGTTAAAGAG ATCACACTGGAAAATAAGGTGGATAAATTAGATGCTTCAGAATCACTGAGAAAAGAAGAAGAACAAGCTACAGAGACACAACCCATTGTT TATGGTCAGCCCCAGTTGATGCTGACAGCAGGACCCAGTGTTGCCGTCCCTCCCCAGGCACCTTTTGGTTATGGTTATACCGCACCACCG TATGGACAGCCACAGCCTGGCTTTGGGTACAGCATGTGAGATGAAGCGCTGATCCTGTAGTCACCTATTTTCGTACTGAAACATCGTCTT TACCCACTTCTCAGTTTATAATGGGGGAAAACAGGCAACGTGTTCTTGTAACCTTTATTTCATGAAGGACTTCTTTTTGTTTCTAACTAT AAACTTGGATCACCTATGTTAAAACCTTATTTCACATTCCACATCATTTTAGAATTTATTTTCGAAGGGGAATAGTTTCAATGTTTTATT CACTTGGGCTTTTTTTCTTCCCCCTCTTTCTTTAAAGAACTGCTCAATATTCAATCTGTTGTGAAGAACCTGATTTGCACTCTGTAGTGT TTAAAGAAACAAAGAAACTCTAATATTGAATCTCTTAAATTTAGTGTATGTAAACAGCTTACAAATACGTATTGTCTAAATGCATTTAAA TCTGTTTTATTCAAAGAAAAGCTAAAGCAAAAACACTGGCATATGACCATGCAAGACTGTCAGTGCCAACAAAGACAACACTAATCAGCA CATCGTACACTGGATTGCAGTGCTTCCCAGATTATTGAAAAATGTTACAGACAACTTGCCTGATTTTTAAATGAGCGTAAAAGGCCCTCT AACCTATGCAGGTTTCCCCATTATGCATATAGAAAATGCTAGTATGTTTTGCTCACTTCATATGTAACAGGTGCCCTTATGTTGTGCTGT ATCCTGTGCTTTTTCTGTGGGACCATTCCATTCAGGAGCAAAGAGCACCATGATTCCAATCTTGTGTGTGTTTACTAACCCTTCCCTGAG GTTTGTGTATGTTGGATATTGTGGTGTTTTAGATCACTGAGTGTACAGAAGAGAGAAATTCAAACAAAATATTGCTGTTCTTCAGTTTTG TTTGTGGAATTTGAAATTACTCAAATTTAAAATAAATTACTGGACTGTGGAAATAACATAGAATTGAAGTTTTAATTAAATACCACTCAA ACGAAAAGAACAGTAGTTTTTGTAGTTTTATATTGGATACTGAGGCATTAGGGAGGCATGAAAGGAAGAGGAATGAGGATTGAGACATGT GAAGACATTGTGCATTATATCAATGTGCATTCCTGTAGTTCATTAACAAGGTACATGCAATAGTCTAAAGAACCAGAGTCACTACTATAG TGGCTTAACATTTAATCTGTCTCCAATATTTTAACCAAGTGACACCGAGGTTTTTATCGAAGCATTTCACTTAAATGAACAAATCATGGC TGTTATATTAACTTGAAATAAAATATATTTAAACATGTAGTTAACATGCTCTTTCTCATCATGAATTATAAAGGTGCCCAAATTATGGGT ATCTTGATTTTTTTCAAAAATTGGTTGTGTGAATCCAGCTCTTGCTTATGGCTGAGAATGTTCCTAAAGTTCATTTCACTATCCCTTAAT CCTCAATAGCTACACATACCCCCTTGCCTAGTTAGTCTTGCACTGCTTTACAACTCTCTTAGGCAAATTACCTAACATAAGCTCCACAGA AGAGTTTTTTAAAACTTAAAATCCCTGTCCCACCAGGTGTGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGGCTGAGGCAGGCGGA TTGCTTGAGGCCAGGAGTTCGAGACAAGCCTGACCAACATGGTGACTCCATCTCTACTAAAAATAAAAAATCAGCCAGGTGTGATGGTGG ATGCCTATAAGATCCTAGCTGCTACTCAGGAGGCAGGAGGCTGAGGCATGAGAATCACTTGAACCTGTGAGGCAAAGGTTGTAGTGAGCC AAGATCACGTCACTGCACTCCATCCTGGGTGACAGAATGAGACTCTTGTCTCAAAAAACAAAAAAACCTATTTAGTTTCATCCAGGACAT TAAGTGAAAGTGTTTAATGTGAGTGCAGGTCAGTGAAGAATACAATGGTGCCCCTGCTTTGATTTGTGGGAGGTGCCATCAGTTTTCCCC ACCACTGCTTTTGCACCCTCAGTGCAAATATCAACACAGAGAAAAAAAAAATAATAATAGTATTATGAAAACATTGACTCTGTGGAGACC CTGTGGGTTCTGCAGAGTATACTTTGAAAACTATAAGATTATGAGTTCTATAAAATACCAAGTACAGATGAACTAGAAAACAAGCATATA TCCACAATTACCAGAATTTTCATAGTATTCCTTTTAGTAGATATGCCATGACAGTTACTTAGCCACCCCATGCCATCCTGTTGGGAATGC CACTACCTTTGAATTTGGAGCCATCAGTCTATCTGAGGCAAACCTCAGAAATTACCTGAGCCAGAATCATGCACTTAGCACTGAGTTCAA CTCAGTCTTAACGTTGCATTTTGTTGCATCACTTTTGCAGCCAGTAAACATTCTGTTTAGAAATAGTTGCTATTGTGGCATCATTCACAG >7558_7558_1_ATG3-CLTC_ATG3_chr3_112262904_ENST00000283290_CLTC_chr17_57767997_ENST00000269122_length(amino acids)=221AA_BP=155 MTPWPLTRLHFPSRRGAGRYSGPRMQNVINTVKGKALEVAEYLTPVLKESKFKETGVITPEEFVAAGDHLVHHCPTWQWATGEELKVKAY LPTGKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDGDGGWVDTYHNTGITGITEAVKEITLENKVDKLDASESLRKEEEQATETQPIVY -------------------------------------------------------------- >7558_7558_2_ATG3-CLTC_ATG3_chr3_112262904_ENST00000283290_CLTC_chr17_57767997_ENST00000579456_length(transcript)=1291nt_BP=828nt TAGTGTCACGTGAGGCCCCGGTGGCGGCGCAGCTACGGCAAGAGAGTGAGAAGGAAGGGAAGCCGGAAGGGGCGCGAGTGAAGCAAAGCG AGGACAGACAGCTCGCAGAGGGCGAGGGGTGCGTGTGCGTCCGCTTCTCACCTCAGGTCTCCCTTCGGCCCCGCTGCCCTCCCTCGCGGC TGGGTGACAGCTGGGTCCGGTCCGTCGCGGGCTGCCTGGGGTGCGAGGATCGCGCACCCCGTCTTCGCGCGCTGTGCCTGCCGCCCCGCC CCCTCGTCCCGCCCGTCCCGTCGCGTCGCGTCCCGTCCCCTCGGGTGCTGCCAGCCGGGTGCTGATGCGAGTCGGTGGCAGCGAGGACAT TTTCTGACTCCCTGGCCCCTGACACGGCTGCACTTTCCATCCCGTCGCGGGGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATT AATACTGTGAAGGGAAAGGCACTGGAAGTGGCTGAGTACCTGACCCCGGTCCTCAAGGAATCAAAGTTTAAGGAAACAGGTGTAATTACC CCAGAAGAGTTTGTGGCAGCTGGAGATCACCTAGTCCACCACTGTCCAACATGGCAATGGGCTACAGGGGAAGAATTGAAAGTGAAGGCA TACCTACCAACAGGCAAACAATTTTTGGTAACCAAAAATGTGCCGTGCTATAAGCGGTGCAAACAGATGGAATATTCAGATGAATTGGAA GCTATCATTGAAGAAGATGATGGTGATGGCGGATGGGTAGATACATATCACAACACAGGTATTACAGGAATAACGGAAGCCGTTAAAGAG ATCACACTGGAAAATAAGGTGGATAAATTAGATGCTTCAGAATCACTGAGAAAAGAAGAAGAACAAGCTACAGAGACACAACCCATTGTT TATGGTCAGCCCCAGTTGATGCTGACAGCAGGACCCAGTGTTGCCGTCCCTCCCCAGGCACCTTTTGGTTATGGTTATACCGCACCACCG TATGGACAGCCACAGCCTGGCTTTGGGTACAGCATGTGAGATGAAGCGCTGATCCTGTAGTCACCTATTTTCGTACTGAAACATCGTCTT TACCCACTTCTCAGTTTATAATGGGGGAAAACAGGCAACGTGTTCTTGTAACCTTTATTTCATGAAGGACTTCTTTTTGTTTCTAACTAT AAACTTGGATCACCTATGTTAAAACCTTATTTCACATTCCACATCATTTTAGAATTTATTTTCGAAGGGGAATAGTTTCAATGTTTTATT >7558_7558_2_ATG3-CLTC_ATG3_chr3_112262904_ENST00000283290_CLTC_chr17_57767997_ENST00000579456_length(amino acids)=221AA_BP=155 MTPWPLTRLHFPSRRGAGRYSGPRMQNVINTVKGKALEVAEYLTPVLKESKFKETGVITPEEFVAAGDHLVHHCPTWQWATGEELKVKAY LPTGKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDGDGGWVDTYHNTGITGITEAVKEITLENKVDKLDASESLRKEEEQATETQPIVY -------------------------------------------------------------- >7558_7558_3_ATG3-CLTC_ATG3_chr3_112262904_ENST00000402314_CLTC_chr17_57767997_ENST00000269122_length(transcript)=3440nt_BP=781nt GAGAAGGAAGGGAAGCCGGAAGGGGCGCGAGTGAAGCAAAGCGAGGACAGACAGCTCGCAGAGGGCGAGGGGTGCGTGTGCGTCCGCTTC TCACCTCAGGTCTCCCTTCGGCCCCGCTGCCCTCCCTCGCGGCTGGGTGACAGCTGGGTCCGGTCCGTCGCGGGCTGCCTGGGGTGCGAG GATCGCGCACCCCGTCTTCGCGCGCTGTGCCTGCCGCCCCGCCCCCTCGTCCCGCCCGTCCCGTCGCGTCGCGTCCCGTCCCCTCGGGTG CTGCCAGCCGGGTGCTGATGCGAGTCGGTGGCAGCGAGGACATTTTCTGACTCCCTGGCCCCTGACACGGCTGCACTTTCCATCCCGTCG CGGGGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATTAATACTGTGAAGGGAAAGGCACTGGAAGTGGCTGAGTACCTGACCCC GGTCCTCAAGGAATCAAAGTTTAAGGAAACAGGTGTAATTACCCCAGAAGAGTTTGTGGCAGCTGGAGATCACCTAGTCCACCACTGTCC AACATGGCAATGGGCTACAGGGGAAGAATTGAAAGTGAAGGCATACCTACCAACAGGCAAACAATTTTTGGTAACCAAAAATGTGCCGTG CTATAAGCGGTGCAAACAGATGGAATATTCAGATGAATTGGAAGCTATCATTGAAGAAGATGATGGTGATGGCGGATGGGTAGATACATA TCACAACACAGGTATTACAGGAATAACGGAAGCCGTTAAAGAGATCACACTGGAAAATAAGGTGGATAAATTAGATGCTTCAGAATCACT GAGAAAAGAAGAAGAACAAGCTACAGAGACACAACCCATTGTTTATGGTCAGCCCCAGTTGATGCTGACAGCAGGACCCAGTGTTGCCGT CCCTCCCCAGGCACCTTTTGGTTATGGTTATACCGCACCACCGTATGGACAGCCACAGCCTGGCTTTGGGTACAGCATGTGAGATGAAGC GCTGATCCTGTAGTCACCTATTTTCGTACTGAAACATCGTCTTTACCCACTTCTCAGTTTATAATGGGGGAAAACAGGCAACGTGTTCTT GTAACCTTTATTTCATGAAGGACTTCTTTTTGTTTCTAACTATAAACTTGGATCACCTATGTTAAAACCTTATTTCACATTCCACATCAT TTTAGAATTTATTTTCGAAGGGGAATAGTTTCAATGTTTTATTCACTTGGGCTTTTTTTCTTCCCCCTCTTTCTTTAAAGAACTGCTCAA TATTCAATCTGTTGTGAAGAACCTGATTTGCACTCTGTAGTGTTTAAAGAAACAAAGAAACTCTAATATTGAATCTCTTAAATTTAGTGT ATGTAAACAGCTTACAAATACGTATTGTCTAAATGCATTTAAATCTGTTTTATTCAAAGAAAAGCTAAAGCAAAAACACTGGCATATGAC CATGCAAGACTGTCAGTGCCAACAAAGACAACACTAATCAGCACATCGTACACTGGATTGCAGTGCTTCCCAGATTATTGAAAAATGTTA CAGACAACTTGCCTGATTTTTAAATGAGCGTAAAAGGCCCTCTAACCTATGCAGGTTTCCCCATTATGCATATAGAAAATGCTAGTATGT TTTGCTCACTTCATATGTAACAGGTGCCCTTATGTTGTGCTGTATCCTGTGCTTTTTCTGTGGGACCATTCCATTCAGGAGCAAAGAGCA CCATGATTCCAATCTTGTGTGTGTTTACTAACCCTTCCCTGAGGTTTGTGTATGTTGGATATTGTGGTGTTTTAGATCACTGAGTGTACA GAAGAGAGAAATTCAAACAAAATATTGCTGTTCTTCAGTTTTGTTTGTGGAATTTGAAATTACTCAAATTTAAAATAAATTACTGGACTG TGGAAATAACATAGAATTGAAGTTTTAATTAAATACCACTCAAACGAAAAGAACAGTAGTTTTTGTAGTTTTATATTGGATACTGAGGCA TTAGGGAGGCATGAAAGGAAGAGGAATGAGGATTGAGACATGTGAAGACATTGTGCATTATATCAATGTGCATTCCTGTAGTTCATTAAC AAGGTACATGCAATAGTCTAAAGAACCAGAGTCACTACTATAGTGGCTTAACATTTAATCTGTCTCCAATATTTTAACCAAGTGACACCG AGGTTTTTATCGAAGCATTTCACTTAAATGAACAAATCATGGCTGTTATATTAACTTGAAATAAAATATATTTAAACATGTAGTTAACAT GCTCTTTCTCATCATGAATTATAAAGGTGCCCAAATTATGGGTATCTTGATTTTTTTCAAAAATTGGTTGTGTGAATCCAGCTCTTGCTT ATGGCTGAGAATGTTCCTAAAGTTCATTTCACTATCCCTTAATCCTCAATAGCTACACATACCCCCTTGCCTAGTTAGTCTTGCACTGCT TTACAACTCTCTTAGGCAAATTACCTAACATAAGCTCCACAGAAGAGTTTTTTAAAACTTAAAATCCCTGTCCCACCAGGTGTGGTGGCT CATGCCTGTAATCCCAGCACTTTGGGAGGCTGAGGCAGGCGGATTGCTTGAGGCCAGGAGTTCGAGACAAGCCTGACCAACATGGTGACT CCATCTCTACTAAAAATAAAAAATCAGCCAGGTGTGATGGTGGATGCCTATAAGATCCTAGCTGCTACTCAGGAGGCAGGAGGCTGAGGC ATGAGAATCACTTGAACCTGTGAGGCAAAGGTTGTAGTGAGCCAAGATCACGTCACTGCACTCCATCCTGGGTGACAGAATGAGACTCTT GTCTCAAAAAACAAAAAAACCTATTTAGTTTCATCCAGGACATTAAGTGAAAGTGTTTAATGTGAGTGCAGGTCAGTGAAGAATACAATG GTGCCCCTGCTTTGATTTGTGGGAGGTGCCATCAGTTTTCCCCACCACTGCTTTTGCACCCTCAGTGCAAATATCAACACAGAGAAAAAA AAAATAATAATAGTATTATGAAAACATTGACTCTGTGGAGACCCTGTGGGTTCTGCAGAGTATACTTTGAAAACTATAAGATTATGAGTT CTATAAAATACCAAGTACAGATGAACTAGAAAACAAGCATATATCCACAATTACCAGAATTTTCATAGTATTCCTTTTAGTAGATATGCC ATGACAGTTACTTAGCCACCCCATGCCATCCTGTTGGGAATGCCACTACCTTTGAATTTGGAGCCATCAGTCTATCTGAGGCAAACCTCA GAAATTACCTGAGCCAGAATCATGCACTTAGCACTGAGTTCAACTCAGTCTTAACGTTGCATTTTGTTGCATCACTTTTGCAGCCAGTAA ACATTCTGTTTAGAAATAGTTGCTATTGTGGCATCATTCACAGTTGCAATTTTTCTCCTGTGCTGTCATATGCCTGTGCGAAGTGTATTA >7558_7558_3_ATG3-CLTC_ATG3_chr3_112262904_ENST00000402314_CLTC_chr17_57767997_ENST00000269122_length(amino acids)=221AA_BP=155 MTPWPLTRLHFPSRRGAGRYSGPRMQNVINTVKGKALEVAEYLTPVLKESKFKETGVITPEEFVAAGDHLVHHCPTWQWATGEELKVKAY LPTGKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDGDGGWVDTYHNTGITGITEAVKEITLENKVDKLDASESLRKEEEQATETQPIVY -------------------------------------------------------------- >7558_7558_4_ATG3-CLTC_ATG3_chr3_112262904_ENST00000402314_CLTC_chr17_57767997_ENST00000579456_length(transcript)=1244nt_BP=781nt GAGAAGGAAGGGAAGCCGGAAGGGGCGCGAGTGAAGCAAAGCGAGGACAGACAGCTCGCAGAGGGCGAGGGGTGCGTGTGCGTCCGCTTC TCACCTCAGGTCTCCCTTCGGCCCCGCTGCCCTCCCTCGCGGCTGGGTGACAGCTGGGTCCGGTCCGTCGCGGGCTGCCTGGGGTGCGAG GATCGCGCACCCCGTCTTCGCGCGCTGTGCCTGCCGCCCCGCCCCCTCGTCCCGCCCGTCCCGTCGCGTCGCGTCCCGTCCCCTCGGGTG CTGCCAGCCGGGTGCTGATGCGAGTCGGTGGCAGCGAGGACATTTTCTGACTCCCTGGCCCCTGACACGGCTGCACTTTCCATCCCGTCG CGGGGCCGGCCGCTACTCCGGCCCCAGGATGCAGAATGTGATTAATACTGTGAAGGGAAAGGCACTGGAAGTGGCTGAGTACCTGACCCC GGTCCTCAAGGAATCAAAGTTTAAGGAAACAGGTGTAATTACCCCAGAAGAGTTTGTGGCAGCTGGAGATCACCTAGTCCACCACTGTCC AACATGGCAATGGGCTACAGGGGAAGAATTGAAAGTGAAGGCATACCTACCAACAGGCAAACAATTTTTGGTAACCAAAAATGTGCCGTG CTATAAGCGGTGCAAACAGATGGAATATTCAGATGAATTGGAAGCTATCATTGAAGAAGATGATGGTGATGGCGGATGGGTAGATACATA TCACAACACAGGTATTACAGGAATAACGGAAGCCGTTAAAGAGATCACACTGGAAAATAAGGTGGATAAATTAGATGCTTCAGAATCACT GAGAAAAGAAGAAGAACAAGCTACAGAGACACAACCCATTGTTTATGGTCAGCCCCAGTTGATGCTGACAGCAGGACCCAGTGTTGCCGT CCCTCCCCAGGCACCTTTTGGTTATGGTTATACCGCACCACCGTATGGACAGCCACAGCCTGGCTTTGGGTACAGCATGTGAGATGAAGC GCTGATCCTGTAGTCACCTATTTTCGTACTGAAACATCGTCTTTACCCACTTCTCAGTTTATAATGGGGGAAAACAGGCAACGTGTTCTT GTAACCTTTATTTCATGAAGGACTTCTTTTTGTTTCTAACTATAAACTTGGATCACCTATGTTAAAACCTTATTTCACATTCCACATCAT >7558_7558_4_ATG3-CLTC_ATG3_chr3_112262904_ENST00000402314_CLTC_chr17_57767997_ENST00000579456_length(amino acids)=221AA_BP=155 MTPWPLTRLHFPSRRGAGRYSGPRMQNVINTVKGKALEVAEYLTPVLKESKFKETGVITPEEFVAAGDHLVHHCPTWQWATGEELKVKAY LPTGKQFLVTKNVPCYKRCKQMEYSDELEAIIEEDDGDGGWVDTYHNTGITGITEAVKEITLENKVDKLDASESLRKEEEQATETQPIVY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ATG3-CLTC |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ATG3-CLTC |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ATG3-CLTC |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
