
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:ATG7-FHIT (FusionGDB2 ID:7612) |
Fusion Gene Summary for ATG7-FHIT |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: ATG7-FHIT | Fusion gene ID: 7612 | Hgene | Tgene | Gene symbol | ATG7 | FHIT | Gene ID | 10533 | 2272 |
| Gene name | autophagy related 7 | fragile histidine triad diadenosine triphosphatase | |
| Synonyms | APG7-LIKE|APG7L|GSA7 | AP3Aase|FRA3B | |
| Cytomap | 3p25.3 | 3p14.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | ubiquitin-like modifier-activating enzyme ATG7APG7 autophagy 7-likeATG12-activating enzyme E1 ATG7hAGP7ubiquitin-activating enzyme E1-like protein | bis(5'-adenosyl)-triphosphataseAP3A hydrolasediadenosine 5',5'''-P1,P3-triphosphate hydrolasedinucleosidetriphosphatase | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | O95352 | P49789 | |
| Ensembl transtripts involved in fusion gene | ENST00000354449, ENST00000354956, ENST00000446450, ENST00000469654, | ENST00000341848, ENST00000466788, ENST00000468189, ENST00000476844, ENST00000492590, | |
| Fusion gene scores | * DoF score | 15 X 16 X 9=2160 | 27 X 20 X 11=5940 |
| # samples | 20 | 32 | |
| ** MAII score | log2(20/2160*10)=-3.43295940727611 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(32/5940*10)=-4.21431912080077 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: ATG7 [Title/Abstract] AND FHIT [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | ATG7(11406208)-FHIT(59908140), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | ATG7 | GO:0006497 | protein lipidation | 12890687 |
| Hgene | ATG7 | GO:0009267 | cellular response to starvation | 20543840 |
| Hgene | ATG7 | GO:0031401 | positive regulation of protein modification process | 12890687 |
| Hgene | ATG7 | GO:0071455 | cellular response to hyperoxia | 20543840 |
| Tgene | FHIT | GO:0006163 | purine nucleotide metabolic process | 9323207 |
 Fusion gene breakpoints across ATG7 (5'-gene) Fusion gene breakpoints across ATG7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
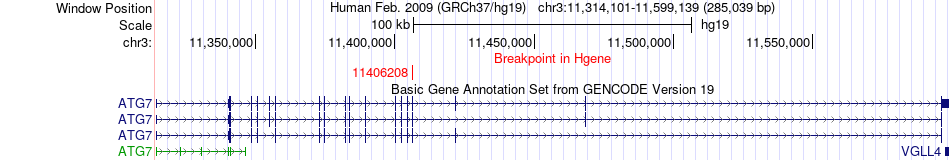 |
 Fusion gene breakpoints across FHIT (3'-gene) Fusion gene breakpoints across FHIT (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
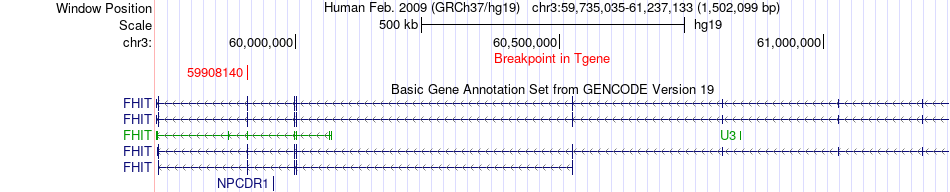 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-XF-A9SK-01A | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
Top |
Fusion Gene ORF analysis for ATG7-FHIT |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000354449 | ENST00000341848 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000354449 | ENST00000466788 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000354449 | ENST00000468189 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000354956 | ENST00000341848 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000354956 | ENST00000466788 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000354956 | ENST00000468189 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000446450 | ENST00000341848 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000446450 | ENST00000466788 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| 5CDS-5UTR | ENST00000446450 | ENST00000468189 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| In-frame | ENST00000354449 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| In-frame | ENST00000354449 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| In-frame | ENST00000354956 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| In-frame | ENST00000354956 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| In-frame | ENST00000446450 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| In-frame | ENST00000446450 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| intron-3CDS | ENST00000469654 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| intron-3CDS | ENST00000469654 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| intron-5UTR | ENST00000469654 | ENST00000341848 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| intron-5UTR | ENST00000469654 | ENST00000466788 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
| intron-5UTR | ENST00000469654 | ENST00000468189 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000446450 | ATG7 | chr3 | 11406208 | - | ENST00000476844 | FHIT | chr3 | 59908140 | - | 2212 | 1783 | 25 | 1947 | 640 |
| ENST00000446450 | ATG7 | chr3 | 11406208 | - | ENST00000492590 | FHIT | chr3 | 59908140 | - | 2223 | 1783 | 25 | 1947 | 640 |
| ENST00000354956 | ATG7 | chr3 | 11406208 | - | ENST00000476844 | FHIT | chr3 | 59908140 | - | 2329 | 1900 | 25 | 2064 | 679 |
| ENST00000354956 | ATG7 | chr3 | 11406208 | - | ENST00000492590 | FHIT | chr3 | 59908140 | - | 2340 | 1900 | 25 | 2064 | 679 |
| ENST00000354449 | ATG7 | chr3 | 11406208 | - | ENST00000476844 | FHIT | chr3 | 59908140 | - | 2329 | 1900 | 25 | 2064 | 679 |
| ENST00000354449 | ATG7 | chr3 | 11406208 | - | ENST00000492590 | FHIT | chr3 | 59908140 | - | 2340 | 1900 | 25 | 2064 | 679 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000446450 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - | 0.004185319 | 0.9958146 |
| ENST00000446450 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - | 0.004170504 | 0.9958295 |
| ENST00000354956 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - | 0.006396486 | 0.9936035 |
| ENST00000354956 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - | 0.006410739 | 0.9935893 |
| ENST00000354449 | ENST00000476844 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - | 0.006396486 | 0.9936035 |
| ENST00000354449 | ENST00000492590 | ATG7 | chr3 | 11406208 | - | FHIT | chr3 | 59908140 | - | 0.006410739 | 0.9935893 |
Top |
Fusion Genomic Features for ATG7-FHIT |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
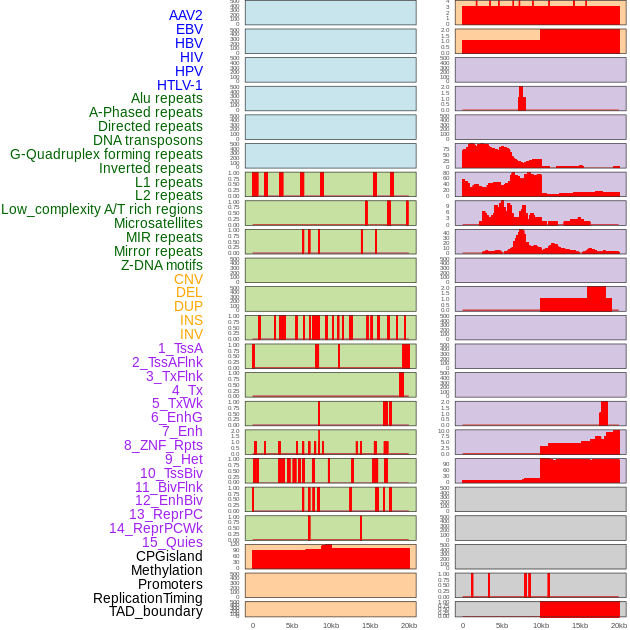 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for ATG7-FHIT |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:11406208/chr3:59908140) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| ATG7 | FHIT |
| FUNCTION: E1-like activating enzyme involved in the 2 ubiquitin-like systems required for cytoplasm to vacuole transport (Cvt) and autophagy. Activates ATG12 for its conjugation with ATG5 as well as the ATG8 family proteins for their conjugation with phosphatidylethanolamine. Both systems are needed for the ATG8 association to Cvt vesicles and autophagosomes membranes. Required for autophagic death induced by caspase-8 inhibition. Required for mitophagy which contributes to regulate mitochondrial quantity and quality by eliminating the mitochondria to a basal level to fulfill cellular energy requirements and preventing excess ROS production. Modulates p53/TP53 activity to regulate cell cycle and survival during metabolic stress. Plays also a key role in the maintenance of axonal homeostasis, the prevention of axonal degeneration, the maintenance of hematopoietic stem cells, the formation of Paneth cell granules, as well as in adipose differentiation. Plays a role in regulating the liver clock and glucose metabolism by mediating the autophagic degradation of CRY1 (clock repressor) in a time-dependent manner (By similarity). {ECO:0000250|UniProtKB:Q9D906, ECO:0000269|PubMed:11096062, ECO:0000269|PubMed:16303767, ECO:0000269|PubMed:22170151}. | FUNCTION: Possesses dinucleoside triphosphate hydrolase activity (PubMed:12574506, PubMed:15182206, PubMed:8794732, PubMed:9323207, PubMed:9576908, PubMed:9543008). Cleaves P(1)-P(3)-bis(5'-adenosyl) triphosphate (Ap3A) to yield AMP and ADP (PubMed:12574506, PubMed:15182206, PubMed:8794732, PubMed:9323207, PubMed:9576908, PubMed:9543008). Can also hydrolyze P(1)-P(4)-bis(5'-adenosyl) tetraphosphate (Ap4A), but has extremely low activity with ATP (PubMed:8794732). Exhibits adenylylsulfatase activity, hydrolyzing adenosine 5'-phosphosulfate to yield AMP and sulfate (PubMed:18694747). Exhibits adenosine 5'-monophosphoramidase activity, hydrolyzing purine nucleotide phosphoramidates with a single phosphate group such as adenosine 5'monophosphoramidate (AMP-NH2) to yield AMP and NH2 (PubMed:18694747). Exhibits adenylylsulfate-ammonia adenylyltransferase, catalyzing the ammonolysis of adenosine 5'-phosphosulfate resulting in the formation of adenosine 5'-phosphoramidate (PubMed:26181368). Also catalyzes the ammonolysis of adenosine 5-phosphorofluoridate and diadenosine triphosphate (PubMed:26181368). Modulates transcriptional activation by CTNNB1 and thereby contributes to regulate the expression of genes essential for cell proliferation and survival, such as CCND1 and BIRC5 (PubMed:18077326). Plays a role in the induction of apoptosis via SRC and AKT1 signaling pathways (PubMed:16407838). Inhibits MDM2-mediated proteasomal degradation of p53/TP53 and thereby plays a role in p53/TP53-mediated apoptosis (PubMed:15313915). Induction of apoptosis depends on the ability of FHIT to bind P(1)-P(3)-bis(5'-adenosyl) triphosphate or related compounds, but does not require its catalytic activity, it may in part come from the mitochondrial form, which sensitizes the low-affinity Ca(2+) transporters, enhancing mitochondrial calcium uptake (PubMed:12574506, PubMed:19622739). Functions as tumor suppressor (By similarity). {ECO:0000250|UniProtKB:O89106, ECO:0000269|PubMed:12574506, ECO:0000269|PubMed:15313915, ECO:0000269|PubMed:16407838, ECO:0000269|PubMed:18077326, ECO:0000269|PubMed:18694747, ECO:0000269|PubMed:19622739, ECO:0000269|PubMed:26181368, ECO:0000269|PubMed:8794732, ECO:0000269|PubMed:9323207, ECO:0000269|PubMed:9543008}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | ATG7 | chr3:11406208 | chr3:59908140 | ENST00000354449 | - | 16 | 19 | 15_17 | 625 | 704.0 | Motif | Note=FAP motif |
| Hgene | ATG7 | chr3:11406208 | chr3:59908140 | ENST00000354956 | - | 16 | 18 | 15_17 | 625 | 677.0 | Motif | Note=FAP motif |
| Hgene | ATG7 | chr3:11406208 | chr3:59908140 | ENST00000446450 | - | 15 | 17 | 15_17 | 586 | 624.0 | Motif | Note=FAP motif |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000341848 | 2 | 5 | 94_98 | 93 | 148.0 | Motif | Histidine triad motif | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000468189 | 6 | 9 | 94_98 | 93 | 148.0 | Motif | Histidine triad motif | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000476844 | 6 | 10 | 94_98 | 93 | 214.0 | Motif | Histidine triad motif | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000492590 | 6 | 10 | 94_98 | 93 | 205.33333333333334 | Motif | Histidine triad motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000341848 | 2 | 5 | 2_109 | 93 | 148.0 | Domain | HIT | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000468189 | 6 | 9 | 2_109 | 93 | 148.0 | Domain | HIT | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000476844 | 6 | 10 | 2_109 | 93 | 214.0 | Domain | HIT | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000492590 | 6 | 10 | 2_109 | 93 | 205.33333333333334 | Domain | HIT | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000341848 | 2 | 5 | 89_92 | 93 | 148.0 | Nucleotide binding | Substrate | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000468189 | 6 | 9 | 89_92 | 93 | 148.0 | Nucleotide binding | Substrate | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000476844 | 6 | 10 | 89_92 | 93 | 214.0 | Nucleotide binding | Substrate | |
| Tgene | FHIT | chr3:11406208 | chr3:59908140 | ENST00000492590 | 6 | 10 | 89_92 | 93 | 205.33333333333334 | Nucleotide binding | Substrate |
Top |
Fusion Gene Sequence for ATG7-FHIT |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >7612_7612_1_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354449_FHIT_chr3_59908140_ENST00000476844_length(transcript)=2329nt_BP=1900nt GGAAGTTGAGCGGCGGCAAGAAATAATGGCGGCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAGTAGTGCCTT GGATGTTGGGTTTTGGCATGAGTTGACCCAGAAGAAGCTGAACGAGTATCGGCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTA CAATGGTGACTCTGCTGGGCTGCCAGCTCGCTTAACATTGGAGTTCAGTGCTTTTGACATGAGTGCTCCCACCCCAGCCCGTTGCTGCCC AGCTATTGGAACACTGTATAACACCAACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGAT ATGGGAATCCATAAAATCAGGCACTGCTCTTGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACATTTGCAGATCTAAAGAAGTA CCACTTCTACTATTGGTTTTGCTATCCTGCCCTCTGTCTTCCAGAGAGTTTACCTCTCATTCAGGGGCCAGTGGGTTTGGATCAAAGGTT TTCACTAAAACAGATTGAAGCACTAGAGTGTGCATATGATAATCTTTGTCAAACAGAAGGAGTCACAGCTCTTCCTTACTTCTTAATCAA GTATGATGAGAACATGGTGCTGGTTTCCTTGCTTAAACACTACAGTGATTTCTTCCAAGGTCAAAGGACGAAGATAACAATTGGTGTATA TGATCCCTGTAACTTAGCCCAGTACCCTGGATGGCCTTTGAGGAATTTTTTGGTCCTAGCAGCCCACAGATGGAGTAGCAGTTTCCAGTC TGTTGAAGTTGTTTGCTTCCGTGACCGTACCATGCAGGGGGCGAGAGACGTTGCCCACAGCATCATCTTCGAAGTGAAGCTTCCAGAAAT GGCATTTAGCCCAGATTGTCCTAAAGCAGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAGTGAATG TATGGACCCTAAAAGGTTAGCTGAGTCATCAGTGGATCTAAATCTCAAACTGATGTGTTGGAGATTGGTTCCTACTTTAGACTTGGACAA GGTTGTGTCTGTCAAATGTCTGCTGCTTGGAGCCGGCACCTTGGGTTGCAATGTAGCTAGGACGTTGATGGGTTGGGGCGTGAGACACAT CACATTTGTGGACAATGCCAAGATCTCCTACTCCAATCCTGTGAGGCAGCCTCTCTATGAGTTTGAAGATTGCCTAGGGGGTGGTAAGCC CAAGGCTCTGGCAGCAGCGGACCGGCTCCAGAAAATATTCCCCGGTGTGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTGGGCA TCCAGTGAACTTCTCCAGTGTCACTCTGGAGCAAGCCCGCAGAGATGTGGAGCAACTGGAGCAGCTCATCGAAAGCCATGATGTCGTCTT CCTATTGATGGACACCAGGGAGAGCCGGTGGCTTCCTGCCGTCATTGCTGCAAGCAAGAGAAAGCTGGTCATCAATGCTGCTTTGGGATT TGACACATTTGTTGTCATGAGACATGGTCTGAAGAAACCAAAGCAGCAAGGAGCTGGGGACTTGTGTCCAAACCACCCTGTGGCATCTGC TGACCTCCTGGGCTCATCGCTTTTTGCCAACATCCCTGGTTACAAGCTTGGCTGCTACTTCTGCAATGATGTGGTGGCCCCAGGAGATTC AACCAGAGACCGGACCTTGGACCAGCAGTGCACTGTGAGTCGTCCAGGACTGGCCGTGATTGCAGGAGCCCTGGCCGTGGAATTGATGGT ATCTGTTTTGCAGCATCCAGAAGGGGGCTATGCCATTGCCAGCAGCAGTGACGATCGGATGAATGAGCCTCCAACCTCTCTTGGGCTTGT GCCTCACCAGCACGTTCACGTCCATGTTCTTCCCAGGAAGGCTGGAGACTTTCACAGGAATGACAGCATCTATGAGGAGCTCCAGAAACA TGACAAGGAGGACTTTCCTGCCTCTTGGAGATCAGAGGAGGAAATGGCAGCAGAAGCCGCAGCTCTGCGGGTCTACTTTCAGTGACACAG ATCCTGAATTCCAGCAAAAGAGCTATTGCCAACCAGTTTGAAGACCGCCCCCCGCCTCTCCCCAAGAGGAACTGAATCAGCATGAAAATG CAGTTTCTTCATCTCACCATCCTGTATTCTTCAACCAGTGATCCCCCACCTCGGTCACTCCAACTCCCTTAAAATACCTAGACCTAAACG >7612_7612_1_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354449_FHIT_chr3_59908140_ENST00000476844_length(amino acids)=679AA_BP=620 MAAATGDPGLSKLQFAPFSSALDVGFWHELTQKKLNEYRLDEAPKDIKGYYYNGDSAGLPARLTLEFSAFDMSAPTPARCCPAIGTLYNT NTLESFKTADKKLLLEQAANEIWESIKSGTALENPVLLNKFLLLTFADLKKYHFYYWFCYPALCLPESLPLIQGPVGLDQRFSLKQIEAL ECAYDNLCQTEGVTALPYFLIKYDENMVLVSLLKHYSDFFQGQRTKITIGVYDPCNLAQYPGWPLRNFLVLAAHRWSSSFQSVEVVCFRD RTMQGARDVAHSIIFEVKLPEMAFSPDCPKAVGWEKNQKGGMGPRMVNLSECMDPKRLAESSVDLNLKLMCWRLVPTLDLDKVVSVKCLL LGAGTLGCNVARTLMGWGVRHITFVDNAKISYSNPVRQPLYEFEDCLGGGKPKALAAADRLQKIFPGVNARGFNMSIPMPGHPVNFSSVT LEQARRDVEQLEQLIESHDVVFLLMDTRESRWLPAVIAASKRKLVINAALGFDTFVVMRHGLKKPKQQGAGDLCPNHPVASADLLGSSLF ANIPGYKLGCYFCNDVVAPGDSTRDRTLDQQCTVSRPGLAVIAGALAVELMVSVLQHPEGGYAIASSSDDRMNEPPTSLGLVPHQHVHVH -------------------------------------------------------------- >7612_7612_2_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354449_FHIT_chr3_59908140_ENST00000492590_length(transcript)=2340nt_BP=1900nt GGAAGTTGAGCGGCGGCAAGAAATAATGGCGGCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAGTAGTGCCTT GGATGTTGGGTTTTGGCATGAGTTGACCCAGAAGAAGCTGAACGAGTATCGGCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTA CAATGGTGACTCTGCTGGGCTGCCAGCTCGCTTAACATTGGAGTTCAGTGCTTTTGACATGAGTGCTCCCACCCCAGCCCGTTGCTGCCC AGCTATTGGAACACTGTATAACACCAACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGAT ATGGGAATCCATAAAATCAGGCACTGCTCTTGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACATTTGCAGATCTAAAGAAGTA CCACTTCTACTATTGGTTTTGCTATCCTGCCCTCTGTCTTCCAGAGAGTTTACCTCTCATTCAGGGGCCAGTGGGTTTGGATCAAAGGTT TTCACTAAAACAGATTGAAGCACTAGAGTGTGCATATGATAATCTTTGTCAAACAGAAGGAGTCACAGCTCTTCCTTACTTCTTAATCAA GTATGATGAGAACATGGTGCTGGTTTCCTTGCTTAAACACTACAGTGATTTCTTCCAAGGTCAAAGGACGAAGATAACAATTGGTGTATA TGATCCCTGTAACTTAGCCCAGTACCCTGGATGGCCTTTGAGGAATTTTTTGGTCCTAGCAGCCCACAGATGGAGTAGCAGTTTCCAGTC TGTTGAAGTTGTTTGCTTCCGTGACCGTACCATGCAGGGGGCGAGAGACGTTGCCCACAGCATCATCTTCGAAGTGAAGCTTCCAGAAAT GGCATTTAGCCCAGATTGTCCTAAAGCAGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAGTGAATG TATGGACCCTAAAAGGTTAGCTGAGTCATCAGTGGATCTAAATCTCAAACTGATGTGTTGGAGATTGGTTCCTACTTTAGACTTGGACAA GGTTGTGTCTGTCAAATGTCTGCTGCTTGGAGCCGGCACCTTGGGTTGCAATGTAGCTAGGACGTTGATGGGTTGGGGCGTGAGACACAT CACATTTGTGGACAATGCCAAGATCTCCTACTCCAATCCTGTGAGGCAGCCTCTCTATGAGTTTGAAGATTGCCTAGGGGGTGGTAAGCC CAAGGCTCTGGCAGCAGCGGACCGGCTCCAGAAAATATTCCCCGGTGTGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTGGGCA TCCAGTGAACTTCTCCAGTGTCACTCTGGAGCAAGCCCGCAGAGATGTGGAGCAACTGGAGCAGCTCATCGAAAGCCATGATGTCGTCTT CCTATTGATGGACACCAGGGAGAGCCGGTGGCTTCCTGCCGTCATTGCTGCAAGCAAGAGAAAGCTGGTCATCAATGCTGCTTTGGGATT TGACACATTTGTTGTCATGAGACATGGTCTGAAGAAACCAAAGCAGCAAGGAGCTGGGGACTTGTGTCCAAACCACCCTGTGGCATCTGC TGACCTCCTGGGCTCATCGCTTTTTGCCAACATCCCTGGTTACAAGCTTGGCTGCTACTTCTGCAATGATGTGGTGGCCCCAGGAGATTC AACCAGAGACCGGACCTTGGACCAGCAGTGCACTGTGAGTCGTCCAGGACTGGCCGTGATTGCAGGAGCCCTGGCCGTGGAATTGATGGT ATCTGTTTTGCAGCATCCAGAAGGGGGCTATGCCATTGCCAGCAGCAGTGACGATCGGATGAATGAGCCTCCAACCTCTCTTGGGCTTGT GCCTCACCAGCACGTTCACGTCCATGTTCTTCCCAGGAAGGCTGGAGACTTTCACAGGAATGACAGCATCTATGAGGAGCTCCAGAAACA TGACAAGGAGGACTTTCCTGCCTCTTGGAGATCAGAGGAGGAAATGGCAGCAGAAGCCGCAGCTCTGCGGGTCTACTTTCAGTGACACAG ATGTTTTTCAGATCCTGAATTCCAGCAAAAGAGCTATTGCCAACCAGTTTGAAGACCGCCCCCCGCCTCTCCCCAAGAGGAACTGAATCA GCATGAAAATGCAGTTTCTTCATCTCACCATCCTGTATTCTTCAACCAGTGATCCCCCACCTCGGTCACTCCAACTCCCTTAAAATACCT AGACCTAAACGGCTCAGACAGGCAGATTTGAGGTTTCCCCCTGTCTCCTTATTCGGCAGCCTTATGATTAAACTTCCTTCTCTGCTGCAA >7612_7612_2_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354449_FHIT_chr3_59908140_ENST00000492590_length(amino acids)=679AA_BP=620 MAAATGDPGLSKLQFAPFSSALDVGFWHELTQKKLNEYRLDEAPKDIKGYYYNGDSAGLPARLTLEFSAFDMSAPTPARCCPAIGTLYNT NTLESFKTADKKLLLEQAANEIWESIKSGTALENPVLLNKFLLLTFADLKKYHFYYWFCYPALCLPESLPLIQGPVGLDQRFSLKQIEAL ECAYDNLCQTEGVTALPYFLIKYDENMVLVSLLKHYSDFFQGQRTKITIGVYDPCNLAQYPGWPLRNFLVLAAHRWSSSFQSVEVVCFRD RTMQGARDVAHSIIFEVKLPEMAFSPDCPKAVGWEKNQKGGMGPRMVNLSECMDPKRLAESSVDLNLKLMCWRLVPTLDLDKVVSVKCLL LGAGTLGCNVARTLMGWGVRHITFVDNAKISYSNPVRQPLYEFEDCLGGGKPKALAAADRLQKIFPGVNARGFNMSIPMPGHPVNFSSVT LEQARRDVEQLEQLIESHDVVFLLMDTRESRWLPAVIAASKRKLVINAALGFDTFVVMRHGLKKPKQQGAGDLCPNHPVASADLLGSSLF ANIPGYKLGCYFCNDVVAPGDSTRDRTLDQQCTVSRPGLAVIAGALAVELMVSVLQHPEGGYAIASSSDDRMNEPPTSLGLVPHQHVHVH -------------------------------------------------------------- >7612_7612_3_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354956_FHIT_chr3_59908140_ENST00000476844_length(transcript)=2329nt_BP=1900nt GGAAGTTGAGCGGCGGCAAGAAATAATGGCGGCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAGTAGTGCCTT GGATGTTGGGTTTTGGCATGAGTTGACCCAGAAGAAGCTGAACGAGTATCGGCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTA CAATGGTGACTCTGCTGGGCTGCCAGCTCGCTTAACATTGGAGTTCAGTGCTTTTGACATGAGTGCTCCCACCCCAGCCCGTTGCTGCCC AGCTATTGGAACACTGTATAACACCAACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGAT ATGGGAATCCATAAAATCAGGCACTGCTCTTGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACATTTGCAGATCTAAAGAAGTA CCACTTCTACTATTGGTTTTGCTATCCTGCCCTCTGTCTTCCAGAGAGTTTACCTCTCATTCAGGGGCCAGTGGGTTTGGATCAAAGGTT TTCACTAAAACAGATTGAAGCACTAGAGTGTGCATATGATAATCTTTGTCAAACAGAAGGAGTCACAGCTCTTCCTTACTTCTTAATCAA GTATGATGAGAACATGGTGCTGGTTTCCTTGCTTAAACACTACAGTGATTTCTTCCAAGGTCAAAGGACGAAGATAACAATTGGTGTATA TGATCCCTGTAACTTAGCCCAGTACCCTGGATGGCCTTTGAGGAATTTTTTGGTCCTAGCAGCCCACAGATGGAGTAGCAGTTTCCAGTC TGTTGAAGTTGTTTGCTTCCGTGACCGTACCATGCAGGGGGCGAGAGACGTTGCCCACAGCATCATCTTCGAAGTGAAGCTTCCAGAAAT GGCATTTAGCCCAGATTGTCCTAAAGCAGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAGTGAATG TATGGACCCTAAAAGGTTAGCTGAGTCATCAGTGGATCTAAATCTCAAACTGATGTGTTGGAGATTGGTTCCTACTTTAGACTTGGACAA GGTTGTGTCTGTCAAATGTCTGCTGCTTGGAGCCGGCACCTTGGGTTGCAATGTAGCTAGGACGTTGATGGGTTGGGGCGTGAGACACAT CACATTTGTGGACAATGCCAAGATCTCCTACTCCAATCCTGTGAGGCAGCCTCTCTATGAGTTTGAAGATTGCCTAGGGGGTGGTAAGCC CAAGGCTCTGGCAGCAGCGGACCGGCTCCAGAAAATATTCCCCGGTGTGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTGGGCA TCCAGTGAACTTCTCCAGTGTCACTCTGGAGCAAGCCCGCAGAGATGTGGAGCAACTGGAGCAGCTCATCGAAAGCCATGATGTCGTCTT CCTATTGATGGACACCAGGGAGAGCCGGTGGCTTCCTGCCGTCATTGCTGCAAGCAAGAGAAAGCTGGTCATCAATGCTGCTTTGGGATT TGACACATTTGTTGTCATGAGACATGGTCTGAAGAAACCAAAGCAGCAAGGAGCTGGGGACTTGTGTCCAAACCACCCTGTGGCATCTGC TGACCTCCTGGGCTCATCGCTTTTTGCCAACATCCCTGGTTACAAGCTTGGCTGCTACTTCTGCAATGATGTGGTGGCCCCAGGAGATTC AACCAGAGACCGGACCTTGGACCAGCAGTGCACTGTGAGTCGTCCAGGACTGGCCGTGATTGCAGGAGCCCTGGCCGTGGAATTGATGGT ATCTGTTTTGCAGCATCCAGAAGGGGGCTATGCCATTGCCAGCAGCAGTGACGATCGGATGAATGAGCCTCCAACCTCTCTTGGGCTTGT GCCTCACCAGCACGTTCACGTCCATGTTCTTCCCAGGAAGGCTGGAGACTTTCACAGGAATGACAGCATCTATGAGGAGCTCCAGAAACA TGACAAGGAGGACTTTCCTGCCTCTTGGAGATCAGAGGAGGAAATGGCAGCAGAAGCCGCAGCTCTGCGGGTCTACTTTCAGTGACACAG ATCCTGAATTCCAGCAAAAGAGCTATTGCCAACCAGTTTGAAGACCGCCCCCCGCCTCTCCCCAAGAGGAACTGAATCAGCATGAAAATG CAGTTTCTTCATCTCACCATCCTGTATTCTTCAACCAGTGATCCCCCACCTCGGTCACTCCAACTCCCTTAAAATACCTAGACCTAAACG >7612_7612_3_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354956_FHIT_chr3_59908140_ENST00000476844_length(amino acids)=679AA_BP=620 MAAATGDPGLSKLQFAPFSSALDVGFWHELTQKKLNEYRLDEAPKDIKGYYYNGDSAGLPARLTLEFSAFDMSAPTPARCCPAIGTLYNT NTLESFKTADKKLLLEQAANEIWESIKSGTALENPVLLNKFLLLTFADLKKYHFYYWFCYPALCLPESLPLIQGPVGLDQRFSLKQIEAL ECAYDNLCQTEGVTALPYFLIKYDENMVLVSLLKHYSDFFQGQRTKITIGVYDPCNLAQYPGWPLRNFLVLAAHRWSSSFQSVEVVCFRD RTMQGARDVAHSIIFEVKLPEMAFSPDCPKAVGWEKNQKGGMGPRMVNLSECMDPKRLAESSVDLNLKLMCWRLVPTLDLDKVVSVKCLL LGAGTLGCNVARTLMGWGVRHITFVDNAKISYSNPVRQPLYEFEDCLGGGKPKALAAADRLQKIFPGVNARGFNMSIPMPGHPVNFSSVT LEQARRDVEQLEQLIESHDVVFLLMDTRESRWLPAVIAASKRKLVINAALGFDTFVVMRHGLKKPKQQGAGDLCPNHPVASADLLGSSLF ANIPGYKLGCYFCNDVVAPGDSTRDRTLDQQCTVSRPGLAVIAGALAVELMVSVLQHPEGGYAIASSSDDRMNEPPTSLGLVPHQHVHVH -------------------------------------------------------------- >7612_7612_4_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354956_FHIT_chr3_59908140_ENST00000492590_length(transcript)=2340nt_BP=1900nt GGAAGTTGAGCGGCGGCAAGAAATAATGGCGGCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAGTAGTGCCTT GGATGTTGGGTTTTGGCATGAGTTGACCCAGAAGAAGCTGAACGAGTATCGGCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTA CAATGGTGACTCTGCTGGGCTGCCAGCTCGCTTAACATTGGAGTTCAGTGCTTTTGACATGAGTGCTCCCACCCCAGCCCGTTGCTGCCC AGCTATTGGAACACTGTATAACACCAACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGAT ATGGGAATCCATAAAATCAGGCACTGCTCTTGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACATTTGCAGATCTAAAGAAGTA CCACTTCTACTATTGGTTTTGCTATCCTGCCCTCTGTCTTCCAGAGAGTTTACCTCTCATTCAGGGGCCAGTGGGTTTGGATCAAAGGTT TTCACTAAAACAGATTGAAGCACTAGAGTGTGCATATGATAATCTTTGTCAAACAGAAGGAGTCACAGCTCTTCCTTACTTCTTAATCAA GTATGATGAGAACATGGTGCTGGTTTCCTTGCTTAAACACTACAGTGATTTCTTCCAAGGTCAAAGGACGAAGATAACAATTGGTGTATA TGATCCCTGTAACTTAGCCCAGTACCCTGGATGGCCTTTGAGGAATTTTTTGGTCCTAGCAGCCCACAGATGGAGTAGCAGTTTCCAGTC TGTTGAAGTTGTTTGCTTCCGTGACCGTACCATGCAGGGGGCGAGAGACGTTGCCCACAGCATCATCTTCGAAGTGAAGCTTCCAGAAAT GGCATTTAGCCCAGATTGTCCTAAAGCAGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAGTGAATG TATGGACCCTAAAAGGTTAGCTGAGTCATCAGTGGATCTAAATCTCAAACTGATGTGTTGGAGATTGGTTCCTACTTTAGACTTGGACAA GGTTGTGTCTGTCAAATGTCTGCTGCTTGGAGCCGGCACCTTGGGTTGCAATGTAGCTAGGACGTTGATGGGTTGGGGCGTGAGACACAT CACATTTGTGGACAATGCCAAGATCTCCTACTCCAATCCTGTGAGGCAGCCTCTCTATGAGTTTGAAGATTGCCTAGGGGGTGGTAAGCC CAAGGCTCTGGCAGCAGCGGACCGGCTCCAGAAAATATTCCCCGGTGTGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTGGGCA TCCAGTGAACTTCTCCAGTGTCACTCTGGAGCAAGCCCGCAGAGATGTGGAGCAACTGGAGCAGCTCATCGAAAGCCATGATGTCGTCTT CCTATTGATGGACACCAGGGAGAGCCGGTGGCTTCCTGCCGTCATTGCTGCAAGCAAGAGAAAGCTGGTCATCAATGCTGCTTTGGGATT TGACACATTTGTTGTCATGAGACATGGTCTGAAGAAACCAAAGCAGCAAGGAGCTGGGGACTTGTGTCCAAACCACCCTGTGGCATCTGC TGACCTCCTGGGCTCATCGCTTTTTGCCAACATCCCTGGTTACAAGCTTGGCTGCTACTTCTGCAATGATGTGGTGGCCCCAGGAGATTC AACCAGAGACCGGACCTTGGACCAGCAGTGCACTGTGAGTCGTCCAGGACTGGCCGTGATTGCAGGAGCCCTGGCCGTGGAATTGATGGT ATCTGTTTTGCAGCATCCAGAAGGGGGCTATGCCATTGCCAGCAGCAGTGACGATCGGATGAATGAGCCTCCAACCTCTCTTGGGCTTGT GCCTCACCAGCACGTTCACGTCCATGTTCTTCCCAGGAAGGCTGGAGACTTTCACAGGAATGACAGCATCTATGAGGAGCTCCAGAAACA TGACAAGGAGGACTTTCCTGCCTCTTGGAGATCAGAGGAGGAAATGGCAGCAGAAGCCGCAGCTCTGCGGGTCTACTTTCAGTGACACAG ATGTTTTTCAGATCCTGAATTCCAGCAAAAGAGCTATTGCCAACCAGTTTGAAGACCGCCCCCCGCCTCTCCCCAAGAGGAACTGAATCA GCATGAAAATGCAGTTTCTTCATCTCACCATCCTGTATTCTTCAACCAGTGATCCCCCACCTCGGTCACTCCAACTCCCTTAAAATACCT AGACCTAAACGGCTCAGACAGGCAGATTTGAGGTTTCCCCCTGTCTCCTTATTCGGCAGCCTTATGATTAAACTTCCTTCTCTGCTGCAA >7612_7612_4_ATG7-FHIT_ATG7_chr3_11406208_ENST00000354956_FHIT_chr3_59908140_ENST00000492590_length(amino acids)=679AA_BP=620 MAAATGDPGLSKLQFAPFSSALDVGFWHELTQKKLNEYRLDEAPKDIKGYYYNGDSAGLPARLTLEFSAFDMSAPTPARCCPAIGTLYNT NTLESFKTADKKLLLEQAANEIWESIKSGTALENPVLLNKFLLLTFADLKKYHFYYWFCYPALCLPESLPLIQGPVGLDQRFSLKQIEAL ECAYDNLCQTEGVTALPYFLIKYDENMVLVSLLKHYSDFFQGQRTKITIGVYDPCNLAQYPGWPLRNFLVLAAHRWSSSFQSVEVVCFRD RTMQGARDVAHSIIFEVKLPEMAFSPDCPKAVGWEKNQKGGMGPRMVNLSECMDPKRLAESSVDLNLKLMCWRLVPTLDLDKVVSVKCLL LGAGTLGCNVARTLMGWGVRHITFVDNAKISYSNPVRQPLYEFEDCLGGGKPKALAAADRLQKIFPGVNARGFNMSIPMPGHPVNFSSVT LEQARRDVEQLEQLIESHDVVFLLMDTRESRWLPAVIAASKRKLVINAALGFDTFVVMRHGLKKPKQQGAGDLCPNHPVASADLLGSSLF ANIPGYKLGCYFCNDVVAPGDSTRDRTLDQQCTVSRPGLAVIAGALAVELMVSVLQHPEGGYAIASSSDDRMNEPPTSLGLVPHQHVHVH -------------------------------------------------------------- >7612_7612_5_ATG7-FHIT_ATG7_chr3_11406208_ENST00000446450_FHIT_chr3_59908140_ENST00000476844_length(transcript)=2212nt_BP=1783nt GGAAGTTGAGCGGCGGCAAGAAATAATGGCGGCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAGTAGTGCCTT GGATGTTGGGTTTTGGCATGAGTTGACCCAGAAGAAGCTGAACGAGTATCGGCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTA CAATGGTGACTCTGCTGGGCTGCCAGCTCGCTTAACATTGGAGTTCAGTGCTTTTGACATGAGTGCTCCCACCCCAGCCCGTTGCTGCCC AGCTATTGGAACACTGTATAACACCAACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGAT ATGGGAATCCATAAAATCAGGCACTGCTCTTGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACATTTGCAATTGAAGCACTAGA GTGTGCATATGATAATCTTTGTCAAACAGAAGGAGTCACAGCTCTTCCTTACTTCTTAATCAAGTATGATGAGAACATGGTGCTGGTTTC CTTGCTTAAACACTACAGTGATTTCTTCCAAGGTCAAAGGACGAAGATAACAATTGGTGTATATGATCCCTGTAACTTAGCCCAGTACCC TGGATGGCCTTTGAGGAATTTTTTGGTCCTAGCAGCCCACAGATGGAGTAGCAGTTTCCAGTCTGTTGAAGTTGTTTGCTTCCGTGACCG TACCATGCAGGGGGCGAGAGACGTTGCCCACAGCATCATCTTCGAAGTGAAGCTTCCAGAAATGGCATTTAGCCCAGATTGTCCTAAAGC AGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAGTGAATGTATGGACCCTAAAAGGTTAGCTGAGTC ATCAGTGGATCTAAATCTCAAACTGATGTGTTGGAGATTGGTTCCTACTTTAGACTTGGACAAGGTTGTGTCTGTCAAATGTCTGCTGCT TGGAGCCGGCACCTTGGGTTGCAATGTAGCTAGGACGTTGATGGGTTGGGGCGTGAGACACATCACATTTGTGGACAATGCCAAGATCTC CTACTCCAATCCTGTGAGGCAGCCTCTCTATGAGTTTGAAGATTGCCTAGGGGGTGGTAAGCCCAAGGCTCTGGCAGCAGCGGACCGGCT CCAGAAAATATTCCCCGGTGTGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTGGGCATCCAGTGAACTTCTCCAGTGTCACTCT GGAGCAAGCCCGCAGAGATGTGGAGCAACTGGAGCAGCTCATCGAAAGCCATGATGTCGTCTTCCTATTGATGGACACCAGGGAGAGCCG GTGGCTTCCTGCCGTCATTGCTGCAAGCAAGAGAAAGCTGGTCATCAATGCTGCTTTGGGATTTGACACATTTGTTGTCATGAGACATGG TCTGAAGAAACCAAAGCAGCAAGGAGCTGGGGACTTGTGTCCAAACCACCCTGTGGCATCTGCTGACCTCCTGGGCTCATCGCTTTTTGC CAACATCCCTGGTTACAAGCTTGGCTGCTACTTCTGCAATGATGTGGTGGCCCCAGGAGATTCAACCAGAGACCGGACCTTGGACCAGCA GTGCACTGTGAGTCGTCCAGGACTGGCCGTGATTGCAGGAGCCCTGGCCGTGGAATTGATGGTATCTGTTTTGCAGCATCCAGAAGGGGG CTATGCCATTGCCAGCAGCAGTGACGATCGGATGAATGAGCCTCCAACCTCTCTTGGGCTTGTGCCTCACCAGCACGTTCACGTCCATGT TCTTCCCAGGAAGGCTGGAGACTTTCACAGGAATGACAGCATCTATGAGGAGCTCCAGAAACATGACAAGGAGGACTTTCCTGCCTCTTG GAGATCAGAGGAGGAAATGGCAGCAGAAGCCGCAGCTCTGCGGGTCTACTTTCAGTGACACAGATCCTGAATTCCAGCAAAAGAGCTATT GCCAACCAGTTTGAAGACCGCCCCCCGCCTCTCCCCAAGAGGAACTGAATCAGCATGAAAATGCAGTTTCTTCATCTCACCATCCTGTAT TCTTCAACCAGTGATCCCCCACCTCGGTCACTCCAACTCCCTTAAAATACCTAGACCTAAACGGCTCAGACAGGCAGATTTGAGGTTTCC >7612_7612_5_ATG7-FHIT_ATG7_chr3_11406208_ENST00000446450_FHIT_chr3_59908140_ENST00000476844_length(amino acids)=640AA_BP=581 MAAATGDPGLSKLQFAPFSSALDVGFWHELTQKKLNEYRLDEAPKDIKGYYYNGDSAGLPARLTLEFSAFDMSAPTPARCCPAIGTLYNT NTLESFKTADKKLLLEQAANEIWESIKSGTALENPVLLNKFLLLTFAIEALECAYDNLCQTEGVTALPYFLIKYDENMVLVSLLKHYSDF FQGQRTKITIGVYDPCNLAQYPGWPLRNFLVLAAHRWSSSFQSVEVVCFRDRTMQGARDVAHSIIFEVKLPEMAFSPDCPKAVGWEKNQK GGMGPRMVNLSECMDPKRLAESSVDLNLKLMCWRLVPTLDLDKVVSVKCLLLGAGTLGCNVARTLMGWGVRHITFVDNAKISYSNPVRQP LYEFEDCLGGGKPKALAAADRLQKIFPGVNARGFNMSIPMPGHPVNFSSVTLEQARRDVEQLEQLIESHDVVFLLMDTRESRWLPAVIAA SKRKLVINAALGFDTFVVMRHGLKKPKQQGAGDLCPNHPVASADLLGSSLFANIPGYKLGCYFCNDVVAPGDSTRDRTLDQQCTVSRPGL AVIAGALAVELMVSVLQHPEGGYAIASSSDDRMNEPPTSLGLVPHQHVHVHVLPRKAGDFHRNDSIYEELQKHDKEDFPASWRSEEEMAA -------------------------------------------------------------- >7612_7612_6_ATG7-FHIT_ATG7_chr3_11406208_ENST00000446450_FHIT_chr3_59908140_ENST00000492590_length(transcript)=2223nt_BP=1783nt GGAAGTTGAGCGGCGGCAAGAAATAATGGCGGCAGCTACGGGGGATCCTGGACTCTCTAAACTGCAGTTTGCCCCTTTTAGTAGTGCCTT GGATGTTGGGTTTTGGCATGAGTTGACCCAGAAGAAGCTGAACGAGTATCGGCTGGATGAAGCTCCCAAGGACATTAAGGGTTATTACTA CAATGGTGACTCTGCTGGGCTGCCAGCTCGCTTAACATTGGAGTTCAGTGCTTTTGACATGAGTGCTCCCACCCCAGCCCGTTGCTGCCC AGCTATTGGAACACTGTATAACACCAACACACTCGAGTCTTTCAAGACTGCAGATAAGAAGCTCCTTTTGGAACAAGCAGCAAATGAGAT ATGGGAATCCATAAAATCAGGCACTGCTCTTGAAAACCCTGTACTCCTCAACAAGTTCCTCCTCTTGACATTTGCAATTGAAGCACTAGA GTGTGCATATGATAATCTTTGTCAAACAGAAGGAGTCACAGCTCTTCCTTACTTCTTAATCAAGTATGATGAGAACATGGTGCTGGTTTC CTTGCTTAAACACTACAGTGATTTCTTCCAAGGTCAAAGGACGAAGATAACAATTGGTGTATATGATCCCTGTAACTTAGCCCAGTACCC TGGATGGCCTTTGAGGAATTTTTTGGTCCTAGCAGCCCACAGATGGAGTAGCAGTTTCCAGTCTGTTGAAGTTGTTTGCTTCCGTGACCG TACCATGCAGGGGGCGAGAGACGTTGCCCACAGCATCATCTTCGAAGTGAAGCTTCCAGAAATGGCATTTAGCCCAGATTGTCCTAAAGC AGTTGGATGGGAAAAGAACCAGAAAGGAGGCATGGGACCAAGGATGGTGAACCTCAGTGAATGTATGGACCCTAAAAGGTTAGCTGAGTC ATCAGTGGATCTAAATCTCAAACTGATGTGTTGGAGATTGGTTCCTACTTTAGACTTGGACAAGGTTGTGTCTGTCAAATGTCTGCTGCT TGGAGCCGGCACCTTGGGTTGCAATGTAGCTAGGACGTTGATGGGTTGGGGCGTGAGACACATCACATTTGTGGACAATGCCAAGATCTC CTACTCCAATCCTGTGAGGCAGCCTCTCTATGAGTTTGAAGATTGCCTAGGGGGTGGTAAGCCCAAGGCTCTGGCAGCAGCGGACCGGCT CCAGAAAATATTCCCCGGTGTGAATGCCAGAGGATTCAACATGAGCATACCTATGCCTGGGCATCCAGTGAACTTCTCCAGTGTCACTCT GGAGCAAGCCCGCAGAGATGTGGAGCAACTGGAGCAGCTCATCGAAAGCCATGATGTCGTCTTCCTATTGATGGACACCAGGGAGAGCCG GTGGCTTCCTGCCGTCATTGCTGCAAGCAAGAGAAAGCTGGTCATCAATGCTGCTTTGGGATTTGACACATTTGTTGTCATGAGACATGG TCTGAAGAAACCAAAGCAGCAAGGAGCTGGGGACTTGTGTCCAAACCACCCTGTGGCATCTGCTGACCTCCTGGGCTCATCGCTTTTTGC CAACATCCCTGGTTACAAGCTTGGCTGCTACTTCTGCAATGATGTGGTGGCCCCAGGAGATTCAACCAGAGACCGGACCTTGGACCAGCA GTGCACTGTGAGTCGTCCAGGACTGGCCGTGATTGCAGGAGCCCTGGCCGTGGAATTGATGGTATCTGTTTTGCAGCATCCAGAAGGGGG CTATGCCATTGCCAGCAGCAGTGACGATCGGATGAATGAGCCTCCAACCTCTCTTGGGCTTGTGCCTCACCAGCACGTTCACGTCCATGT TCTTCCCAGGAAGGCTGGAGACTTTCACAGGAATGACAGCATCTATGAGGAGCTCCAGAAACATGACAAGGAGGACTTTCCTGCCTCTTG GAGATCAGAGGAGGAAATGGCAGCAGAAGCCGCAGCTCTGCGGGTCTACTTTCAGTGACACAGATGTTTTTCAGATCCTGAATTCCAGCA AAAGAGCTATTGCCAACCAGTTTGAAGACCGCCCCCCGCCTCTCCCCAAGAGGAACTGAATCAGCATGAAAATGCAGTTTCTTCATCTCA CCATCCTGTATTCTTCAACCAGTGATCCCCCACCTCGGTCACTCCAACTCCCTTAAAATACCTAGACCTAAACGGCTCAGACAGGCAGAT >7612_7612_6_ATG7-FHIT_ATG7_chr3_11406208_ENST00000446450_FHIT_chr3_59908140_ENST00000492590_length(amino acids)=640AA_BP=581 MAAATGDPGLSKLQFAPFSSALDVGFWHELTQKKLNEYRLDEAPKDIKGYYYNGDSAGLPARLTLEFSAFDMSAPTPARCCPAIGTLYNT NTLESFKTADKKLLLEQAANEIWESIKSGTALENPVLLNKFLLLTFAIEALECAYDNLCQTEGVTALPYFLIKYDENMVLVSLLKHYSDF FQGQRTKITIGVYDPCNLAQYPGWPLRNFLVLAAHRWSSSFQSVEVVCFRDRTMQGARDVAHSIIFEVKLPEMAFSPDCPKAVGWEKNQK GGMGPRMVNLSECMDPKRLAESSVDLNLKLMCWRLVPTLDLDKVVSVKCLLLGAGTLGCNVARTLMGWGVRHITFVDNAKISYSNPVRQP LYEFEDCLGGGKPKALAAADRLQKIFPGVNARGFNMSIPMPGHPVNFSSVTLEQARRDVEQLEQLIESHDVVFLLMDTRESRWLPAVIAA SKRKLVINAALGFDTFVVMRHGLKKPKQQGAGDLCPNHPVASADLLGSSLFANIPGYKLGCYFCNDVVAPGDSTRDRTLDQQCTVSRPGL AVIAGALAVELMVSVLQHPEGGYAIASSSDDRMNEPPTSLGLVPHQHVHVHVLPRKAGDFHRNDSIYEELQKHDKEDFPASWRSEEEMAA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for ATG7-FHIT |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for ATG7-FHIT |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for ATG7-FHIT |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
