
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RPRD2-CA14 (FusionGDB2 ID:77255) |
Fusion Gene Summary for RPRD2-CA14 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RPRD2-CA14 | Fusion gene ID: 77255 | Hgene | Tgene | Gene symbol | RPRD2 | CA14 | Gene ID | 23248 | 23632 |
| Gene name | regulation of nuclear pre-mRNA domain containing 2 | carbonic anhydrase 14 | |
| Synonyms | HSPC099|KIAA0460 | CAXiV | |
| Cytomap | 1q21.2 | 1q21.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | regulation of nuclear pre-mRNA domain-containing protein 2 | carbonic anhydrase 14CA-XIVcarbonate dehydratase XIVcarbonic anhydrase XIVcarbonic dehydratase | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9ULX7 | |
| Ensembl transtripts involved in fusion gene | ENST00000492220, ENST00000369067, ENST00000369068, ENST00000401000, ENST00000539519, | ENST00000369111, | |
| Fusion gene scores | * DoF score | 11 X 8 X 8=704 | 3 X 3 X 4=36 |
| # samples | 13 | 5 | |
| ** MAII score | log2(13/704*10)=-2.43706380560884 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/36*10)=0.473931188332412 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: RPRD2 [Title/Abstract] AND CA14 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RPRD2(150337395)-CA14(150236993), # samples:2 RPRD2(150337395)-CA14(150232546), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across RPRD2 (5'-gene) Fusion gene breakpoints across RPRD2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
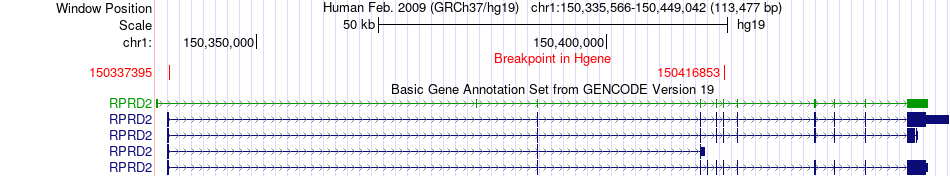 |
 Fusion gene breakpoints across CA14 (3'-gene) Fusion gene breakpoints across CA14 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
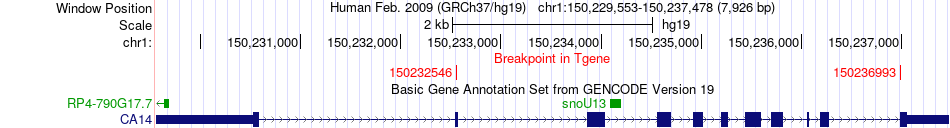 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BRCA | TCGA-A7-A13D-01A | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + |
| ChimerDB4 | LIHC | TCGA-G3-A6UC-01A | RPRD2 | chr1 | 150337395 | - | CA14 | chr1 | 150236993 | + |
| ChimerDB4 | LIHC | TCGA-G3-A6UC-01A | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + |
| ChimerDB4 | LUAD | TCGA-55-7227-01A | RPRD2 | chr1 | 150337395 | - | CA14 | chr1 | 150232546 | + |
| ChimerDB4 | LUAD | TCGA-55-7227-01A | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + |
Top |
Fusion Gene ORF analysis for RPRD2-CA14 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000492220 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000369067 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + |
| In-frame | ENST00000369067 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000369068 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000369068 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + |
| In-frame | ENST00000369068 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000401000 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000401000 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + |
| In-frame | ENST00000401000 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000539519 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + |
| In-frame | ENST00000539519 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + |
| In-frame | ENST00000539519 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + |
| intron-3CDS | ENST00000369067 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + |
| intron-3CDS | ENST00000492220 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + |
| intron-3CDS | ENST00000492220 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000401000 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150236993 | + | 756 | 270 | 247 | 2 | 82 |
| ENST00000539519 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150236993 | + | 713 | 227 | 494 | 267 | 75 |
| ENST00000369067 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150236993 | + | 698 | 212 | 479 | 252 | 75 |
| ENST00000369068 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150236993 | + | 695 | 209 | 476 | 249 | 75 |
| ENST00000401000 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150232546 | + | 1648 | 270 | 65 | 1228 | 387 |
| ENST00000539519 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150232546 | + | 1605 | 227 | 22 | 1185 | 387 |
| ENST00000369067 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150232546 | + | 1590 | 212 | 7 | 1170 | 387 |
| ENST00000369068 | RPRD2 | chr1 | 150337395 | + | ENST00000369111 | CA14 | chr1 | 150232546 | + | 1587 | 209 | 4 | 1167 | 387 |
| ENST00000401000 | RPRD2 | chr1 | 150416853 | - | ENST00000369111 | CA14 | chr1 | 150232546 | + | 2059 | 681 | 65 | 1639 | 524 |
| ENST00000539519 | RPRD2 | chr1 | 150416853 | - | ENST00000369111 | CA14 | chr1 | 150232546 | + | 2016 | 638 | 22 | 1596 | 524 |
| ENST00000369068 | RPRD2 | chr1 | 150416853 | - | ENST00000369111 | CA14 | chr1 | 150232546 | + | 2076 | 698 | 4 | 1656 | 550 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000401000 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + | 0.083627656 | 0.91637236 |
| ENST00000539519 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + | 0.10403539 | 0.89596456 |
| ENST00000369067 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + | 0.084253475 | 0.9157465 |
| ENST00000369068 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236993 | + | 0.087323114 | 0.91267693 |
| ENST00000401000 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + | 0.01328586 | 0.98671407 |
| ENST00000539519 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + | 0.012323059 | 0.987677 |
| ENST00000369067 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + | 0.012416498 | 0.98758346 |
| ENST00000369068 | ENST00000369111 | RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232546 | + | 0.012476333 | 0.98752373 |
| ENST00000401000 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + | 0.006912969 | 0.993087 |
| ENST00000539519 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + | 0.005718798 | 0.9942812 |
| ENST00000369068 | ENST00000369111 | RPRD2 | chr1 | 150416853 | - | CA14 | chr1 | 150232546 | + | 0.011080507 | 0.98891944 |
Top |
Fusion Genomic Features for RPRD2-CA14 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232545 | + | 0.000175958 | 0.9998241 |
| RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236992 | + | 3.94E-07 | 0.99999964 |
| RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150232545 | + | 0.000175958 | 0.9998241 |
| RPRD2 | chr1 | 150337395 | + | CA14 | chr1 | 150236992 | + | 3.94E-07 | 0.99999964 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for RPRD2-CA14 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:150337395/chr1:150236993) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CA14 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Reversible hydration of carbon dioxide. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369067 | + | 1 | 3 | 4_19 | 68 | 153.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369068 | + | 1 | 11 | 4_19 | 68 | 1462.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000401000 | + | 1 | 10 | 4_19 | 68 | 1436.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369067 | + | 1 | 3 | 4_19 | 68 | 153.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369068 | + | 1 | 11 | 4_19 | 68 | 1462.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000401000 | + | 1 | 10 | 4_19 | 68 | 1436.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369068 | - | 6 | 11 | 4_19 | 231 | 1462.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000401000 | - | 5 | 10 | 4_19 | 205 | 1436.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369068 | - | 6 | 11 | 19_149 | 231 | 1462.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000401000 | - | 5 | 10 | 19_149 | 205 | 1436.0 | Domain | CID |
| Tgene | CA14 | chr1:150337395 | chr1:150232546 | ENST00000369111 | 0 | 11 | 20_278 | 18 | 338.0 | Domain | Alpha-carbonic anhydrase | |
| Tgene | CA14 | chr1:150416853 | chr1:150232546 | ENST00000369111 | 0 | 11 | 20_278 | 18 | 338.0 | Domain | Alpha-carbonic anhydrase | |
| Tgene | CA14 | chr1:150337395 | chr1:150232546 | ENST00000369111 | 0 | 11 | 217_218 | 18 | 338.0 | Region | Substrate binding | |
| Tgene | CA14 | chr1:150416853 | chr1:150232546 | ENST00000369111 | 0 | 11 | 217_218 | 18 | 338.0 | Region | Substrate binding | |
| Tgene | CA14 | chr1:150337395 | chr1:150232546 | ENST00000369111 | 0 | 11 | 16_290 | 18 | 338.0 | Topological domain | Extracellular | |
| Tgene | CA14 | chr1:150337395 | chr1:150232546 | ENST00000369111 | 0 | 11 | 312_337 | 18 | 338.0 | Topological domain | Cytoplasmic | |
| Tgene | CA14 | chr1:150416853 | chr1:150232546 | ENST00000369111 | 0 | 11 | 16_290 | 18 | 338.0 | Topological domain | Extracellular | |
| Tgene | CA14 | chr1:150416853 | chr1:150232546 | ENST00000369111 | 0 | 11 | 312_337 | 18 | 338.0 | Topological domain | Cytoplasmic | |
| Tgene | CA14 | chr1:150337395 | chr1:150232546 | ENST00000369111 | 0 | 11 | 291_311 | 18 | 338.0 | Transmembrane | Helical | |
| Tgene | CA14 | chr1:150416853 | chr1:150232546 | ENST00000369111 | 0 | 11 | 291_311 | 18 | 338.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369067 | + | 1 | 3 | 1054_1057 | 68 | 153.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369067 | + | 1 | 3 | 1152_1158 | 68 | 153.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369067 | + | 1 | 3 | 1195_1459 | 68 | 153.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369067 | + | 1 | 3 | 457_950 | 68 | 153.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369068 | + | 1 | 11 | 1054_1057 | 68 | 1462.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369068 | + | 1 | 11 | 1152_1158 | 68 | 1462.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369068 | + | 1 | 11 | 1195_1459 | 68 | 1462.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369068 | + | 1 | 11 | 457_950 | 68 | 1462.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000401000 | + | 1 | 10 | 1054_1057 | 68 | 1436.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000401000 | + | 1 | 10 | 1152_1158 | 68 | 1436.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000401000 | + | 1 | 10 | 1195_1459 | 68 | 1436.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000401000 | + | 1 | 10 | 457_950 | 68 | 1436.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369067 | + | 1 | 3 | 1054_1057 | 68 | 153.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369067 | + | 1 | 3 | 1152_1158 | 68 | 153.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369067 | + | 1 | 3 | 1195_1459 | 68 | 153.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369067 | + | 1 | 3 | 457_950 | 68 | 153.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369068 | + | 1 | 11 | 1054_1057 | 68 | 1462.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369068 | + | 1 | 11 | 1152_1158 | 68 | 1462.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369068 | + | 1 | 11 | 1195_1459 | 68 | 1462.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369068 | + | 1 | 11 | 457_950 | 68 | 1462.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000401000 | + | 1 | 10 | 1054_1057 | 68 | 1436.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000401000 | + | 1 | 10 | 1152_1158 | 68 | 1436.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000401000 | + | 1 | 10 | 1195_1459 | 68 | 1436.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000401000 | + | 1 | 10 | 457_950 | 68 | 1436.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369067 | - | 1 | 3 | 1054_1057 | 0 | 153.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369067 | - | 1 | 3 | 1152_1158 | 0 | 153.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369067 | - | 1 | 3 | 1195_1459 | 0 | 153.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369067 | - | 1 | 3 | 457_950 | 0 | 153.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369067 | - | 1 | 3 | 4_19 | 0 | 153.0 | Compositional bias | Note=Gly/Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369068 | - | 6 | 11 | 1054_1057 | 231 | 1462.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369068 | - | 6 | 11 | 1152_1158 | 231 | 1462.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369068 | - | 6 | 11 | 1195_1459 | 231 | 1462.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369068 | - | 6 | 11 | 457_950 | 231 | 1462.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000401000 | - | 5 | 10 | 1054_1057 | 205 | 1436.0 | Compositional bias | Note=Poly-Gln |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000401000 | - | 5 | 10 | 1152_1158 | 205 | 1436.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000401000 | - | 5 | 10 | 1195_1459 | 205 | 1436.0 | Compositional bias | Note=Pro-rich |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000401000 | - | 5 | 10 | 457_950 | 205 | 1436.0 | Compositional bias | Note=Ser-rich |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369067 | + | 1 | 3 | 19_149 | 68 | 153.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000369068 | + | 1 | 11 | 19_149 | 68 | 1462.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150337395 | chr1:150232546 | ENST00000401000 | + | 1 | 10 | 19_149 | 68 | 1436.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369067 | + | 1 | 3 | 19_149 | 68 | 153.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000369068 | + | 1 | 11 | 19_149 | 68 | 1462.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150337395 | chr1:150236993 | ENST00000401000 | + | 1 | 10 | 19_149 | 68 | 1436.0 | Domain | CID |
| Hgene | RPRD2 | chr1:150416853 | chr1:150232546 | ENST00000369067 | - | 1 | 3 | 19_149 | 0 | 153.0 | Domain | CID |
| Tgene | CA14 | chr1:150337395 | chr1:150236993 | ENST00000369111 | 9 | 11 | 20_278 | 315 | 338.0 | Domain | Alpha-carbonic anhydrase | |
| Tgene | CA14 | chr1:150337395 | chr1:150236993 | ENST00000369111 | 9 | 11 | 217_218 | 315 | 338.0 | Region | Substrate binding | |
| Tgene | CA14 | chr1:150337395 | chr1:150236993 | ENST00000369111 | 9 | 11 | 16_290 | 315 | 338.0 | Topological domain | Extracellular | |
| Tgene | CA14 | chr1:150337395 | chr1:150236993 | ENST00000369111 | 9 | 11 | 312_337 | 315 | 338.0 | Topological domain | Cytoplasmic | |
| Tgene | CA14 | chr1:150337395 | chr1:150236993 | ENST00000369111 | 9 | 11 | 291_311 | 315 | 338.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for RPRD2-CA14 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >77255_77255_1_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369067_CA14_chr1_150232546_ENST00000369111_length(transcript)=1590nt_BP=212nt CGGGAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGA TCGAAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTAT CGTCTATCATTGGATGAAGTGGCTCCGGAGATGTCAACACTGGACGTATGAGGGCCCACATGGTCAGGACCATTGGCCAGCCTCTTACCC TGAGTGTGGAAACAATGCCCAGTCGCCCATCGATATTCAGACAGACAGTGTGACATTTGACCCTGATTTGCCTGCTCTGCAGCCCCACGG ATATGACCAGCCTGGCACCGAGCCTTTGGACCTGCACAACAATGGCCACACAGTGCAACTCTCTCTGCCCTCTACCCTGTATCTGGGTGG ACTTCCCCGAAAATATGTAGCTGCCCAGCTCCACCTGCACTGGGGTCAGAAAGGATCCCCAGGGGGGTCAGAACACCAGATCAACAGTGA AGCCACATTTGCAGAGCTCCACATTGTACATTATGACTCTGATTCCTATGACAGCTTGAGTGAGGCTGCTGAGAGGCCTCAGGGCCTGGC TGTCCTGGGCATCCTAATTGAGGTGGGTGAGACTAAGAATATAGCTTATGAACACATTCTGAGTCACTTGCATGAAGTCAGGCATAAAGA TCAGAAGACCTCAGTGCCTCCCTTCAACCTAAGAGAGCTGCTCCCCAAACAGCTGGGGCAGTACTTCCGCTACAATGGCTCGCTCACAAC TCCCCCTTGCTACCAGAGTGTGCTCTGGACAGTTTTTTATAGAAGGTCCCAGATTTCAATGGAACAGCTGGAAAAGCTTCAGGGGACATT GTTCTCCACAGAAGAGGAGCCCTCTAAGCTTCTGGTACAGAACTACCGAGCCCTTCAGCCTCTCAATCAGCGCATGGTCTTTGCTTCTTT CATCCAAGCAGGATCCTCGTATACCACAGGTGAAATGCTGAGTCTAGGTGTAGGAATCTTGGTTGGCTGTCTCTGCCTTCTCCTGGCTGT TTATTTCATTGCTAGAAAGATTCGGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACTGAGGCATA AATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGGCCAGAAAC ACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAGAACTGCAG AGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTGCAGGGGAA GTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGATAAAGAGT >77255_77255_1_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369067_CA14_chr1_150232546_ENST00000369111_length(amino acids)=387AA_BP=69 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRCQHWTYEGPHGQDHWPASYPEC GNNAQSPIDIQTDSVTFDPDLPALQPHGYDQPGTEPLDLHNNGHTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEAT FAELHIVHYDSDSYDSLSEAAERPQGLAVLGILIEVGETKNIAYEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPP CYQSVLWTVFYRRSQISMEQLEKLQGTLFSTEEEPSKLLVQNYRALQPLNQRMVFASFIQAGSSYTTGEMLSLGVGILVGCLCLLLAVYF -------------------------------------------------------------- >77255_77255_2_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369067_CA14_chr1_150236993_ENST00000369111_length(transcript)=698nt_BP=212nt CGGGAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGA TCGAAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTAT CGTCTATCATTGGATGAAGTGGCTCCGGAGATGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACT GAGGCATAAATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGG CCAGAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAG AACTGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTG CAGGGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGA >77255_77255_2_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369067_CA14_chr1_150236993_ENST00000369111_length(amino acids)=75AA_BP= -------------------------------------------------------------- >77255_77255_3_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369068_CA14_chr1_150232546_ENST00000369111_length(transcript)=1587nt_BP=209nt GAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGATCG AAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTATCGT CTATCATTGGATGAAGTGGCTCCGGAGATGTCAACACTGGACGTATGAGGGCCCACATGGTCAGGACCATTGGCCAGCCTCTTACCCTGA GTGTGGAAACAATGCCCAGTCGCCCATCGATATTCAGACAGACAGTGTGACATTTGACCCTGATTTGCCTGCTCTGCAGCCCCACGGATA TGACCAGCCTGGCACCGAGCCTTTGGACCTGCACAACAATGGCCACACAGTGCAACTCTCTCTGCCCTCTACCCTGTATCTGGGTGGACT TCCCCGAAAATATGTAGCTGCCCAGCTCCACCTGCACTGGGGTCAGAAAGGATCCCCAGGGGGGTCAGAACACCAGATCAACAGTGAAGC CACATTTGCAGAGCTCCACATTGTACATTATGACTCTGATTCCTATGACAGCTTGAGTGAGGCTGCTGAGAGGCCTCAGGGCCTGGCTGT CCTGGGCATCCTAATTGAGGTGGGTGAGACTAAGAATATAGCTTATGAACACATTCTGAGTCACTTGCATGAAGTCAGGCATAAAGATCA GAAGACCTCAGTGCCTCCCTTCAACCTAAGAGAGCTGCTCCCCAAACAGCTGGGGCAGTACTTCCGCTACAATGGCTCGCTCACAACTCC CCCTTGCTACCAGAGTGTGCTCTGGACAGTTTTTTATAGAAGGTCCCAGATTTCAATGGAACAGCTGGAAAAGCTTCAGGGGACATTGTT CTCCACAGAAGAGGAGCCCTCTAAGCTTCTGGTACAGAACTACCGAGCCCTTCAGCCTCTCAATCAGCGCATGGTCTTTGCTTCTTTCAT CCAAGCAGGATCCTCGTATACCACAGGTGAAATGCTGAGTCTAGGTGTAGGAATCTTGGTTGGCTGTCTCTGCCTTCTCCTGGCTGTTTA TTTCATTGCTAGAAAGATTCGGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACTGAGGCATAAAT TCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGGCCAGAAACACT GTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAGAACTGCAGAGC CTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTGCAGGGGAAGTT TGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGATAAAGAGTTGC >77255_77255_3_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369068_CA14_chr1_150232546_ENST00000369111_length(amino acids)=387AA_BP=69 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRCQHWTYEGPHGQDHWPASYPEC GNNAQSPIDIQTDSVTFDPDLPALQPHGYDQPGTEPLDLHNNGHTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEAT FAELHIVHYDSDSYDSLSEAAERPQGLAVLGILIEVGETKNIAYEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPP CYQSVLWTVFYRRSQISMEQLEKLQGTLFSTEEEPSKLLVQNYRALQPLNQRMVFASFIQAGSSYTTGEMLSLGVGILVGCLCLLLAVYF -------------------------------------------------------------- >77255_77255_4_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369068_CA14_chr1_150236993_ENST00000369111_length(transcript)=695nt_BP=209nt GAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGATCG AAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTATCGT CTATCATTGGATGAAGTGGCTCCGGAGATGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACTGAG GCATAAATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGGCCA GAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAGAAC TGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTGCAG GGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGATAA >77255_77255_4_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000369068_CA14_chr1_150236993_ENST00000369111_length(amino acids)=75AA_BP= -------------------------------------------------------------- >77255_77255_5_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000401000_CA14_chr1_150232546_ENST00000369111_length(transcript)=1648nt_BP=270nt CCGCTCCCGCCGCCGCCGCCGCCGCCGCCGCCAGAGGAGCAGCAGCGCTTGTGCAAACCGGGAAGATGGCGGCCGGCGGCGGCGGAGGCA GCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGATCGAAAATTCCAGTCGGTAACCAACACCATGG AGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTATCGTCTATCATTGGATGAAGTGGCTCCGGAGAT GTCAACACTGGACGTATGAGGGCCCACATGGTCAGGACCATTGGCCAGCCTCTTACCCTGAGTGTGGAAACAATGCCCAGTCGCCCATCG ATATTCAGACAGACAGTGTGACATTTGACCCTGATTTGCCTGCTCTGCAGCCCCACGGATATGACCAGCCTGGCACCGAGCCTTTGGACC TGCACAACAATGGCCACACAGTGCAACTCTCTCTGCCCTCTACCCTGTATCTGGGTGGACTTCCCCGAAAATATGTAGCTGCCCAGCTCC ACCTGCACTGGGGTCAGAAAGGATCCCCAGGGGGGTCAGAACACCAGATCAACAGTGAAGCCACATTTGCAGAGCTCCACATTGTACATT ATGACTCTGATTCCTATGACAGCTTGAGTGAGGCTGCTGAGAGGCCTCAGGGCCTGGCTGTCCTGGGCATCCTAATTGAGGTGGGTGAGA CTAAGAATATAGCTTATGAACACATTCTGAGTCACTTGCATGAAGTCAGGCATAAAGATCAGAAGACCTCAGTGCCTCCCTTCAACCTAA GAGAGCTGCTCCCCAAACAGCTGGGGCAGTACTTCCGCTACAATGGCTCGCTCACAACTCCCCCTTGCTACCAGAGTGTGCTCTGGACAG TTTTTTATAGAAGGTCCCAGATTTCAATGGAACAGCTGGAAAAGCTTCAGGGGACATTGTTCTCCACAGAAGAGGAGCCCTCTAAGCTTC TGGTACAGAACTACCGAGCCCTTCAGCCTCTCAATCAGCGCATGGTCTTTGCTTCTTTCATCCAAGCAGGATCCTCGTATACCACAGGTG AAATGCTGAGTCTAGGTGTAGGAATCTTGGTTGGCTGTCTCTGCCTTCTCCTGGCTGTTTATTTCATTGCTAGAAAGATTCGGAAGAAGA GGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACTGAGGCATAAATTCCTTCTCAGATACCATGGATGTGGATGA CTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTC CCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAA ATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCC TCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGATAAAGAGTTGCTGTTGAAGTTGTATATTTTTGATCAATAT >77255_77255_5_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000401000_CA14_chr1_150232546_ENST00000369111_length(amino acids)=387AA_BP=69 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRCQHWTYEGPHGQDHWPASYPEC GNNAQSPIDIQTDSVTFDPDLPALQPHGYDQPGTEPLDLHNNGHTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEAT FAELHIVHYDSDSYDSLSEAAERPQGLAVLGILIEVGETKNIAYEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPP CYQSVLWTVFYRRSQISMEQLEKLQGTLFSTEEEPSKLLVQNYRALQPLNQRMVFASFIQAGSSYTTGEMLSLGVGILVGCLCLLLAVYF -------------------------------------------------------------- >77255_77255_6_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000401000_CA14_chr1_150236993_ENST00000369111_length(transcript)=756nt_BP=270nt CCGCTCCCGCCGCCGCCGCCGCCGCCGCCGCCAGAGGAGCAGCAGCGCTTGTGCAAACCGGGAAGATGGCGGCCGGCGGCGGCGGAGGCA GCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGATCGAAAATTCCAGTCGGTAACCAACACCATGG AGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTATCGTCTATCATTGGATGAAGTGGCTCCGGAGAT GAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACTGAGGCATAAATTCCTTCTCAGATACCATGGAT GTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGT CCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGT AGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCT CTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGATAAAGAGTTGCTGTTGAAGTTGTATATTTTTG >77255_77255_6_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000401000_CA14_chr1_150236993_ENST00000369111_length(amino acids)=82AA_BP= -------------------------------------------------------------- >77255_77255_7_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000539519_CA14_chr1_150232546_ENST00000369111_length(transcript)=1605nt_BP=227nt AGCGCTTGTGCAAACCGGGAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCT GGAGTCCTCGTTGGATCGAAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAA ACACCACAGTACTATCGTCTATCATTGGATGAAGTGGCTCCGGAGATGTCAACACTGGACGTATGAGGGCCCACATGGTCAGGACCATTG GCCAGCCTCTTACCCTGAGTGTGGAAACAATGCCCAGTCGCCCATCGATATTCAGACAGACAGTGTGACATTTGACCCTGATTTGCCTGC TCTGCAGCCCCACGGATATGACCAGCCTGGCACCGAGCCTTTGGACCTGCACAACAATGGCCACACAGTGCAACTCTCTCTGCCCTCTAC CCTGTATCTGGGTGGACTTCCCCGAAAATATGTAGCTGCCCAGCTCCACCTGCACTGGGGTCAGAAAGGATCCCCAGGGGGGTCAGAACA CCAGATCAACAGTGAAGCCACATTTGCAGAGCTCCACATTGTACATTATGACTCTGATTCCTATGACAGCTTGAGTGAGGCTGCTGAGAG GCCTCAGGGCCTGGCTGTCCTGGGCATCCTAATTGAGGTGGGTGAGACTAAGAATATAGCTTATGAACACATTCTGAGTCACTTGCATGA AGTCAGGCATAAAGATCAGAAGACCTCAGTGCCTCCCTTCAACCTAAGAGAGCTGCTCCCCAAACAGCTGGGGCAGTACTTCCGCTACAA TGGCTCGCTCACAACTCCCCCTTGCTACCAGAGTGTGCTCTGGACAGTTTTTTATAGAAGGTCCCAGATTTCAATGGAACAGCTGGAAAA GCTTCAGGGGACATTGTTCTCCACAGAAGAGGAGCCCTCTAAGCTTCTGGTACAGAACTACCGAGCCCTTCAGCCTCTCAATCAGCGCAT GGTCTTTGCTTCTTTCATCCAAGCAGGATCCTCGTATACCACAGGTGAAATGCTGAGTCTAGGTGTAGGAATCTTGGTTGGCTGTCTCTG CCTTCTCCTGGCTGTTTATTTCATTGCTAGAAAGATTCGGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGC CACGACTGAGGCATAAATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAG GATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTC CAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGT TTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTC >77255_77255_7_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000539519_CA14_chr1_150232546_ENST00000369111_length(amino acids)=387AA_BP=69 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRCQHWTYEGPHGQDHWPASYPEC GNNAQSPIDIQTDSVTFDPDLPALQPHGYDQPGTEPLDLHNNGHTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEAT FAELHIVHYDSDSYDSLSEAAERPQGLAVLGILIEVGETKNIAYEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPP CYQSVLWTVFYRRSQISMEQLEKLQGTLFSTEEEPSKLLVQNYRALQPLNQRMVFASFIQAGSSYTTGEMLSLGVGILVGCLCLLLAVYF -------------------------------------------------------------- >77255_77255_8_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000539519_CA14_chr1_150236993_ENST00000369111_length(transcript)=713nt_BP=227nt AGCGCTTGTGCAAACCGGGAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCT GGAGTCCTCGTTGGATCGAAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAA ACACCACAGTACTATCGTCTATCATTGGATGAAGTGGCTCCGGAGATGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCA GCACAAGCCACGACTGAGGCATAAATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATG GGGTGTAGGATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGC TGTCATTCCAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACA AACTCTGTTTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGG >77255_77255_8_RPRD2-CA14_RPRD2_chr1_150337395_ENST00000539519_CA14_chr1_150236993_ENST00000369111_length(amino acids)=75AA_BP= -------------------------------------------------------------- >77255_77255_9_RPRD2-CA14_RPRD2_chr1_150416853_ENST00000369068_CA14_chr1_150232546_ENST00000369111_length(transcript)=2076nt_BP=698nt GAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGATCG AAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTATCGT CTATCATTGGATGAAGTGGCTCCGGAGATCTGCATATCCCCACCGTTTGAATCTCTTTTACCTTGCCAATGATGTCATACAGAACTGTAA AAGGAAAAATGCAATCATATTCCGTGAATCATTTGCTGATGTACTTCCTGAAGCAGCTGCTCTAGTGAAGGATCCATCTGTCTCTAAGTC TGTAGAACGAATCTTTAAAATCTGGGAAGATAGAAATGTATACCCAGAAGAAATGATTGTGGCATTGAGAGAAGCTTTGAGTACCACTTT CAAAACTCAGAAGCAGCTGAAAGAAAATCTGAACAAACAACCGAATAAGCAGTGGAAGAAATCACAAACATCTACAAATCCAAAAGCTGC TCTCAAGTCTAAGATAGTTGCTGAATTTCGATCTCAGGCCCTAATTGAAGAGCTGTTGCTATACAAGCGCTCAGAAGATCAGATAGAACT GAAGGAAAAGCAGTTGTCAACTATGAGGGTGGATGTGTGCAGCACAGAAACTCTCAAATGCTTAAAAGGTCAACACTGGACGTATGAGGG CCCACATGGTCAGGACCATTGGCCAGCCTCTTACCCTGAGTGTGGAAACAATGCCCAGTCGCCCATCGATATTCAGACAGACAGTGTGAC ATTTGACCCTGATTTGCCTGCTCTGCAGCCCCACGGATATGACCAGCCTGGCACCGAGCCTTTGGACCTGCACAACAATGGCCACACAGT GCAACTCTCTCTGCCCTCTACCCTGTATCTGGGTGGACTTCCCCGAAAATATGTAGCTGCCCAGCTCCACCTGCACTGGGGTCAGAAAGG ATCCCCAGGGGGGTCAGAACACCAGATCAACAGTGAAGCCACATTTGCAGAGCTCCACATTGTACATTATGACTCTGATTCCTATGACAG CTTGAGTGAGGCTGCTGAGAGGCCTCAGGGCCTGGCTGTCCTGGGCATCCTAATTGAGGTGGGTGAGACTAAGAATATAGCTTATGAACA CATTCTGAGTCACTTGCATGAAGTCAGGCATAAAGATCAGAAGACCTCAGTGCCTCCCTTCAACCTAAGAGAGCTGCTCCCCAAACAGCT GGGGCAGTACTTCCGCTACAATGGCTCGCTCACAACTCCCCCTTGCTACCAGAGTGTGCTCTGGACAGTTTTTTATAGAAGGTCCCAGAT TTCAATGGAACAGCTGGAAAAGCTTCAGGGGACATTGTTCTCCACAGAAGAGGAGCCCTCTAAGCTTCTGGTACAGAACTACCGAGCCCT TCAGCCTCTCAATCAGCGCATGGTCTTTGCTTCTTTCATCCAAGCAGGATCCTCGTATACCACAGGTGAAATGCTGAGTCTAGGTGTAGG AATCTTGGTTGGCTGTCTCTGCCTTCTCCTGGCTGTTTATTTCATTGCTAGAAAGATTCGGAAGAAGAGGCTGGAAAACCGAAAGAGTGT GGTCTTCACCTCAGCACAAGCCACGACTGAGGCATAAATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGA AGCCTCTAAAATGGGGTGTAGGATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGG AATGGACCCAGGCTGTCATTCCAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTT AATGCAGAGAACAAACTCTGTTTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCT AGATATACTGCGGGATCTCTCCTTAGGATAAAGAGTTGCTGTTGAAGTTGTATATTTTTGATCAATATATTTGGAAATTAAAGTTTCTGA >77255_77255_9_RPRD2-CA14_RPRD2_chr1_150416853_ENST00000369068_CA14_chr1_150232546_ENST00000369111_length(amino acids)=550AA_BP=226 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRSAYPHRLNLFYLANDVIQNCKR KNAIIFRESFADVLPEAAALVKDPSVSKSVERIFKIWEDRNVYPEEMIVALREALSTTFKTQKQLKENLNKQPNKQWKKSQTSTNPKAAL KSKIVAEFRSQALIEELLLYKRSEDQIELKEKQLSTMRVDVCSTETLKCLKGQHWTYEGPHGQDHWPASYPECGNNAQSPIDIQTDSVTF DPDLPALQPHGYDQPGTEPLDLHNNGHTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEATFAELHIVHYDSDSYDSL SEAAERPQGLAVLGILIEVGETKNIAYEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPPCYQSVLWTVFYRRSQIS MEQLEKLQGTLFSTEEEPSKLLVQNYRALQPLNQRMVFASFIQAGSSYTTGEMLSLGVGILVGCLCLLLAVYFIARKIRKKRLENRKSVV -------------------------------------------------------------- >77255_77255_10_RPRD2-CA14_RPRD2_chr1_150416853_ENST00000401000_CA14_chr1_150232546_ENST00000369111_length(transcript)=2059nt_BP=681nt CCGCTCCCGCCGCCGCCGCCGCCGCCGCCGCCAGAGGAGCAGCAGCGCTTGTGCAAACCGGGAAGATGGCGGCCGGCGGCGGCGGAGGCA GCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCTGGAGTCCTCGTTGGATCGAAAATTCCAGTCGGTAACCAACACCATGG AGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAAACACCACAGTACTATCGTCTATCATTGGATGAAGTGGCTCCGGAGAT CTGCATATCCCCACCGTTTGAATCTCTTTTACCTTGCCAATGATGTCATACAGAACTGTAAAAGGAAAAATGCAATCATATTCCGTGAAT CATTTGCTGATGTACTTCCTGAAGCAGCTGCTCTAGTGAAGGATCCATCTGTCTCTAAGTCTGTAGAACGAATCTTTAAAATCTGGGAAG ATAGAAATGTATACCCAGAAGAAATGATTGTGGCATTGAGAGAAGCTTTGACATCTACAAATCCAAAAGCTGCTCTCAAGTCTAAGATAG TTGCTGAATTTCGATCTCAGGCCCTAATTGAAGAGCTGTTGCTATACAAGCGCTCAGAAGATCAGATAGAACTGAAGGAAAAGCAGTTGT CAACTATGAGGGTGGATGTGTGCAGCACAGAAACTCTCAAATGCTTAAAAGGTCAACACTGGACGTATGAGGGCCCACATGGTCAGGACC ATTGGCCAGCCTCTTACCCTGAGTGTGGAAACAATGCCCAGTCGCCCATCGATATTCAGACAGACAGTGTGACATTTGACCCTGATTTGC CTGCTCTGCAGCCCCACGGATATGACCAGCCTGGCACCGAGCCTTTGGACCTGCACAACAATGGCCACACAGTGCAACTCTCTCTGCCCT CTACCCTGTATCTGGGTGGACTTCCCCGAAAATATGTAGCTGCCCAGCTCCACCTGCACTGGGGTCAGAAAGGATCCCCAGGGGGGTCAG AACACCAGATCAACAGTGAAGCCACATTTGCAGAGCTCCACATTGTACATTATGACTCTGATTCCTATGACAGCTTGAGTGAGGCTGCTG AGAGGCCTCAGGGCCTGGCTGTCCTGGGCATCCTAATTGAGGTGGGTGAGACTAAGAATATAGCTTATGAACACATTCTGAGTCACTTGC ATGAAGTCAGGCATAAAGATCAGAAGACCTCAGTGCCTCCCTTCAACCTAAGAGAGCTGCTCCCCAAACAGCTGGGGCAGTACTTCCGCT ACAATGGCTCGCTCACAACTCCCCCTTGCTACCAGAGTGTGCTCTGGACAGTTTTTTATAGAAGGTCCCAGATTTCAATGGAACAGCTGG AAAAGCTTCAGGGGACATTGTTCTCCACAGAAGAGGAGCCCTCTAAGCTTCTGGTACAGAACTACCGAGCCCTTCAGCCTCTCAATCAGC GCATGGTCTTTGCTTCTTTCATCCAAGCAGGATCCTCGTATACCACAGGTGAAATGCTGAGTCTAGGTGTAGGAATCTTGGTTGGCTGTC TCTGCCTTCTCCTGGCTGTTTATTTCATTGCTAGAAAGATTCGGAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCAC AAGCCACGACTGAGGCATAAATTCCTTCTCAGATACCATGGATGTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGT GTAGGATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGTCCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTC ATTCCAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGTAGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACT CTGTTTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCTCTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATC >77255_77255_10_RPRD2-CA14_RPRD2_chr1_150416853_ENST00000401000_CA14_chr1_150232546_ENST00000369111_length(amino acids)=524AA_BP=200 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRSAYPHRLNLFYLANDVIQNCKR KNAIIFRESFADVLPEAAALVKDPSVSKSVERIFKIWEDRNVYPEEMIVALREALTSTNPKAALKSKIVAEFRSQALIEELLLYKRSEDQ IELKEKQLSTMRVDVCSTETLKCLKGQHWTYEGPHGQDHWPASYPECGNNAQSPIDIQTDSVTFDPDLPALQPHGYDQPGTEPLDLHNNG HTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEATFAELHIVHYDSDSYDSLSEAAERPQGLAVLGILIEVGETKNIA YEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPPCYQSVLWTVFYRRSQISMEQLEKLQGTLFSTEEEPSKLLVQNY -------------------------------------------------------------- >77255_77255_11_RPRD2-CA14_RPRD2_chr1_150416853_ENST00000539519_CA14_chr1_150232546_ENST00000369111_length(transcript)=2016nt_BP=638nt AGCGCTTGTGCAAACCGGGAAGATGGCGGCCGGCGGCGGCGGAGGCAGCAGTAAGGCCTCCTCCTCGTCGGCCTCTTCGGCAGGGGCTCT GGAGTCCTCGTTGGATCGAAAATTCCAGTCGGTAACCAACACCATGGAGTCCATTCAAGGCTTGTCGTCTTGGTGTATAGAGAACAAAAA ACACCACAGTACTATCGTCTATCATTGGATGAAGTGGCTCCGGAGATCTGCATATCCCCACCGTTTGAATCTCTTTTACCTTGCCAATGA TGTCATACAGAACTGTAAAAGGAAAAATGCAATCATATTCCGTGAATCATTTGCTGATGTACTTCCTGAAGCAGCTGCTCTAGTGAAGGA TCCATCTGTCTCTAAGTCTGTAGAACGAATCTTTAAAATCTGGGAAGATAGAAATGTATACCCAGAAGAAATGATTGTGGCATTGAGAGA AGCTTTGACATCTACAAATCCAAAAGCTGCTCTCAAGTCTAAGATAGTTGCTGAATTTCGATCTCAGGCCCTAATTGAAGAGCTGTTGCT ATACAAGCGCTCAGAAGATCAGATAGAACTGAAGGAAAAGCAGTTGTCAACTATGAGGGTGGATGTGTGCAGCACAGAAACTCTCAAATG CTTAAAAGGTCAACACTGGACGTATGAGGGCCCACATGGTCAGGACCATTGGCCAGCCTCTTACCCTGAGTGTGGAAACAATGCCCAGTC GCCCATCGATATTCAGACAGACAGTGTGACATTTGACCCTGATTTGCCTGCTCTGCAGCCCCACGGATATGACCAGCCTGGCACCGAGCC TTTGGACCTGCACAACAATGGCCACACAGTGCAACTCTCTCTGCCCTCTACCCTGTATCTGGGTGGACTTCCCCGAAAATATGTAGCTGC CCAGCTCCACCTGCACTGGGGTCAGAAAGGATCCCCAGGGGGGTCAGAACACCAGATCAACAGTGAAGCCACATTTGCAGAGCTCCACAT TGTACATTATGACTCTGATTCCTATGACAGCTTGAGTGAGGCTGCTGAGAGGCCTCAGGGCCTGGCTGTCCTGGGCATCCTAATTGAGGT GGGTGAGACTAAGAATATAGCTTATGAACACATTCTGAGTCACTTGCATGAAGTCAGGCATAAAGATCAGAAGACCTCAGTGCCTCCCTT CAACCTAAGAGAGCTGCTCCCCAAACAGCTGGGGCAGTACTTCCGCTACAATGGCTCGCTCACAACTCCCCCTTGCTACCAGAGTGTGCT CTGGACAGTTTTTTATAGAAGGTCCCAGATTTCAATGGAACAGCTGGAAAAGCTTCAGGGGACATTGTTCTCCACAGAAGAGGAGCCCTC TAAGCTTCTGGTACAGAACTACCGAGCCCTTCAGCCTCTCAATCAGCGCATGGTCTTTGCTTCTTTCATCCAAGCAGGATCCTCGTATAC CACAGGTGAAATGCTGAGTCTAGGTGTAGGAATCTTGGTTGGCTGTCTCTGCCTTCTCCTGGCTGTTTATTTCATTGCTAGAAAGATTCG GAAGAAGAGGCTGGAAAACCGAAAGAGTGTGGTCTTCACCTCAGCACAAGCCACGACTGAGGCATAAATTCCTTCTCAGATACCATGGAT GTGGATGACTTCCCTTCATGCCTATCAGGAAGCCTCTAAAATGGGGTGTAGGATCTGGCCAGAAACACTGTAGGAGTAGTAAGCAGATGT CCTCCTTCCCCTGGACATCTCCTAGAGAGGAATGGACCCAGGCTGTCATTCCAGGAAGAACTGCAGAGCCTTCAGCCTCTCCAAACATGT AGGAGGAAATGAGGAAATCGCTGTGTTGTTAATGCAGAGAACAAACTCTGTTTAGTTGCAGGGGAAGTTTGGGATATACCCCAAAGTCCT CTACCCCCTCACTTTTATGGCCCTTTCCCTAGATATACTGCGGGATCTCTCCTTAGGATAAAGAGTTGCTGTTGAAGTTGTATATTTTTG >77255_77255_11_RPRD2-CA14_RPRD2_chr1_150416853_ENST00000539519_CA14_chr1_150232546_ENST00000369111_length(amino acids)=524AA_BP=200 MAAGGGGGSSKASSSSASSAGALESSLDRKFQSVTNTMESIQGLSSWCIENKKHHSTIVYHWMKWLRRSAYPHRLNLFYLANDVIQNCKR KNAIIFRESFADVLPEAAALVKDPSVSKSVERIFKIWEDRNVYPEEMIVALREALTSTNPKAALKSKIVAEFRSQALIEELLLYKRSEDQ IELKEKQLSTMRVDVCSTETLKCLKGQHWTYEGPHGQDHWPASYPECGNNAQSPIDIQTDSVTFDPDLPALQPHGYDQPGTEPLDLHNNG HTVQLSLPSTLYLGGLPRKYVAAQLHLHWGQKGSPGGSEHQINSEATFAELHIVHYDSDSYDSLSEAAERPQGLAVLGILIEVGETKNIA YEHILSHLHEVRHKDQKTSVPPFNLRELLPKQLGQYFRYNGSLTTPPCYQSVLWTVFYRRSQISMEQLEKLQGTLFSTEEEPSKLLVQNY -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RPRD2-CA14 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RPRD2-CA14 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RPRD2-CA14 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
