
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RPS20-POMT2 (FusionGDB2 ID:77582) |
Fusion Gene Summary for RPS20-POMT2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RPS20-POMT2 | Fusion gene ID: 77582 | Hgene | Tgene | Gene symbol | RPS20 | POMT2 | Gene ID | 6224 | 29954 |
| Gene name | ribosomal protein S20 | protein O-mannosyltransferase 2 | |
| Synonyms | S20|uS10 | LGMD2N|LGMDR14|MDDGA2|MDDGB2|MDDGC2 | |
| Cytomap | 8q12.1 | 14q24.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | 40S ribosomal protein S20small ribosomal subunit protein uS10 | protein O-mannosyl-transferase 2dolichyl-phosphate-mannose--protein mannosyltransferase 2 | |
| Modification date | 20200327 | 20200328 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000009589, ENST00000518875, ENST00000519606, ENST00000519807, ENST00000521262, ENST00000523936, ENST00000520490, ENST00000524349, ENST00000520627, | ENST00000556880, ENST00000261534, | |
| Fusion gene scores | * DoF score | 25 X 13 X 7=2275 | 7 X 8 X 4=224 |
| # samples | 26 | 7 | |
| ** MAII score | log2(26/2275*10)=-3.12928301694497 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/224*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: RPS20 [Title/Abstract] AND POMT2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RPS20(56986618)-POMT2(77750216), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | RPS20-POMT2 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. RPS20-POMT2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across RPS20 (5'-gene) Fusion gene breakpoints across RPS20 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
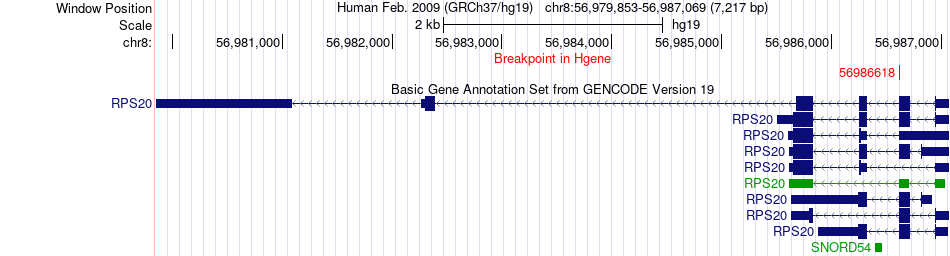 |
 Fusion gene breakpoints across POMT2 (3'-gene) Fusion gene breakpoints across POMT2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
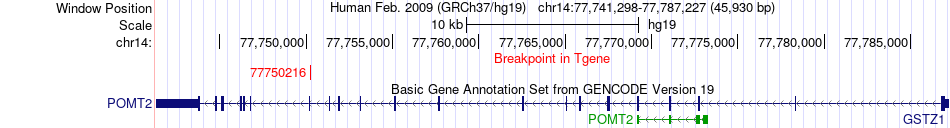 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CR-6467 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
Top |
Fusion Gene ORF analysis for RPS20-POMT2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000009589 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5CDS-intron | ENST00000518875 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5CDS-intron | ENST00000519606 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5CDS-intron | ENST00000519807 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5CDS-intron | ENST00000521262 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5CDS-intron | ENST00000523936 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5UTR-3CDS | ENST00000520490 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5UTR-3CDS | ENST00000524349 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5UTR-intron | ENST00000520490 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| 5UTR-intron | ENST00000524349 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| Frame-shift | ENST00000519807 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| In-frame | ENST00000009589 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| In-frame | ENST00000518875 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| In-frame | ENST00000519606 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| In-frame | ENST00000521262 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| In-frame | ENST00000523936 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| intron-3CDS | ENST00000520627 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
| intron-intron | ENST00000520627 | ENST00000556880 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000009589 | RPS20 | chr8 | 56986618 | - | ENST00000261534 | POMT2 | chr14 | 77750216 | - | 3323 | 226 | 123 | 902 | 259 |
| ENST00000521262 | RPS20 | chr8 | 56986618 | - | ENST00000261534 | POMT2 | chr14 | 77750216 | - | 3454 | 357 | 254 | 1033 | 259 |
| ENST00000523936 | RPS20 | chr8 | 56986618 | - | ENST00000261534 | POMT2 | chr14 | 77750216 | - | 3300 | 203 | 100 | 879 | 259 |
| ENST00000519606 | RPS20 | chr8 | 56986618 | - | ENST00000261534 | POMT2 | chr14 | 77750216 | - | 3323 | 226 | 123 | 902 | 259 |
| ENST00000518875 | RPS20 | chr8 | 56986618 | - | ENST00000261534 | POMT2 | chr14 | 77750216 | - | 3315 | 218 | 115 | 894 | 259 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000009589 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - | 0.086247556 | 0.9137524 |
| ENST00000521262 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - | 0.082839824 | 0.91716015 |
| ENST00000523936 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - | 0.08997406 | 0.9100259 |
| ENST00000519606 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - | 0.086247556 | 0.9137524 |
| ENST00000518875 | ENST00000261534 | RPS20 | chr8 | 56986618 | - | POMT2 | chr14 | 77750216 | - | 0.087432906 | 0.91256714 |
Top |
Fusion Genomic Features for RPS20-POMT2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
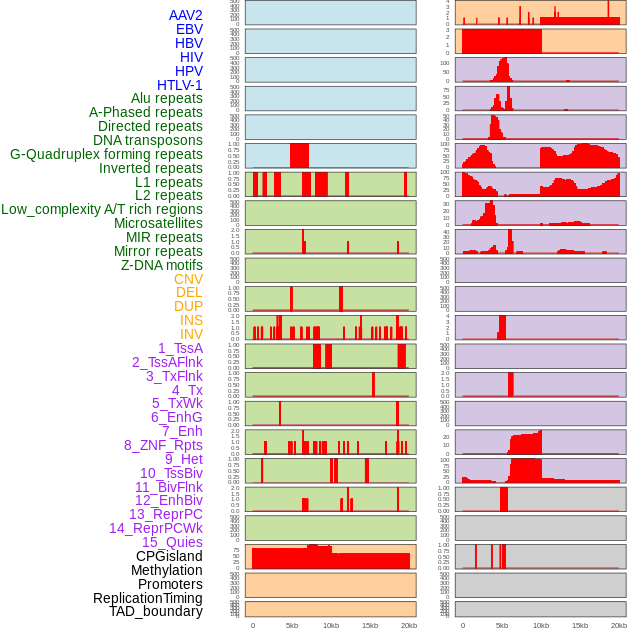 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for RPS20-POMT2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr8:56986618/chr14:77750216) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 596_616 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 643_663 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 665_685 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 700_720 | 525 | 751.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 334_390 | 525 | 751.0 | Domain | MIR 1 | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 403_459 | 525 | 751.0 | Domain | MIR 2 | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 464_521 | 525 | 751.0 | Domain | MIR 3 | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 100_120 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 146_166 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 191_211 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 231_251 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 283_303 | 525 | 751.0 | Transmembrane | Helical | |
| Tgene | POMT2 | chr8:56986618 | chr14:77750216 | ENST00000261534 | 13 | 21 | 54_74 | 525 | 751.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for RPS20-POMT2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >77582_77582_1_RPS20-POMT2_RPS20_chr8_56986618_ENST00000009589_POMT2_chr14_77750216_ENST00000261534_length(transcript)=3323nt_BP=226nt CTTTTTGAGGAAGACGCGGTCGTAAGGGCTGAGGATTTTTGGTCCGCACGCTCCTGCTCCTGACTCACCGCTGTTCGCTCTCGCCGAGGA ACAAGTCGGTCAGGAAGCCCGCGCGCAACAGCCATGGCTTTTAAGGATACCGGAAAAACACCCGTGGAGCCGGAGGTGGCAATTCACCGA ATTCGAATCACCCTAACAAGCCGCAACGTAAAATCCTTGGAAAAGGTGCCAAACATCAGCCTGGATGTGCTACAGCCCAGTTTTCCTGAG ATCTTGCTGGAATCCCACATGGTCATGATCCGGGGGAACAGTGGCCTCAAACCCAAGGACAATGAGTTCACGTCCAAACCCTGGCACTGG CCTATCAACTATCAGGGCCTACGCTTCTCAGGGGTCAATGACACAGATTTCCGAGTCTATCTGCTTGGCAACCCGGTGGTTTGGTGGCTG AATCTGTTGAGCATCGCCCTCTACCTCCTCTCAGGGAGCATCATTGCTGTAGCCATGCAGAGAGGGGCACGGCTGCCAGCGGAGGTTGCA GGGTTGTCCCAGGTCCTGCTTCGAGGAGGCGGCCAGGTCCTGCTCGGCTGGACACTCCATTACTTCCCGTTTTTCCTGATGGGCCGGGTC CTCTACTTCCACCACTACTTCCCAGCCATGCTCTTCTCAAGCATGTTGACAGGCATTCTGTGGGACACCCTCCTGCGGCTCTGTGCCTGG GGCTTGGCCTCATGGCCCCTGGCGAGGGGCATACATGTGGCGGGAATCCTGAGCCTGCTCCTGGGAACTGCCTACAGCTTCTACCTCTTC CACCCTCTGGCTTACGGGATGGTTGGTCCCCTGGCCCAGGACCCCCAAAGTCCAATGGCAGGACTAAGGTGGCTGGACTCATGGGACTTT TGAGGCCACTGCAAAGATTCCAGCCTGGGTCCAGGAACTTCCCAGGAGAGCCAGCTGGGAAGCAGTACTGGACCATCCGGCCATGAGGAA GCTGCCTAAGGAGCCCGAAGAATTCTGCTGCTCACCAGGACCCAGCTCTGGGAGTGCAGCTGGAATGGAACCCAGCATTGGAGAGAGCCG GGAGAGAGCCGCAGCTCCGTCCACAGCACCACGGAGAGGACAGCGCCTCTTGGGCTGCTCCCTCGTGCACAGCGCAGGGGAGGATGGGTG CCAGAGGGTGGGAGTGGGTGCGCGTGTGTGTATGTGAGGGCATGTTGTCCATATCCCTGTGGAATACAGACCGTGTAACTGGACAGCTGA GTGTCATAAACTCTAAAATAGTCCAGGCTATAGTAATATTTTCTGGTATTTTTTTCTGTTGGATTTCTTTGGTTTTGGTTTGTAAGATCC ATCAAAAATTCTCAGACCTCTGAGAGGGCCTTGAGGGCAGAGAATACTGACCTCACCTTGGGATGTCACCAAAGGCAGAGCAGATGGGAC TGTCAAAGTTGTGCTGCCCTCCCTCCCACAAACAAATGAATCCAGCGTGGAGGAAGGCGCGCCGCCCCGAGCTGGAGGGCCTGGCTCCCA CCATGCCCCCCAGTGGCTGCAGGTAGCTGGACTTCCTGCCTGTGACGCACAGAGAGCAGCGGACTCCGGAGCGTCAGCCTGCCTTAGCCC TTGGTTCCTCGTTTTTTGGGATTGTCACACTGTGACACTCGGGAGGAACGATGTCCTATCCTGGGCTGCTCAGTGTTCCCAGGCCCTGTG TGCCAGAGGCCAGGTCTCTCCAAGTGCATCTAGCCTGGCCTGGAGGGCAAGGCAGATGCATTCTCCTGGCTGCTGGGAGAGCCCATGGGA ACCTTGCTTAGCGCCCAAGGTAGATGAGAGACAGGGCAGGGAATCTGCGTGGTGCTGGGATGTGGGGACTCTGGCCTCTCCAGACTAATG TTCCTGTCACCTGCTGAGCTTTCCCAGTGTCACTTCTGCAGTCCTCCTCTGCTCACCCCTCCCTCACTCCGTGGGCAATTCCCTTTCTCT CTGTCACTGCTTCAGCAGTGAAGACCCATGGCACCTAGAAATGCATGCCAGCTTCCTGCGTCTACCACTCTGCAGCTCAAACAAGAAGCT CAGAGCTGCCCCTTCCACTTCCTCCCCAAAAGGGATGAAGAAAACCCTGTGGTAGAACCAAACCACCCAAGGCCAGCCCTTAAGCCTGCC TCCACCTTCCTGGTGCATGACAAAATATTTCTTTTGTCTCAGAGGACTTTGATGCTTCAATATTTAATGATGAATCCGCCACACTCCATT TTCCCTCCTCTGAAGCTCCCAACAACTCTGACAGGGTCAGATCTCTTTAAAGCATCTTCAGGAGGCGGTTGCCACGGAGACTGGATATTG GTCCCAGGGACAGGCTGTGCTACCTACTGGGTGGTGGGGAGGGAAGCTTCTGGCCATGCTTGGGCATTCAGACCAACCATCATTAATGTT AATAGAGGAAATGCAGCAACGTCAAAAGCAAGGCAGGTTGGTTAATTAGATCTGGGGCCCGATGACCTCCCTTTCCCACTGCTACAGCCC TTTTGACTTCCCCCAGCAGGAAGCGCTGTCTGTGCGGCTGAGTCTGGGCCACTTGCTTAATTGCATTTCTGGCAGGGAAGGAGAGGAGGG ATGGAGAAGCTCTGCTCGCTGCTTCCACCTCACCGTGTGTCCTCCCCTCCCCTCCCTTCCTCTCCTCCCCACAGGCGGAATTGCTTTCTT CTGAGCCGGGTGGGCTCTCCTGCAGGGAGAATGCAGGGGTCTGTGCTTGGCTCTTAGCAAAGCAGGTGGCCCACAGTGACCTCCAGGAGG TGGCTGATTTTCCTTTGTGCCTTCTTATTTCTCAGTAGGAACTTGGAAGGGAAGAGCTCTGAAGGAGCCAGAGGGGCAGCCACCTTGCAT GAGGGTGAGGAAGGAGAAGCCCAGCCAGCCTCTACCCATCCCTCCCTCCCAATTAGGCTGCACTGGGGCCGAACGCTTCCCTGGGCTCCC ACTGCCTTCTAAGGAGCCAAGGAGAATTCCCTGCCCACTTTCTGTTCCTCCATGCCCAAACTGGCAGATGCTTCTCTGGGAGCAAGGCCA CATGGGCTGATGCAGCTTTGAGTGCTCAGGTTCTGTTTTCTGGAAGTTGCTGGAGAAGGGCTGTCAGGGTGCTTCTATGAATATGGGCTT CAGCTGTAGCACTTGGCAGAAACGTGATGTGTCCCACAAACTGCTGGTGGAGGAAGAAGATGTTCGTCAGCCAATTCAACCTCCAGCCAG >77582_77582_1_RPS20-POMT2_RPS20_chr8_56986618_ENST00000009589_POMT2_chr14_77750216_ENST00000261534_length(amino acids)=259AA_BP=34 MAFKDTGKTPVEPEVAIHRIRITLTSRNVKSLEKVPNISLDVLQPSFPEILLESHMVMIRGNSGLKPKDNEFTSKPWHWPINYQGLRFSG VNDTDFRVYLLGNPVVWWLNLLSIALYLLSGSIIAVAMQRGARLPAEVAGLSQVLLRGGGQVLLGWTLHYFPFFLMGRVLYFHHYFPAML -------------------------------------------------------------- >77582_77582_2_RPS20-POMT2_RPS20_chr8_56986618_ENST00000518875_POMT2_chr14_77750216_ENST00000261534_length(transcript)=3315nt_BP=218nt GGAAGACGCGGTCGTAAGGGCTGAGGATTTTTGGTCCGCACGCTCCTGCTCCTGACTCACCGCTGTTCGCTCTCGCCGAGGAACAAGTCG GTCAGGAAGCCCGCGCGCAACAGCCATGGCTTTTAAGGATACCGGAAAAACACCCGTGGAGCCGGAGGTGGCAATTCACCGAATTCGAAT CACCCTAACAAGCCGCAACGTAAAATCCTTGGAAAAGGTGCCAAACATCAGCCTGGATGTGCTACAGCCCAGTTTTCCTGAGATCTTGCT GGAATCCCACATGGTCATGATCCGGGGGAACAGTGGCCTCAAACCCAAGGACAATGAGTTCACGTCCAAACCCTGGCACTGGCCTATCAA CTATCAGGGCCTACGCTTCTCAGGGGTCAATGACACAGATTTCCGAGTCTATCTGCTTGGCAACCCGGTGGTTTGGTGGCTGAATCTGTT GAGCATCGCCCTCTACCTCCTCTCAGGGAGCATCATTGCTGTAGCCATGCAGAGAGGGGCACGGCTGCCAGCGGAGGTTGCAGGGTTGTC CCAGGTCCTGCTTCGAGGAGGCGGCCAGGTCCTGCTCGGCTGGACACTCCATTACTTCCCGTTTTTCCTGATGGGCCGGGTCCTCTACTT CCACCACTACTTCCCAGCCATGCTCTTCTCAAGCATGTTGACAGGCATTCTGTGGGACACCCTCCTGCGGCTCTGTGCCTGGGGCTTGGC CTCATGGCCCCTGGCGAGGGGCATACATGTGGCGGGAATCCTGAGCCTGCTCCTGGGAACTGCCTACAGCTTCTACCTCTTCCACCCTCT GGCTTACGGGATGGTTGGTCCCCTGGCCCAGGACCCCCAAAGTCCAATGGCAGGACTAAGGTGGCTGGACTCATGGGACTTTTGAGGCCA CTGCAAAGATTCCAGCCTGGGTCCAGGAACTTCCCAGGAGAGCCAGCTGGGAAGCAGTACTGGACCATCCGGCCATGAGGAAGCTGCCTA AGGAGCCCGAAGAATTCTGCTGCTCACCAGGACCCAGCTCTGGGAGTGCAGCTGGAATGGAACCCAGCATTGGAGAGAGCCGGGAGAGAG CCGCAGCTCCGTCCACAGCACCACGGAGAGGACAGCGCCTCTTGGGCTGCTCCCTCGTGCACAGCGCAGGGGAGGATGGGTGCCAGAGGG TGGGAGTGGGTGCGCGTGTGTGTATGTGAGGGCATGTTGTCCATATCCCTGTGGAATACAGACCGTGTAACTGGACAGCTGAGTGTCATA AACTCTAAAATAGTCCAGGCTATAGTAATATTTTCTGGTATTTTTTTCTGTTGGATTTCTTTGGTTTTGGTTTGTAAGATCCATCAAAAA TTCTCAGACCTCTGAGAGGGCCTTGAGGGCAGAGAATACTGACCTCACCTTGGGATGTCACCAAAGGCAGAGCAGATGGGACTGTCAAAG TTGTGCTGCCCTCCCTCCCACAAACAAATGAATCCAGCGTGGAGGAAGGCGCGCCGCCCCGAGCTGGAGGGCCTGGCTCCCACCATGCCC CCCAGTGGCTGCAGGTAGCTGGACTTCCTGCCTGTGACGCACAGAGAGCAGCGGACTCCGGAGCGTCAGCCTGCCTTAGCCCTTGGTTCC TCGTTTTTTGGGATTGTCACACTGTGACACTCGGGAGGAACGATGTCCTATCCTGGGCTGCTCAGTGTTCCCAGGCCCTGTGTGCCAGAG GCCAGGTCTCTCCAAGTGCATCTAGCCTGGCCTGGAGGGCAAGGCAGATGCATTCTCCTGGCTGCTGGGAGAGCCCATGGGAACCTTGCT TAGCGCCCAAGGTAGATGAGAGACAGGGCAGGGAATCTGCGTGGTGCTGGGATGTGGGGACTCTGGCCTCTCCAGACTAATGTTCCTGTC ACCTGCTGAGCTTTCCCAGTGTCACTTCTGCAGTCCTCCTCTGCTCACCCCTCCCTCACTCCGTGGGCAATTCCCTTTCTCTCTGTCACT GCTTCAGCAGTGAAGACCCATGGCACCTAGAAATGCATGCCAGCTTCCTGCGTCTACCACTCTGCAGCTCAAACAAGAAGCTCAGAGCTG CCCCTTCCACTTCCTCCCCAAAAGGGATGAAGAAAACCCTGTGGTAGAACCAAACCACCCAAGGCCAGCCCTTAAGCCTGCCTCCACCTT CCTGGTGCATGACAAAATATTTCTTTTGTCTCAGAGGACTTTGATGCTTCAATATTTAATGATGAATCCGCCACACTCCATTTTCCCTCC TCTGAAGCTCCCAACAACTCTGACAGGGTCAGATCTCTTTAAAGCATCTTCAGGAGGCGGTTGCCACGGAGACTGGATATTGGTCCCAGG GACAGGCTGTGCTACCTACTGGGTGGTGGGGAGGGAAGCTTCTGGCCATGCTTGGGCATTCAGACCAACCATCATTAATGTTAATAGAGG AAATGCAGCAACGTCAAAAGCAAGGCAGGTTGGTTAATTAGATCTGGGGCCCGATGACCTCCCTTTCCCACTGCTACAGCCCTTTTGACT TCCCCCAGCAGGAAGCGCTGTCTGTGCGGCTGAGTCTGGGCCACTTGCTTAATTGCATTTCTGGCAGGGAAGGAGAGGAGGGATGGAGAA GCTCTGCTCGCTGCTTCCACCTCACCGTGTGTCCTCCCCTCCCCTCCCTTCCTCTCCTCCCCACAGGCGGAATTGCTTTCTTCTGAGCCG GGTGGGCTCTCCTGCAGGGAGAATGCAGGGGTCTGTGCTTGGCTCTTAGCAAAGCAGGTGGCCCACAGTGACCTCCAGGAGGTGGCTGAT TTTCCTTTGTGCCTTCTTATTTCTCAGTAGGAACTTGGAAGGGAAGAGCTCTGAAGGAGCCAGAGGGGCAGCCACCTTGCATGAGGGTGA GGAAGGAGAAGCCCAGCCAGCCTCTACCCATCCCTCCCTCCCAATTAGGCTGCACTGGGGCCGAACGCTTCCCTGGGCTCCCACTGCCTT CTAAGGAGCCAAGGAGAATTCCCTGCCCACTTTCTGTTCCTCCATGCCCAAACTGGCAGATGCTTCTCTGGGAGCAAGGCCACATGGGCT GATGCAGCTTTGAGTGCTCAGGTTCTGTTTTCTGGAAGTTGCTGGAGAAGGGCTGTCAGGGTGCTTCTATGAATATGGGCTTCAGCTGTA GCACTTGGCAGAAACGTGATGTGTCCCACAAACTGCTGGTGGAGGAAGAAGATGTTCGTCAGCCAATTCAACCTCCAGCCAGCTTCTTGA >77582_77582_2_RPS20-POMT2_RPS20_chr8_56986618_ENST00000518875_POMT2_chr14_77750216_ENST00000261534_length(amino acids)=259AA_BP=34 MAFKDTGKTPVEPEVAIHRIRITLTSRNVKSLEKVPNISLDVLQPSFPEILLESHMVMIRGNSGLKPKDNEFTSKPWHWPINYQGLRFSG VNDTDFRVYLLGNPVVWWLNLLSIALYLLSGSIIAVAMQRGARLPAEVAGLSQVLLRGGGQVLLGWTLHYFPFFLMGRVLYFHHYFPAML -------------------------------------------------------------- >77582_77582_3_RPS20-POMT2_RPS20_chr8_56986618_ENST00000519606_POMT2_chr14_77750216_ENST00000261534_length(transcript)=3323nt_BP=226nt CTTTTTGAGGAAGACGCGGTCGTAAGGGCTGAGGATTTTTGGTCCGCACGCTCCTGCTCCTGACTCACCGCTGTTCGCTCTCGCCGAGGA ACAAGTCGGTCAGGAAGCCCGCGCGCAACAGCCATGGCTTTTAAGGATACCGGAAAAACACCCGTGGAGCCGGAGGTGGCAATTCACCGA ATTCGAATCACCCTAACAAGCCGCAACGTAAAATCCTTGGAAAAGGTGCCAAACATCAGCCTGGATGTGCTACAGCCCAGTTTTCCTGAG ATCTTGCTGGAATCCCACATGGTCATGATCCGGGGGAACAGTGGCCTCAAACCCAAGGACAATGAGTTCACGTCCAAACCCTGGCACTGG CCTATCAACTATCAGGGCCTACGCTTCTCAGGGGTCAATGACACAGATTTCCGAGTCTATCTGCTTGGCAACCCGGTGGTTTGGTGGCTG AATCTGTTGAGCATCGCCCTCTACCTCCTCTCAGGGAGCATCATTGCTGTAGCCATGCAGAGAGGGGCACGGCTGCCAGCGGAGGTTGCA GGGTTGTCCCAGGTCCTGCTTCGAGGAGGCGGCCAGGTCCTGCTCGGCTGGACACTCCATTACTTCCCGTTTTTCCTGATGGGCCGGGTC CTCTACTTCCACCACTACTTCCCAGCCATGCTCTTCTCAAGCATGTTGACAGGCATTCTGTGGGACACCCTCCTGCGGCTCTGTGCCTGG GGCTTGGCCTCATGGCCCCTGGCGAGGGGCATACATGTGGCGGGAATCCTGAGCCTGCTCCTGGGAACTGCCTACAGCTTCTACCTCTTC CACCCTCTGGCTTACGGGATGGTTGGTCCCCTGGCCCAGGACCCCCAAAGTCCAATGGCAGGACTAAGGTGGCTGGACTCATGGGACTTT TGAGGCCACTGCAAAGATTCCAGCCTGGGTCCAGGAACTTCCCAGGAGAGCCAGCTGGGAAGCAGTACTGGACCATCCGGCCATGAGGAA GCTGCCTAAGGAGCCCGAAGAATTCTGCTGCTCACCAGGACCCAGCTCTGGGAGTGCAGCTGGAATGGAACCCAGCATTGGAGAGAGCCG GGAGAGAGCCGCAGCTCCGTCCACAGCACCACGGAGAGGACAGCGCCTCTTGGGCTGCTCCCTCGTGCACAGCGCAGGGGAGGATGGGTG CCAGAGGGTGGGAGTGGGTGCGCGTGTGTGTATGTGAGGGCATGTTGTCCATATCCCTGTGGAATACAGACCGTGTAACTGGACAGCTGA GTGTCATAAACTCTAAAATAGTCCAGGCTATAGTAATATTTTCTGGTATTTTTTTCTGTTGGATTTCTTTGGTTTTGGTTTGTAAGATCC ATCAAAAATTCTCAGACCTCTGAGAGGGCCTTGAGGGCAGAGAATACTGACCTCACCTTGGGATGTCACCAAAGGCAGAGCAGATGGGAC TGTCAAAGTTGTGCTGCCCTCCCTCCCACAAACAAATGAATCCAGCGTGGAGGAAGGCGCGCCGCCCCGAGCTGGAGGGCCTGGCTCCCA CCATGCCCCCCAGTGGCTGCAGGTAGCTGGACTTCCTGCCTGTGACGCACAGAGAGCAGCGGACTCCGGAGCGTCAGCCTGCCTTAGCCC TTGGTTCCTCGTTTTTTGGGATTGTCACACTGTGACACTCGGGAGGAACGATGTCCTATCCTGGGCTGCTCAGTGTTCCCAGGCCCTGTG TGCCAGAGGCCAGGTCTCTCCAAGTGCATCTAGCCTGGCCTGGAGGGCAAGGCAGATGCATTCTCCTGGCTGCTGGGAGAGCCCATGGGA ACCTTGCTTAGCGCCCAAGGTAGATGAGAGACAGGGCAGGGAATCTGCGTGGTGCTGGGATGTGGGGACTCTGGCCTCTCCAGACTAATG TTCCTGTCACCTGCTGAGCTTTCCCAGTGTCACTTCTGCAGTCCTCCTCTGCTCACCCCTCCCTCACTCCGTGGGCAATTCCCTTTCTCT CTGTCACTGCTTCAGCAGTGAAGACCCATGGCACCTAGAAATGCATGCCAGCTTCCTGCGTCTACCACTCTGCAGCTCAAACAAGAAGCT CAGAGCTGCCCCTTCCACTTCCTCCCCAAAAGGGATGAAGAAAACCCTGTGGTAGAACCAAACCACCCAAGGCCAGCCCTTAAGCCTGCC TCCACCTTCCTGGTGCATGACAAAATATTTCTTTTGTCTCAGAGGACTTTGATGCTTCAATATTTAATGATGAATCCGCCACACTCCATT TTCCCTCCTCTGAAGCTCCCAACAACTCTGACAGGGTCAGATCTCTTTAAAGCATCTTCAGGAGGCGGTTGCCACGGAGACTGGATATTG GTCCCAGGGACAGGCTGTGCTACCTACTGGGTGGTGGGGAGGGAAGCTTCTGGCCATGCTTGGGCATTCAGACCAACCATCATTAATGTT AATAGAGGAAATGCAGCAACGTCAAAAGCAAGGCAGGTTGGTTAATTAGATCTGGGGCCCGATGACCTCCCTTTCCCACTGCTACAGCCC TTTTGACTTCCCCCAGCAGGAAGCGCTGTCTGTGCGGCTGAGTCTGGGCCACTTGCTTAATTGCATTTCTGGCAGGGAAGGAGAGGAGGG ATGGAGAAGCTCTGCTCGCTGCTTCCACCTCACCGTGTGTCCTCCCCTCCCCTCCCTTCCTCTCCTCCCCACAGGCGGAATTGCTTTCTT CTGAGCCGGGTGGGCTCTCCTGCAGGGAGAATGCAGGGGTCTGTGCTTGGCTCTTAGCAAAGCAGGTGGCCCACAGTGACCTCCAGGAGG TGGCTGATTTTCCTTTGTGCCTTCTTATTTCTCAGTAGGAACTTGGAAGGGAAGAGCTCTGAAGGAGCCAGAGGGGCAGCCACCTTGCAT GAGGGTGAGGAAGGAGAAGCCCAGCCAGCCTCTACCCATCCCTCCCTCCCAATTAGGCTGCACTGGGGCCGAACGCTTCCCTGGGCTCCC ACTGCCTTCTAAGGAGCCAAGGAGAATTCCCTGCCCACTTTCTGTTCCTCCATGCCCAAACTGGCAGATGCTTCTCTGGGAGCAAGGCCA CATGGGCTGATGCAGCTTTGAGTGCTCAGGTTCTGTTTTCTGGAAGTTGCTGGAGAAGGGCTGTCAGGGTGCTTCTATGAATATGGGCTT CAGCTGTAGCACTTGGCAGAAACGTGATGTGTCCCACAAACTGCTGGTGGAGGAAGAAGATGTTCGTCAGCCAATTCAACCTCCAGCCAG >77582_77582_3_RPS20-POMT2_RPS20_chr8_56986618_ENST00000519606_POMT2_chr14_77750216_ENST00000261534_length(amino acids)=259AA_BP=34 MAFKDTGKTPVEPEVAIHRIRITLTSRNVKSLEKVPNISLDVLQPSFPEILLESHMVMIRGNSGLKPKDNEFTSKPWHWPINYQGLRFSG VNDTDFRVYLLGNPVVWWLNLLSIALYLLSGSIIAVAMQRGARLPAEVAGLSQVLLRGGGQVLLGWTLHYFPFFLMGRVLYFHHYFPAML -------------------------------------------------------------- >77582_77582_4_RPS20-POMT2_RPS20_chr8_56986618_ENST00000521262_POMT2_chr14_77750216_ENST00000261534_length(transcript)=3454nt_BP=357nt CTTTCTTTTTGAGGAAGACGCGGTCGTAAGGGCTGAGGATTTTTGGTCCGCACGCTCCTGCTCCTGACTCACCGCTGTTCGCTCTCGCCG AGGAACAAGTCGGTCAGGAAGCCCGCGCGCAACAGCCATGGTGAGGCGGCAGTCGGCGGCGGGCGGCGCAACGCAGGGCTCGGGCGCCGG GGGGACCCCGGTTTCGAGTTCGTGCTCCTGAGGGCAGATGCCGGGGCGTGTAGGGGCGGAAAGGCGCTCCTGAAATGGCTTTTAAGGATA CCGGAAAAACACCCGTGGAGCCGGAGGTGGCAATTCACCGAATTCGAATCACCCTAACAAGCCGCAACGTAAAATCCTTGGAAAAGGTGC CAAACATCAGCCTGGATGTGCTACAGCCCAGTTTTCCTGAGATCTTGCTGGAATCCCACATGGTCATGATCCGGGGGAACAGTGGCCTCA AACCCAAGGACAATGAGTTCACGTCCAAACCCTGGCACTGGCCTATCAACTATCAGGGCCTACGCTTCTCAGGGGTCAATGACACAGATT TCCGAGTCTATCTGCTTGGCAACCCGGTGGTTTGGTGGCTGAATCTGTTGAGCATCGCCCTCTACCTCCTCTCAGGGAGCATCATTGCTG TAGCCATGCAGAGAGGGGCACGGCTGCCAGCGGAGGTTGCAGGGTTGTCCCAGGTCCTGCTTCGAGGAGGCGGCCAGGTCCTGCTCGGCT GGACACTCCATTACTTCCCGTTTTTCCTGATGGGCCGGGTCCTCTACTTCCACCACTACTTCCCAGCCATGCTCTTCTCAAGCATGTTGA CAGGCATTCTGTGGGACACCCTCCTGCGGCTCTGTGCCTGGGGCTTGGCCTCATGGCCCCTGGCGAGGGGCATACATGTGGCGGGAATCC TGAGCCTGCTCCTGGGAACTGCCTACAGCTTCTACCTCTTCCACCCTCTGGCTTACGGGATGGTTGGTCCCCTGGCCCAGGACCCCCAAA GTCCAATGGCAGGACTAAGGTGGCTGGACTCATGGGACTTTTGAGGCCACTGCAAAGATTCCAGCCTGGGTCCAGGAACTTCCCAGGAGA GCCAGCTGGGAAGCAGTACTGGACCATCCGGCCATGAGGAAGCTGCCTAAGGAGCCCGAAGAATTCTGCTGCTCACCAGGACCCAGCTCT GGGAGTGCAGCTGGAATGGAACCCAGCATTGGAGAGAGCCGGGAGAGAGCCGCAGCTCCGTCCACAGCACCACGGAGAGGACAGCGCCTC TTGGGCTGCTCCCTCGTGCACAGCGCAGGGGAGGATGGGTGCCAGAGGGTGGGAGTGGGTGCGCGTGTGTGTATGTGAGGGCATGTTGTC CATATCCCTGTGGAATACAGACCGTGTAACTGGACAGCTGAGTGTCATAAACTCTAAAATAGTCCAGGCTATAGTAATATTTTCTGGTAT TTTTTTCTGTTGGATTTCTTTGGTTTTGGTTTGTAAGATCCATCAAAAATTCTCAGACCTCTGAGAGGGCCTTGAGGGCAGAGAATACTG ACCTCACCTTGGGATGTCACCAAAGGCAGAGCAGATGGGACTGTCAAAGTTGTGCTGCCCTCCCTCCCACAAACAAATGAATCCAGCGTG GAGGAAGGCGCGCCGCCCCGAGCTGGAGGGCCTGGCTCCCACCATGCCCCCCAGTGGCTGCAGGTAGCTGGACTTCCTGCCTGTGACGCA CAGAGAGCAGCGGACTCCGGAGCGTCAGCCTGCCTTAGCCCTTGGTTCCTCGTTTTTTGGGATTGTCACACTGTGACACTCGGGAGGAAC GATGTCCTATCCTGGGCTGCTCAGTGTTCCCAGGCCCTGTGTGCCAGAGGCCAGGTCTCTCCAAGTGCATCTAGCCTGGCCTGGAGGGCA AGGCAGATGCATTCTCCTGGCTGCTGGGAGAGCCCATGGGAACCTTGCTTAGCGCCCAAGGTAGATGAGAGACAGGGCAGGGAATCTGCG TGGTGCTGGGATGTGGGGACTCTGGCCTCTCCAGACTAATGTTCCTGTCACCTGCTGAGCTTTCCCAGTGTCACTTCTGCAGTCCTCCTC TGCTCACCCCTCCCTCACTCCGTGGGCAATTCCCTTTCTCTCTGTCACTGCTTCAGCAGTGAAGACCCATGGCACCTAGAAATGCATGCC AGCTTCCTGCGTCTACCACTCTGCAGCTCAAACAAGAAGCTCAGAGCTGCCCCTTCCACTTCCTCCCCAAAAGGGATGAAGAAAACCCTG TGGTAGAACCAAACCACCCAAGGCCAGCCCTTAAGCCTGCCTCCACCTTCCTGGTGCATGACAAAATATTTCTTTTGTCTCAGAGGACTT TGATGCTTCAATATTTAATGATGAATCCGCCACACTCCATTTTCCCTCCTCTGAAGCTCCCAACAACTCTGACAGGGTCAGATCTCTTTA AAGCATCTTCAGGAGGCGGTTGCCACGGAGACTGGATATTGGTCCCAGGGACAGGCTGTGCTACCTACTGGGTGGTGGGGAGGGAAGCTT CTGGCCATGCTTGGGCATTCAGACCAACCATCATTAATGTTAATAGAGGAAATGCAGCAACGTCAAAAGCAAGGCAGGTTGGTTAATTAG ATCTGGGGCCCGATGACCTCCCTTTCCCACTGCTACAGCCCTTTTGACTTCCCCCAGCAGGAAGCGCTGTCTGTGCGGCTGAGTCTGGGC CACTTGCTTAATTGCATTTCTGGCAGGGAAGGAGAGGAGGGATGGAGAAGCTCTGCTCGCTGCTTCCACCTCACCGTGTGTCCTCCCCTC CCCTCCCTTCCTCTCCTCCCCACAGGCGGAATTGCTTTCTTCTGAGCCGGGTGGGCTCTCCTGCAGGGAGAATGCAGGGGTCTGTGCTTG GCTCTTAGCAAAGCAGGTGGCCCACAGTGACCTCCAGGAGGTGGCTGATTTTCCTTTGTGCCTTCTTATTTCTCAGTAGGAACTTGGAAG GGAAGAGCTCTGAAGGAGCCAGAGGGGCAGCCACCTTGCATGAGGGTGAGGAAGGAGAAGCCCAGCCAGCCTCTACCCATCCCTCCCTCC CAATTAGGCTGCACTGGGGCCGAACGCTTCCCTGGGCTCCCACTGCCTTCTAAGGAGCCAAGGAGAATTCCCTGCCCACTTTCTGTTCCT CCATGCCCAAACTGGCAGATGCTTCTCTGGGAGCAAGGCCACATGGGCTGATGCAGCTTTGAGTGCTCAGGTTCTGTTTTCTGGAAGTTG CTGGAGAAGGGCTGTCAGGGTGCTTCTATGAATATGGGCTTCAGCTGTAGCACTTGGCAGAAACGTGATGTGTCCCACAAACTGCTGGTG GAGGAAGAAGATGTTCGTCAGCCAATTCAACCTCCAGCCAGCTTCTTGAGCCTATTTGGTTTAAAAAAAAAAAGTCTTCTAATCAGAAGC >77582_77582_4_RPS20-POMT2_RPS20_chr8_56986618_ENST00000521262_POMT2_chr14_77750216_ENST00000261534_length(amino acids)=259AA_BP=34 MAFKDTGKTPVEPEVAIHRIRITLTSRNVKSLEKVPNISLDVLQPSFPEILLESHMVMIRGNSGLKPKDNEFTSKPWHWPINYQGLRFSG VNDTDFRVYLLGNPVVWWLNLLSIALYLLSGSIIAVAMQRGARLPAEVAGLSQVLLRGGGQVLLGWTLHYFPFFLMGRVLYFHHYFPAML -------------------------------------------------------------- >77582_77582_5_RPS20-POMT2_RPS20_chr8_56986618_ENST00000523936_POMT2_chr14_77750216_ENST00000261534_length(transcript)=3300nt_BP=203nt GGCGCAACGCAGGGCTCGGGCGCCGGGGGGACCCCGGTTTCGAGTTCGTGCTCCTGAGGGCAGATGCCGGGGCGTGTAGGGGCGGAAAGG CGCTCCTGAAATGGCTTTTAAGGATACCGGAAAAACACCCGTGGAGCCGGAGGTGGCAATTCACCGAATTCGAATCACCCTAACAAGCCG CAACGTAAAATCCTTGGAAAAGGTGCCAAACATCAGCCTGGATGTGCTACAGCCCAGTTTTCCTGAGATCTTGCTGGAATCCCACATGGT CATGATCCGGGGGAACAGTGGCCTCAAACCCAAGGACAATGAGTTCACGTCCAAACCCTGGCACTGGCCTATCAACTATCAGGGCCTACG CTTCTCAGGGGTCAATGACACAGATTTCCGAGTCTATCTGCTTGGCAACCCGGTGGTTTGGTGGCTGAATCTGTTGAGCATCGCCCTCTA CCTCCTCTCAGGGAGCATCATTGCTGTAGCCATGCAGAGAGGGGCACGGCTGCCAGCGGAGGTTGCAGGGTTGTCCCAGGTCCTGCTTCG AGGAGGCGGCCAGGTCCTGCTCGGCTGGACACTCCATTACTTCCCGTTTTTCCTGATGGGCCGGGTCCTCTACTTCCACCACTACTTCCC AGCCATGCTCTTCTCAAGCATGTTGACAGGCATTCTGTGGGACACCCTCCTGCGGCTCTGTGCCTGGGGCTTGGCCTCATGGCCCCTGGC GAGGGGCATACATGTGGCGGGAATCCTGAGCCTGCTCCTGGGAACTGCCTACAGCTTCTACCTCTTCCACCCTCTGGCTTACGGGATGGT TGGTCCCCTGGCCCAGGACCCCCAAAGTCCAATGGCAGGACTAAGGTGGCTGGACTCATGGGACTTTTGAGGCCACTGCAAAGATTCCAG CCTGGGTCCAGGAACTTCCCAGGAGAGCCAGCTGGGAAGCAGTACTGGACCATCCGGCCATGAGGAAGCTGCCTAAGGAGCCCGAAGAAT TCTGCTGCTCACCAGGACCCAGCTCTGGGAGTGCAGCTGGAATGGAACCCAGCATTGGAGAGAGCCGGGAGAGAGCCGCAGCTCCGTCCA CAGCACCACGGAGAGGACAGCGCCTCTTGGGCTGCTCCCTCGTGCACAGCGCAGGGGAGGATGGGTGCCAGAGGGTGGGAGTGGGTGCGC GTGTGTGTATGTGAGGGCATGTTGTCCATATCCCTGTGGAATACAGACCGTGTAACTGGACAGCTGAGTGTCATAAACTCTAAAATAGTC CAGGCTATAGTAATATTTTCTGGTATTTTTTTCTGTTGGATTTCTTTGGTTTTGGTTTGTAAGATCCATCAAAAATTCTCAGACCTCTGA GAGGGCCTTGAGGGCAGAGAATACTGACCTCACCTTGGGATGTCACCAAAGGCAGAGCAGATGGGACTGTCAAAGTTGTGCTGCCCTCCC TCCCACAAACAAATGAATCCAGCGTGGAGGAAGGCGCGCCGCCCCGAGCTGGAGGGCCTGGCTCCCACCATGCCCCCCAGTGGCTGCAGG TAGCTGGACTTCCTGCCTGTGACGCACAGAGAGCAGCGGACTCCGGAGCGTCAGCCTGCCTTAGCCCTTGGTTCCTCGTTTTTTGGGATT GTCACACTGTGACACTCGGGAGGAACGATGTCCTATCCTGGGCTGCTCAGTGTTCCCAGGCCCTGTGTGCCAGAGGCCAGGTCTCTCCAA GTGCATCTAGCCTGGCCTGGAGGGCAAGGCAGATGCATTCTCCTGGCTGCTGGGAGAGCCCATGGGAACCTTGCTTAGCGCCCAAGGTAG ATGAGAGACAGGGCAGGGAATCTGCGTGGTGCTGGGATGTGGGGACTCTGGCCTCTCCAGACTAATGTTCCTGTCACCTGCTGAGCTTTC CCAGTGTCACTTCTGCAGTCCTCCTCTGCTCACCCCTCCCTCACTCCGTGGGCAATTCCCTTTCTCTCTGTCACTGCTTCAGCAGTGAAG ACCCATGGCACCTAGAAATGCATGCCAGCTTCCTGCGTCTACCACTCTGCAGCTCAAACAAGAAGCTCAGAGCTGCCCCTTCCACTTCCT CCCCAAAAGGGATGAAGAAAACCCTGTGGTAGAACCAAACCACCCAAGGCCAGCCCTTAAGCCTGCCTCCACCTTCCTGGTGCATGACAA AATATTTCTTTTGTCTCAGAGGACTTTGATGCTTCAATATTTAATGATGAATCCGCCACACTCCATTTTCCCTCCTCTGAAGCTCCCAAC AACTCTGACAGGGTCAGATCTCTTTAAAGCATCTTCAGGAGGCGGTTGCCACGGAGACTGGATATTGGTCCCAGGGACAGGCTGTGCTAC CTACTGGGTGGTGGGGAGGGAAGCTTCTGGCCATGCTTGGGCATTCAGACCAACCATCATTAATGTTAATAGAGGAAATGCAGCAACGTC AAAAGCAAGGCAGGTTGGTTAATTAGATCTGGGGCCCGATGACCTCCCTTTCCCACTGCTACAGCCCTTTTGACTTCCCCCAGCAGGAAG CGCTGTCTGTGCGGCTGAGTCTGGGCCACTTGCTTAATTGCATTTCTGGCAGGGAAGGAGAGGAGGGATGGAGAAGCTCTGCTCGCTGCT TCCACCTCACCGTGTGTCCTCCCCTCCCCTCCCTTCCTCTCCTCCCCACAGGCGGAATTGCTTTCTTCTGAGCCGGGTGGGCTCTCCTGC AGGGAGAATGCAGGGGTCTGTGCTTGGCTCTTAGCAAAGCAGGTGGCCCACAGTGACCTCCAGGAGGTGGCTGATTTTCCTTTGTGCCTT CTTATTTCTCAGTAGGAACTTGGAAGGGAAGAGCTCTGAAGGAGCCAGAGGGGCAGCCACCTTGCATGAGGGTGAGGAAGGAGAAGCCCA GCCAGCCTCTACCCATCCCTCCCTCCCAATTAGGCTGCACTGGGGCCGAACGCTTCCCTGGGCTCCCACTGCCTTCTAAGGAGCCAAGGA GAATTCCCTGCCCACTTTCTGTTCCTCCATGCCCAAACTGGCAGATGCTTCTCTGGGAGCAAGGCCACATGGGCTGATGCAGCTTTGAGT GCTCAGGTTCTGTTTTCTGGAAGTTGCTGGAGAAGGGCTGTCAGGGTGCTTCTATGAATATGGGCTTCAGCTGTAGCACTTGGCAGAAAC GTGATGTGTCCCACAAACTGCTGGTGGAGGAAGAAGATGTTCGTCAGCCAATTCAACCTCCAGCCAGCTTCTTGAGCCTATTTGGTTTAA >77582_77582_5_RPS20-POMT2_RPS20_chr8_56986618_ENST00000523936_POMT2_chr14_77750216_ENST00000261534_length(amino acids)=259AA_BP=34 MAFKDTGKTPVEPEVAIHRIRITLTSRNVKSLEKVPNISLDVLQPSFPEILLESHMVMIRGNSGLKPKDNEFTSKPWHWPINYQGLRFSG VNDTDFRVYLLGNPVVWWLNLLSIALYLLSGSIIAVAMQRGARLPAEVAGLSQVLLRGGGQVLLGWTLHYFPFFLMGRVLYFHHYFPAML -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RPS20-POMT2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RPS20-POMT2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RPS20-POMT2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
