
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RPS3-ZNF536 (FusionGDB2 ID:77917) |
Fusion Gene Summary for RPS3-ZNF536 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RPS3-ZNF536 | Fusion gene ID: 77917 | Hgene | Tgene | Gene symbol | RPS3 | ZNF536 | Gene ID | 6188 | 9745 |
| Gene name | ribosomal protein S3 | zinc finger protein 536 | |
| Synonyms | S3 | - | |
| Cytomap | 11q13.4 | 19q12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | 40S ribosomal protein S3IMR-90 ribosomal protein S3small ribosomal subunit protein uS3 | zinc finger protein 536 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000278572, ENST00000524851, ENST00000526608, ENST00000527446, ENST00000531188, ENST00000529285, ENST00000530164, ENST00000534440, | ENST00000590564, ENST00000355537, | |
| Fusion gene scores | * DoF score | 17 X 19 X 5=1615 | 21 X 13 X 13=3549 |
| # samples | 22 | 25 | |
| ** MAII score | log2(22/1615*10)=-2.87595873605663 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(25/3549*10)=-3.82741257439886 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: RPS3 [Title/Abstract] AND ZNF536 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RPS3(75115227)-ZNF536(31039220), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | RPS3 | GO:0006979 | response to oxidative stress | 23911537 |
| Hgene | RPS3 | GO:0017148 | negative regulation of translation | 20217897 |
| Hgene | RPS3 | GO:0031397 | negative regulation of protein ubiquitination | 19656744 |
| Hgene | RPS3 | GO:0032079 | positive regulation of endodeoxyribonuclease activity | 18973764 |
| Hgene | RPS3 | GO:0042769 | DNA damage response, detection of DNA damage | 23911537 |
| Hgene | RPS3 | GO:0045739 | positive regulation of DNA repair | 23911537 |
| Hgene | RPS3 | GO:0061481 | response to TNF agonist | 20041225 |
| Hgene | RPS3 | GO:0070301 | cellular response to hydrogen peroxide | 23911537 |
| Hgene | RPS3 | GO:1901224 | positive regulation of NIK/NF-kappaB signaling | 20041225 |
| Hgene | RPS3 | GO:1902546 | positive regulation of DNA N-glycosylase activity | 15518571 |
| Hgene | RPS3 | GO:1905053 | positive regulation of base-excision repair | 18973764 |
| Hgene | RPS3 | GO:2001235 | positive regulation of apoptotic signaling pathway | 14988002 |
| Tgene | ZNF536 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 19398580 |
 Fusion gene breakpoints across RPS3 (5'-gene) Fusion gene breakpoints across RPS3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
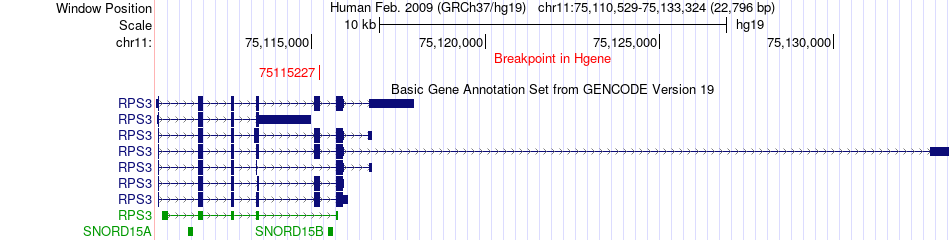 |
 Fusion gene breakpoints across ZNF536 (3'-gene) Fusion gene breakpoints across ZNF536 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
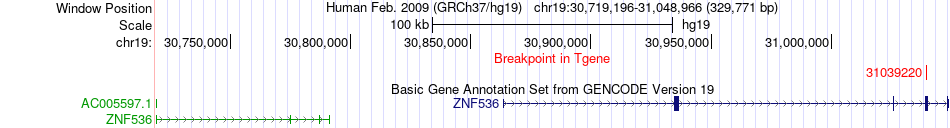 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | READ | TCGA-AG-3582 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
Top |
Fusion Gene ORF analysis for RPS3-ZNF536 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000278572 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| 5CDS-intron | ENST00000524851 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| 5CDS-intron | ENST00000526608 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| 5CDS-intron | ENST00000527446 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| 5CDS-intron | ENST00000531188 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| In-frame | ENST00000278572 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| In-frame | ENST00000524851 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| In-frame | ENST00000526608 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| In-frame | ENST00000527446 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| In-frame | ENST00000531188 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| intron-3CDS | ENST00000529285 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| intron-3CDS | ENST00000530164 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| intron-3CDS | ENST00000534440 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| intron-intron | ENST00000529285 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| intron-intron | ENST00000530164 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
| intron-intron | ENST00000534440 | ENST00000590564 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000531188 | RPS3 | chr11 | 75115227 | + | ENST00000355537 | ZNF536 | chr19 | 31039220 | + | 2540 | 436 | 493 | 1644 | 383 |
| ENST00000278572 | RPS3 | chr11 | 75115227 | + | ENST00000355537 | ZNF536 | chr19 | 31039220 | + | 2544 | 440 | 497 | 1648 | 383 |
| ENST00000527446 | RPS3 | chr11 | 75115227 | + | ENST00000355537 | ZNF536 | chr19 | 31039220 | + | 2495 | 391 | 448 | 1599 | 383 |
| ENST00000526608 | RPS3 | chr11 | 75115227 | + | ENST00000355537 | ZNF536 | chr19 | 31039220 | + | 2453 | 349 | 406 | 1557 | 383 |
| ENST00000524851 | RPS3 | chr11 | 75115227 | + | ENST00000355537 | ZNF536 | chr19 | 31039220 | + | 2483 | 379 | 436 | 1587 | 383 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000531188 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + | 0.001193389 | 0.9988066 |
| ENST00000278572 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + | 0.001086421 | 0.99891365 |
| ENST00000527446 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + | 0.001224153 | 0.9987758 |
| ENST00000526608 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + | 0.001168023 | 0.9988319 |
| ENST00000524851 | ENST00000355537 | RPS3 | chr11 | 75115227 | + | ZNF536 | chr19 | 31039220 | + | 0.001224397 | 0.99877554 |
Top |
Fusion Genomic Features for RPS3-ZNF536 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
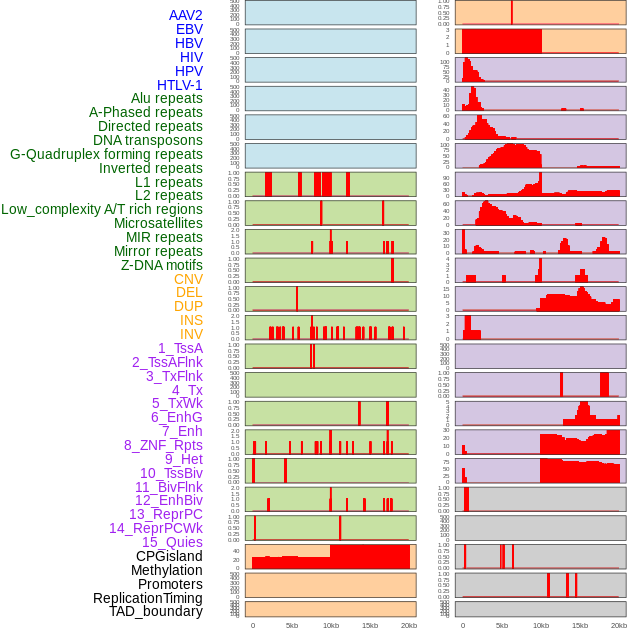 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for RPS3-ZNF536 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:75115227/chr19:31039220) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 130_152 | 0 | 1301.0 | Zinc finger | C2H2-type 1 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 158_180 | 0 | 1301.0 | Zinc finger | C2H2-type 2 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 274_297 | 0 | 1301.0 | Zinc finger | C2H2-type 3 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 300_323 | 0 | 1301.0 | Zinc finger | C2H2-type 4 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 345_367 | 0 | 1301.0 | Zinc finger | C2H2-type 5 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 373_395 | 0 | 1301.0 | Zinc finger | C2H2-type 6 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 631_653 | 0 | 1301.0 | Zinc finger | C2H2-type 7 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 751_773 | 0 | 1301.0 | Zinc finger | C2H2-type 8 | |
| Tgene | ZNF536 | chr11:75115227 | chr19:31039220 | ENST00000355537 | 0 | 5 | 779_801 | 0 | 1301.0 | Zinc finger | C2H2-type 9 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RPS3 | chr11:75115227 | chr19:31039220 | ENST00000278572 | + | 1 | 7 | 21_92 | 0 | 279.3333333333333 | Domain | KH type-2 |
| Hgene | RPS3 | chr11:75115227 | chr19:31039220 | ENST00000524851 | + | 1 | 6 | 21_92 | 0 | 244.0 | Domain | KH type-2 |
| Hgene | RPS3 | chr11:75115227 | chr19:31039220 | ENST00000527446 | + | 1 | 7 | 21_92 | 0 | 413.0 | Domain | KH type-2 |
| Hgene | RPS3 | chr11:75115227 | chr19:31039220 | ENST00000531188 | + | 1 | 7 | 21_92 | 0 | 646.6666666666666 | Domain | KH type-2 |
Top |
Fusion Gene Sequence for RPS3-ZNF536 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >77917_77917_1_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000278572_ZNF536_chr19_31039220_ENST00000355537_length(transcript)=2544nt_BP=440nt GCGGAGCGCGGCGGCAAGATGGCAGTGCAAATATCCAAGAAGAGGAAGTTTGTCGCTGATGGCATCTTCAAAGCTGAACTGAATGAGTTT CTTACTCGGGAGCTGGCTGAAGATGGCTACTCTGGAGTTGAGGTGCGAGTTACACCAACCAGGACAGAAATCATTATCTTAGCCACCAGA ACACAGAATGTTCTTGGTGAGAAGGGCCGGCGGATTCGGGAACTGACTGCTGTAGTTCAGAAGAGGTTTGGCTTTCCAGAGGGCAGTGTA GAGTTGAAAATAATGGTGATGGTAACAGGATATCCCTTGCTTCCTTTAAAGCTTTATGCTGAAAAGGTGGCCACTAGAGGTCTGTGTGCC ATTGCCCAGGCAGAGTCTCTGCGTTACAAACTCCTAGGAGGGCTTGCTGTGCGGAGTGCGCCACGTGTTGCTCAGACAGGGAGATCGGAA GAGCTTATCAAAGCATTGTGAGCAACGGTGTGAATTTCCAAGGGTCCTTGCAAGCTTTCATGGACAGTTTTGTCCTCAGTTCCTTGAAGA AGGAGAAGGACATGAAGGACAAAGCCCTGGCTGACCCCCCTTCCATGAAAGTCCACGGAGTGGATGGTGGTGAGGAGAAACCCAGTGGCA AGTCCTCCCAGAGGAAGTCCGAGAAATCTCAGTATGAACCCCTGGACTTGTCTGTGCGGCCAGATGCCGCCTCCCTCCCGGGCTCCTCGG TAACTGTGCAGGACAGCATTGCATGGCACGGCTGCTTGTTTTGTGCTTTCACAACGTCCTCCATGGAGCTCATGGCCCTTCATCTCCAGG CCAACCACCTGGGCAAAGCGAAACGCAAAGATAACACCATCGGGGTCACAGTCAACTGCAAAGACCAAGCCCGGGAGGCGAGTAAGATGG CCCTGCTGCCCTCGTTACAATCAAACAAAGACCTGGGCCTCTCCAATATGATCAGCTCTCTAGACTCTGCTTCTGAGAAGATGGCCCAAG GTCAGCTCAAGGAGACTCTGGGAGAGCAGAAGAGCGGTGCATGGACCGGCCACGTGGACCCTGCATTTTGTAACTTCCCATCAGACTTCT ACAAGCAGTTTGGTGTTTACCCAGGCATGGTTGGCTCAGGGGCCTCCAGTTCCTGCCCCAACAAGGAGCCTGATGGAAAGGCCCACTCTG AAGAGGATGTCCCCATCCTGATCCCCGAAACCACGAGTAAGAACACTACTGATGACCTCTCTGACATTGCCTCCTCAGAGGACATGGACT CCTCCAAGGGGGAGAACAACGATGAAGAGGATGTTGAAACCGAACCGGAAATGATGACCAAGCCACTGTCTGCCCTCAGCAAAGACAGCA GCAGCGATGGCGGGGACAGCCTGCAGCCCACAGGCACCTCCCAGCCCGTCCAGGGACTGGTCTCACCTTTATCCCAAGCACCGGAGAAGC AGTGGCACAGCCAGGGTCTTCTCCAAGCCCAGGACCCCTTGGCGGGCCTGCCAAAGCCGGAGCGGGGGCCCCAGAGCCTGGACAAGCCGA TGAACATGCTGTCGGTCCTCAGGGCCTACAGTTCTGATGGCTTAGCAGCCTTTAACGGACTTGCAAGTAGCACAGCAAATTCTGGATGTA TCAAGAGGCCAGACTTGTGTGGTAAGTGACACTCCCTGTCCTAGTCGGTCTATCTGGACTTGCCCTTGTCTGTTCGTGGTCCTCGGTGGT TATCTGCAGCTTGTTAATCGTGTAAAGTCAAGAGAAGAATGTATACACATATGTGTGTTGAATAATTACTATTGGCATAGGTATGTGTAT ACACACGGTGCACCAATCTACAGTATATATAGCAGAGAATCAGAGGCTAAAAATATTACCCCATATGTTCCAGTATTAGTCATGGATTGC AAAGGCTTAGTAACTTGAGCAGGAGAGAAAACTCCCTCAAAGTCATAAATCCTGAGTGACAACTGCTGCTGGATGACAGATCCCTTCACC TGTGGACAACCTGGCTGGGGGTGGGGGGCTGTTCCACCAGCTCACCTGAGCATGTAGAGGTGGGTCCTGCAGTGGTCTCGTGGGTATTAC TGCTTGTGTCTGATTGTCCTGTATTTTGTAACACTTTAGAAGAATACAGAAAAGTGCAGTAATTCTCTTTCTCCATAGTATTTAAGCAGA AATATTGCTAGTTTAATATTGTGTCAGGTCGTCCTATTAACCAGGAGCAGATGACAGTAAAATTTCAGTGAATAGCACCTTGACATCTAC AACTTAAAAATGGTGATTGAAGCAAAATATGTAAACTTGTACGGGGTGATCGTGTGCTTTGGAACAGAGTATTGTTGAAGTAATTAGAAG ATATATTAAGGTGTTCCTGGTAATGAAGGCATGTAAGTTATAATAATTGTAGCTTTCTGAATAAGTGTCAAACTATATCTTTAAGTGTGC TGTATGCTGAGTTACAAGTTAGGTCATTTATGAATGGAATGTAAAATAATACTAAAAATGCTTCAATAACTTATCTTGGTATTGCTAATA >77917_77917_1_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000278572_ZNF536_chr19_31039220_ENST00000355537_length(amino acids)=383AA_BP= MQAFMDSFVLSSLKKEKDMKDKALADPPSMKVHGVDGGEEKPSGKSSQRKSEKSQYEPLDLSVRPDAASLPGSSVTVQDSIAWHGCLFCA FTTSSMELMALHLQANHLGKAKRKDNTIGVTVNCKDQAREASKMALLPSLQSNKDLGLSNMISSLDSASEKMAQGQLKETLGEQKSGAWT GHVDPAFCNFPSDFYKQFGVYPGMVGSGASSSCPNKEPDGKAHSEEDVPILIPETTSKNTTDDLSDIASSEDMDSSKGENNDEEDVETEP EMMTKPLSALSKDSSSDGGDSLQPTGTSQPVQGLVSPLSQAPEKQWHSQGLLQAQDPLAGLPKPERGPQSLDKPMNMLSVLRAYSSDGLA -------------------------------------------------------------- >77917_77917_2_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000524851_ZNF536_chr19_31039220_ENST00000355537_length(transcript)=2483nt_BP=379nt GCAAGATGGCAGTGCAAATATCCAAGAAGAGGAAGTTTGTCGCTGATGGCATCTTCAAAGCTGAACTGAATGAGTTTCTTACTCGGGAGC TGGCTGAAGATGGCTACTCTGGAGTTGAGGTGCGAGTTACACCAACCAGGACAGAAATCATTATCTTAGCCACCAGAACACAGAATGTTC TTGGTGAGAAGGGCCGGCGGATTCGGGAACTGACTGCTGTAGTTCAGAAGAGGTTTGGCTTTCCAGAGGGCAGTGTAGAGCTTTATGCTG AAAAGGTGGCCACTAGAGGTCTGTGTGCCATTGCCCAGGCAGAGTCTCTGCGTTACAAACTCCTAGGAGGGCTTGCTGTGCGGAGTGCGC CACGTGTTGCTCAGACAGGGAGATCGGAAGAGCTTATCAAAGCATTGTGAGCAACGGTGTGAATTTCCAAGGGTCCTTGCAAGCTTTCAT GGACAGTTTTGTCCTCAGTTCCTTGAAGAAGGAGAAGGACATGAAGGACAAAGCCCTGGCTGACCCCCCTTCCATGAAAGTCCACGGAGT GGATGGTGGTGAGGAGAAACCCAGTGGCAAGTCCTCCCAGAGGAAGTCCGAGAAATCTCAGTATGAACCCCTGGACTTGTCTGTGCGGCC AGATGCCGCCTCCCTCCCGGGCTCCTCGGTAACTGTGCAGGACAGCATTGCATGGCACGGCTGCTTGTTTTGTGCTTTCACAACGTCCTC CATGGAGCTCATGGCCCTTCATCTCCAGGCCAACCACCTGGGCAAAGCGAAACGCAAAGATAACACCATCGGGGTCACAGTCAACTGCAA AGACCAAGCCCGGGAGGCGAGTAAGATGGCCCTGCTGCCCTCGTTACAATCAAACAAAGACCTGGGCCTCTCCAATATGATCAGCTCTCT AGACTCTGCTTCTGAGAAGATGGCCCAAGGTCAGCTCAAGGAGACTCTGGGAGAGCAGAAGAGCGGTGCATGGACCGGCCACGTGGACCC TGCATTTTGTAACTTCCCATCAGACTTCTACAAGCAGTTTGGTGTTTACCCAGGCATGGTTGGCTCAGGGGCCTCCAGTTCCTGCCCCAA CAAGGAGCCTGATGGAAAGGCCCACTCTGAAGAGGATGTCCCCATCCTGATCCCCGAAACCACGAGTAAGAACACTACTGATGACCTCTC TGACATTGCCTCCTCAGAGGACATGGACTCCTCCAAGGGGGAGAACAACGATGAAGAGGATGTTGAAACCGAACCGGAAATGATGACCAA GCCACTGTCTGCCCTCAGCAAAGACAGCAGCAGCGATGGCGGGGACAGCCTGCAGCCCACAGGCACCTCCCAGCCCGTCCAGGGACTGGT CTCACCTTTATCCCAAGCACCGGAGAAGCAGTGGCACAGCCAGGGTCTTCTCCAAGCCCAGGACCCCTTGGCGGGCCTGCCAAAGCCGGA GCGGGGGCCCCAGAGCCTGGACAAGCCGATGAACATGCTGTCGGTCCTCAGGGCCTACAGTTCTGATGGCTTAGCAGCCTTTAACGGACT TGCAAGTAGCACAGCAAATTCTGGATGTATCAAGAGGCCAGACTTGTGTGGTAAGTGACACTCCCTGTCCTAGTCGGTCTATCTGGACTT GCCCTTGTCTGTTCGTGGTCCTCGGTGGTTATCTGCAGCTTGTTAATCGTGTAAAGTCAAGAGAAGAATGTATACACATATGTGTGTTGA ATAATTACTATTGGCATAGGTATGTGTATACACACGGTGCACCAATCTACAGTATATATAGCAGAGAATCAGAGGCTAAAAATATTACCC CATATGTTCCAGTATTAGTCATGGATTGCAAAGGCTTAGTAACTTGAGCAGGAGAGAAAACTCCCTCAAAGTCATAAATCCTGAGTGACA ACTGCTGCTGGATGACAGATCCCTTCACCTGTGGACAACCTGGCTGGGGGTGGGGGGCTGTTCCACCAGCTCACCTGAGCATGTAGAGGT GGGTCCTGCAGTGGTCTCGTGGGTATTACTGCTTGTGTCTGATTGTCCTGTATTTTGTAACACTTTAGAAGAATACAGAAAAGTGCAGTA ATTCTCTTTCTCCATAGTATTTAAGCAGAAATATTGCTAGTTTAATATTGTGTCAGGTCGTCCTATTAACCAGGAGCAGATGACAGTAAA ATTTCAGTGAATAGCACCTTGACATCTACAACTTAAAAATGGTGATTGAAGCAAAATATGTAAACTTGTACGGGGTGATCGTGTGCTTTG GAACAGAGTATTGTTGAAGTAATTAGAAGATATATTAAGGTGTTCCTGGTAATGAAGGCATGTAAGTTATAATAATTGTAGCTTTCTGAA TAAGTGTCAAACTATATCTTTAAGTGTGCTGTATGCTGAGTTACAAGTTAGGTCATTTATGAATGGAATGTAAAATAATACTAAAAATGC >77917_77917_2_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000524851_ZNF536_chr19_31039220_ENST00000355537_length(amino acids)=383AA_BP= MQAFMDSFVLSSLKKEKDMKDKALADPPSMKVHGVDGGEEKPSGKSSQRKSEKSQYEPLDLSVRPDAASLPGSSVTVQDSIAWHGCLFCA FTTSSMELMALHLQANHLGKAKRKDNTIGVTVNCKDQAREASKMALLPSLQSNKDLGLSNMISSLDSASEKMAQGQLKETLGEQKSGAWT GHVDPAFCNFPSDFYKQFGVYPGMVGSGASSSCPNKEPDGKAHSEEDVPILIPETTSKNTTDDLSDIASSEDMDSSKGENNDEEDVETEP EMMTKPLSALSKDSSSDGGDSLQPTGTSQPVQGLVSPLSQAPEKQWHSQGLLQAQDPLAGLPKPERGPQSLDKPMNMLSVLRAYSSDGLA -------------------------------------------------------------- >77917_77917_3_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000526608_ZNF536_chr19_31039220_ENST00000355537_length(transcript)=2453nt_BP=349nt GCGGCGGCAAGATGGCAGTGCAAATATCCAAGAAGAGGAAGTTTGTCGCTGATGGCATCTTCAAAGCTGAACTGAATGAGTTTCTTACTC GGGAGCTGGCTGAAGATGGCTACTCTGGAGTTGAGGTGCGAGTTACACCAACCAGGACAGAAATCATTATCTTAGCCACCAGAACACAGA ATGTTCTTGGTGAGAAGGGCCGGCGGATTCGGGAACTGACTGCTGTAGTTCAGAAGAGGTTTGGCTTTCCAGAGGGCAGTCTGTGTGCCA TTGCCCAGGCAGAGTCTCTGCGTTACAAACTCCTAGGAGGGCTTGCTGTGCGGAGTGCGCCACGTGTTGCTCAGACAGGGAGATCGGAAG AGCTTATCAAAGCATTGTGAGCAACGGTGTGAATTTCCAAGGGTCCTTGCAAGCTTTCATGGACAGTTTTGTCCTCAGTTCCTTGAAGAA GGAGAAGGACATGAAGGACAAAGCCCTGGCTGACCCCCCTTCCATGAAAGTCCACGGAGTGGATGGTGGTGAGGAGAAACCCAGTGGCAA GTCCTCCCAGAGGAAGTCCGAGAAATCTCAGTATGAACCCCTGGACTTGTCTGTGCGGCCAGATGCCGCCTCCCTCCCGGGCTCCTCGGT AACTGTGCAGGACAGCATTGCATGGCACGGCTGCTTGTTTTGTGCTTTCACAACGTCCTCCATGGAGCTCATGGCCCTTCATCTCCAGGC CAACCACCTGGGCAAAGCGAAACGCAAAGATAACACCATCGGGGTCACAGTCAACTGCAAAGACCAAGCCCGGGAGGCGAGTAAGATGGC CCTGCTGCCCTCGTTACAATCAAACAAAGACCTGGGCCTCTCCAATATGATCAGCTCTCTAGACTCTGCTTCTGAGAAGATGGCCCAAGG TCAGCTCAAGGAGACTCTGGGAGAGCAGAAGAGCGGTGCATGGACCGGCCACGTGGACCCTGCATTTTGTAACTTCCCATCAGACTTCTA CAAGCAGTTTGGTGTTTACCCAGGCATGGTTGGCTCAGGGGCCTCCAGTTCCTGCCCCAACAAGGAGCCTGATGGAAAGGCCCACTCTGA AGAGGATGTCCCCATCCTGATCCCCGAAACCACGAGTAAGAACACTACTGATGACCTCTCTGACATTGCCTCCTCAGAGGACATGGACTC CTCCAAGGGGGAGAACAACGATGAAGAGGATGTTGAAACCGAACCGGAAATGATGACCAAGCCACTGTCTGCCCTCAGCAAAGACAGCAG CAGCGATGGCGGGGACAGCCTGCAGCCCACAGGCACCTCCCAGCCCGTCCAGGGACTGGTCTCACCTTTATCCCAAGCACCGGAGAAGCA GTGGCACAGCCAGGGTCTTCTCCAAGCCCAGGACCCCTTGGCGGGCCTGCCAAAGCCGGAGCGGGGGCCCCAGAGCCTGGACAAGCCGAT GAACATGCTGTCGGTCCTCAGGGCCTACAGTTCTGATGGCTTAGCAGCCTTTAACGGACTTGCAAGTAGCACAGCAAATTCTGGATGTAT CAAGAGGCCAGACTTGTGTGGTAAGTGACACTCCCTGTCCTAGTCGGTCTATCTGGACTTGCCCTTGTCTGTTCGTGGTCCTCGGTGGTT ATCTGCAGCTTGTTAATCGTGTAAAGTCAAGAGAAGAATGTATACACATATGTGTGTTGAATAATTACTATTGGCATAGGTATGTGTATA CACACGGTGCACCAATCTACAGTATATATAGCAGAGAATCAGAGGCTAAAAATATTACCCCATATGTTCCAGTATTAGTCATGGATTGCA AAGGCTTAGTAACTTGAGCAGGAGAGAAAACTCCCTCAAAGTCATAAATCCTGAGTGACAACTGCTGCTGGATGACAGATCCCTTCACCT GTGGACAACCTGGCTGGGGGTGGGGGGCTGTTCCACCAGCTCACCTGAGCATGTAGAGGTGGGTCCTGCAGTGGTCTCGTGGGTATTACT GCTTGTGTCTGATTGTCCTGTATTTTGTAACACTTTAGAAGAATACAGAAAAGTGCAGTAATTCTCTTTCTCCATAGTATTTAAGCAGAA ATATTGCTAGTTTAATATTGTGTCAGGTCGTCCTATTAACCAGGAGCAGATGACAGTAAAATTTCAGTGAATAGCACCTTGACATCTACA ACTTAAAAATGGTGATTGAAGCAAAATATGTAAACTTGTACGGGGTGATCGTGTGCTTTGGAACAGAGTATTGTTGAAGTAATTAGAAGA TATATTAAGGTGTTCCTGGTAATGAAGGCATGTAAGTTATAATAATTGTAGCTTTCTGAATAAGTGTCAAACTATATCTTTAAGTGTGCT GTATGCTGAGTTACAAGTTAGGTCATTTATGAATGGAATGTAAAATAATACTAAAAATGCTTCAATAACTTATCTTGGTATTGCTAATAA >77917_77917_3_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000526608_ZNF536_chr19_31039220_ENST00000355537_length(amino acids)=383AA_BP= MQAFMDSFVLSSLKKEKDMKDKALADPPSMKVHGVDGGEEKPSGKSSQRKSEKSQYEPLDLSVRPDAASLPGSSVTVQDSIAWHGCLFCA FTTSSMELMALHLQANHLGKAKRKDNTIGVTVNCKDQAREASKMALLPSLQSNKDLGLSNMISSLDSASEKMAQGQLKETLGEQKSGAWT GHVDPAFCNFPSDFYKQFGVYPGMVGSGASSSCPNKEPDGKAHSEEDVPILIPETTSKNTTDDLSDIASSEDMDSSKGENNDEEDVETEP EMMTKPLSALSKDSSSDGGDSLQPTGTSQPVQGLVSPLSQAPEKQWHSQGLLQAQDPLAGLPKPERGPQSLDKPMNMLSVLRAYSSDGLA -------------------------------------------------------------- >77917_77917_4_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000527446_ZNF536_chr19_31039220_ENST00000355537_length(transcript)=2495nt_BP=391nt CGGAGCGCGGCGGCAAGATGGCAGTGCAAATATCCAAGAAGAGGAAGTTTGTCGCTGATGGCATCTTCAAAGCTGAACTGAATGAGTTTC TTACTCGGGAGCTGGCTGAAGATGGCTACTCTGGAGTTGAGGTGCGAGTTACACCAACCAGGACAGAAATCATTATCTTAGCCACCAGAA CACAGAATGTTCTTGGTGAGAAGGGCCGGCGGATTCGGGAACTGACTGCTGTAGTTCAGAAGAGGTTTGGCTTTCCAGAGGGCAGTGTAG AGCTTTATGCTGAAAAGGTGGCCACTAGAGGTCTGTGTGCCATTGCCCAGGCAGAGTCTCTGCGTTACAAACTCCTAGGAGGGCTTGCTG TGCGGAGTGCGCCACGTGTTGCTCAGACAGGGAGATCGGAAGAGCTTATCAAAGCATTGTGAGCAACGGTGTGAATTTCCAAGGGTCCTT GCAAGCTTTCATGGACAGTTTTGTCCTCAGTTCCTTGAAGAAGGAGAAGGACATGAAGGACAAAGCCCTGGCTGACCCCCCTTCCATGAA AGTCCACGGAGTGGATGGTGGTGAGGAGAAACCCAGTGGCAAGTCCTCCCAGAGGAAGTCCGAGAAATCTCAGTATGAACCCCTGGACTT GTCTGTGCGGCCAGATGCCGCCTCCCTCCCGGGCTCCTCGGTAACTGTGCAGGACAGCATTGCATGGCACGGCTGCTTGTTTTGTGCTTT CACAACGTCCTCCATGGAGCTCATGGCCCTTCATCTCCAGGCCAACCACCTGGGCAAAGCGAAACGCAAAGATAACACCATCGGGGTCAC AGTCAACTGCAAAGACCAAGCCCGGGAGGCGAGTAAGATGGCCCTGCTGCCCTCGTTACAATCAAACAAAGACCTGGGCCTCTCCAATAT GATCAGCTCTCTAGACTCTGCTTCTGAGAAGATGGCCCAAGGTCAGCTCAAGGAGACTCTGGGAGAGCAGAAGAGCGGTGCATGGACCGG CCACGTGGACCCTGCATTTTGTAACTTCCCATCAGACTTCTACAAGCAGTTTGGTGTTTACCCAGGCATGGTTGGCTCAGGGGCCTCCAG TTCCTGCCCCAACAAGGAGCCTGATGGAAAGGCCCACTCTGAAGAGGATGTCCCCATCCTGATCCCCGAAACCACGAGTAAGAACACTAC TGATGACCTCTCTGACATTGCCTCCTCAGAGGACATGGACTCCTCCAAGGGGGAGAACAACGATGAAGAGGATGTTGAAACCGAACCGGA AATGATGACCAAGCCACTGTCTGCCCTCAGCAAAGACAGCAGCAGCGATGGCGGGGACAGCCTGCAGCCCACAGGCACCTCCCAGCCCGT CCAGGGACTGGTCTCACCTTTATCCCAAGCACCGGAGAAGCAGTGGCACAGCCAGGGTCTTCTCCAAGCCCAGGACCCCTTGGCGGGCCT GCCAAAGCCGGAGCGGGGGCCCCAGAGCCTGGACAAGCCGATGAACATGCTGTCGGTCCTCAGGGCCTACAGTTCTGATGGCTTAGCAGC CTTTAACGGACTTGCAAGTAGCACAGCAAATTCTGGATGTATCAAGAGGCCAGACTTGTGTGGTAAGTGACACTCCCTGTCCTAGTCGGT CTATCTGGACTTGCCCTTGTCTGTTCGTGGTCCTCGGTGGTTATCTGCAGCTTGTTAATCGTGTAAAGTCAAGAGAAGAATGTATACACA TATGTGTGTTGAATAATTACTATTGGCATAGGTATGTGTATACACACGGTGCACCAATCTACAGTATATATAGCAGAGAATCAGAGGCTA AAAATATTACCCCATATGTTCCAGTATTAGTCATGGATTGCAAAGGCTTAGTAACTTGAGCAGGAGAGAAAACTCCCTCAAAGTCATAAA TCCTGAGTGACAACTGCTGCTGGATGACAGATCCCTTCACCTGTGGACAACCTGGCTGGGGGTGGGGGGCTGTTCCACCAGCTCACCTGA GCATGTAGAGGTGGGTCCTGCAGTGGTCTCGTGGGTATTACTGCTTGTGTCTGATTGTCCTGTATTTTGTAACACTTTAGAAGAATACAG AAAAGTGCAGTAATTCTCTTTCTCCATAGTATTTAAGCAGAAATATTGCTAGTTTAATATTGTGTCAGGTCGTCCTATTAACCAGGAGCA GATGACAGTAAAATTTCAGTGAATAGCACCTTGACATCTACAACTTAAAAATGGTGATTGAAGCAAAATATGTAAACTTGTACGGGGTGA TCGTGTGCTTTGGAACAGAGTATTGTTGAAGTAATTAGAAGATATATTAAGGTGTTCCTGGTAATGAAGGCATGTAAGTTATAATAATTG TAGCTTTCTGAATAAGTGTCAAACTATATCTTTAAGTGTGCTGTATGCTGAGTTACAAGTTAGGTCATTTATGAATGGAATGTAAAATAA >77917_77917_4_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000527446_ZNF536_chr19_31039220_ENST00000355537_length(amino acids)=383AA_BP= MQAFMDSFVLSSLKKEKDMKDKALADPPSMKVHGVDGGEEKPSGKSSQRKSEKSQYEPLDLSVRPDAASLPGSSVTVQDSIAWHGCLFCA FTTSSMELMALHLQANHLGKAKRKDNTIGVTVNCKDQAREASKMALLPSLQSNKDLGLSNMISSLDSASEKMAQGQLKETLGEQKSGAWT GHVDPAFCNFPSDFYKQFGVYPGMVGSGASSSCPNKEPDGKAHSEEDVPILIPETTSKNTTDDLSDIASSEDMDSSKGENNDEEDVETEP EMMTKPLSALSKDSSSDGGDSLQPTGTSQPVQGLVSPLSQAPEKQWHSQGLLQAQDPLAGLPKPERGPQSLDKPMNMLSVLRAYSSDGLA -------------------------------------------------------------- >77917_77917_5_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000531188_ZNF536_chr19_31039220_ENST00000355537_length(transcript)=2540nt_BP=436nt CCTTTATACTCACTTCCGCCCGCGAGCCACTTCCTTTCCTTTCAGCGGAGCGCGGCGGCAAGATGGCAGTGCAAATATCCAAGAAGAGGA AGTTTGTCGCTGATGGCATCTTCAAAGCTGAACTGAATGAGTTTCTTACTCGGGAGCTGGCTGAAGATGGCTACTCTGGAGTTGAGGTGC GAGTTACACCAACCAGGACAGAAATCATTATCTTAGCCACCAGAACACAGAATGTTCTTGGTGAGAAGGGCCGGCGGATTCGGGAACTGA CTGCTGTAGTTCAGAAGAGGTTTGGCTTTCCAGAGGGCAGTGTAGAGCTTTATGCTGAAAAGGTGGCCACTAGAGGTCTGTGTGCCATTG CCCAGGCAGAGTCTCTGCGTTACAAACTCCTAGGAGGGCTTGCTGTGCGGAGTGCGCCACGTGTTGCTCAGACAGGGAGATCGGAAGAGC TTATCAAAGCATTGTGAGCAACGGTGTGAATTTCCAAGGGTCCTTGCAAGCTTTCATGGACAGTTTTGTCCTCAGTTCCTTGAAGAAGGA GAAGGACATGAAGGACAAAGCCCTGGCTGACCCCCCTTCCATGAAAGTCCACGGAGTGGATGGTGGTGAGGAGAAACCCAGTGGCAAGTC CTCCCAGAGGAAGTCCGAGAAATCTCAGTATGAACCCCTGGACTTGTCTGTGCGGCCAGATGCCGCCTCCCTCCCGGGCTCCTCGGTAAC TGTGCAGGACAGCATTGCATGGCACGGCTGCTTGTTTTGTGCTTTCACAACGTCCTCCATGGAGCTCATGGCCCTTCATCTCCAGGCCAA CCACCTGGGCAAAGCGAAACGCAAAGATAACACCATCGGGGTCACAGTCAACTGCAAAGACCAAGCCCGGGAGGCGAGTAAGATGGCCCT GCTGCCCTCGTTACAATCAAACAAAGACCTGGGCCTCTCCAATATGATCAGCTCTCTAGACTCTGCTTCTGAGAAGATGGCCCAAGGTCA GCTCAAGGAGACTCTGGGAGAGCAGAAGAGCGGTGCATGGACCGGCCACGTGGACCCTGCATTTTGTAACTTCCCATCAGACTTCTACAA GCAGTTTGGTGTTTACCCAGGCATGGTTGGCTCAGGGGCCTCCAGTTCCTGCCCCAACAAGGAGCCTGATGGAAAGGCCCACTCTGAAGA GGATGTCCCCATCCTGATCCCCGAAACCACGAGTAAGAACACTACTGATGACCTCTCTGACATTGCCTCCTCAGAGGACATGGACTCCTC CAAGGGGGAGAACAACGATGAAGAGGATGTTGAAACCGAACCGGAAATGATGACCAAGCCACTGTCTGCCCTCAGCAAAGACAGCAGCAG CGATGGCGGGGACAGCCTGCAGCCCACAGGCACCTCCCAGCCCGTCCAGGGACTGGTCTCACCTTTATCCCAAGCACCGGAGAAGCAGTG GCACAGCCAGGGTCTTCTCCAAGCCCAGGACCCCTTGGCGGGCCTGCCAAAGCCGGAGCGGGGGCCCCAGAGCCTGGACAAGCCGATGAA CATGCTGTCGGTCCTCAGGGCCTACAGTTCTGATGGCTTAGCAGCCTTTAACGGACTTGCAAGTAGCACAGCAAATTCTGGATGTATCAA GAGGCCAGACTTGTGTGGTAAGTGACACTCCCTGTCCTAGTCGGTCTATCTGGACTTGCCCTTGTCTGTTCGTGGTCCTCGGTGGTTATC TGCAGCTTGTTAATCGTGTAAAGTCAAGAGAAGAATGTATACACATATGTGTGTTGAATAATTACTATTGGCATAGGTATGTGTATACAC ACGGTGCACCAATCTACAGTATATATAGCAGAGAATCAGAGGCTAAAAATATTACCCCATATGTTCCAGTATTAGTCATGGATTGCAAAG GCTTAGTAACTTGAGCAGGAGAGAAAACTCCCTCAAAGTCATAAATCCTGAGTGACAACTGCTGCTGGATGACAGATCCCTTCACCTGTG GACAACCTGGCTGGGGGTGGGGGGCTGTTCCACCAGCTCACCTGAGCATGTAGAGGTGGGTCCTGCAGTGGTCTCGTGGGTATTACTGCT TGTGTCTGATTGTCCTGTATTTTGTAACACTTTAGAAGAATACAGAAAAGTGCAGTAATTCTCTTTCTCCATAGTATTTAAGCAGAAATA TTGCTAGTTTAATATTGTGTCAGGTCGTCCTATTAACCAGGAGCAGATGACAGTAAAATTTCAGTGAATAGCACCTTGACATCTACAACT TAAAAATGGTGATTGAAGCAAAATATGTAAACTTGTACGGGGTGATCGTGTGCTTTGGAACAGAGTATTGTTGAAGTAATTAGAAGATAT ATTAAGGTGTTCCTGGTAATGAAGGCATGTAAGTTATAATAATTGTAGCTTTCTGAATAAGTGTCAAACTATATCTTTAAGTGTGCTGTA TGCTGAGTTACAAGTTAGGTCATTTATGAATGGAATGTAAAATAATACTAAAAATGCTTCAATAACTTATCTTGGTATTGCTAATAAAAA >77917_77917_5_RPS3-ZNF536_RPS3_chr11_75115227_ENST00000531188_ZNF536_chr19_31039220_ENST00000355537_length(amino acids)=383AA_BP= MQAFMDSFVLSSLKKEKDMKDKALADPPSMKVHGVDGGEEKPSGKSSQRKSEKSQYEPLDLSVRPDAASLPGSSVTVQDSIAWHGCLFCA FTTSSMELMALHLQANHLGKAKRKDNTIGVTVNCKDQAREASKMALLPSLQSNKDLGLSNMISSLDSASEKMAQGQLKETLGEQKSGAWT GHVDPAFCNFPSDFYKQFGVYPGMVGSGASSSCPNKEPDGKAHSEEDVPILIPETTSKNTTDDLSDIASSEDMDSSKGENNDEEDVETEP EMMTKPLSALSKDSSSDGGDSLQPTGTSQPVQGLVSPLSQAPEKQWHSQGLLQAQDPLAGLPKPERGPQSLDKPMNMLSVLRAYSSDGLA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RPS3-ZNF536 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RPS3-ZNF536 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RPS3-ZNF536 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
