
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:RUNX3-LDLRAP1 (FusionGDB2 ID:78714) |
Fusion Gene Summary for RUNX3-LDLRAP1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: RUNX3-LDLRAP1 | Fusion gene ID: 78714 | Hgene | Tgene | Gene symbol | RUNX3 | LDLRAP1 | Gene ID | 864 | 26119 |
| Gene name | RUNX family transcription factor 3 | low density lipoprotein receptor adaptor protein 1 | |
| Synonyms | AML2|CBFA3|PEBP2aC | ARH|ARH1|ARH2|FHCB1|FHCB2|FHCL4 | |
| Cytomap | 1p36.11 | 1p36.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | runt-related transcription factor 3CBF-alpha-3PEA2 alpha CPEBP2 alpha CSL3-3 enhancer factor 1 alpha C subunitSL3/AKV core-binding factor alpha C subunitacute myeloid leukemia 2 proteinacute myeloid leukemia gene 2core-binding factor subunit alpha | low density lipoprotein receptor adapter protein 1LDL receptor adaptor proteinautosomal recessive hypercholesterolemia protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q5SW96 | |
| Ensembl transtripts involved in fusion gene | ENST00000308873, ENST00000338888, ENST00000399916, ENST00000540420, ENST00000496967, | ENST00000488127, ENST00000374338, | |
| Fusion gene scores | * DoF score | 3 X 4 X 3=36 | 1 X 1 X 1=1 |
| # samples | 4 | 1 | |
| ** MAII score | log2(4/36*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: RUNX3 [Title/Abstract] AND LDLRAP1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | RUNX3(25245730)-LDLRAP1(25880412), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | RUNX3-LDLRAP1 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. RUNX3-LDLRAP1 seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. RUNX3-LDLRAP1 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | RUNX3 | GO:0006468 | protein phosphorylation | 20100835 |
| Hgene | RUNX3 | GO:0045893 | positive regulation of transcription, DNA-templated | 20599712 |
| Hgene | RUNX3 | GO:0071559 | response to transforming growth factor beta | 20599712 |
| Tgene | LDLRAP1 | GO:0006898 | receptor-mediated endocytosis | 14528014 |
 Fusion gene breakpoints across RUNX3 (5'-gene) Fusion gene breakpoints across RUNX3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
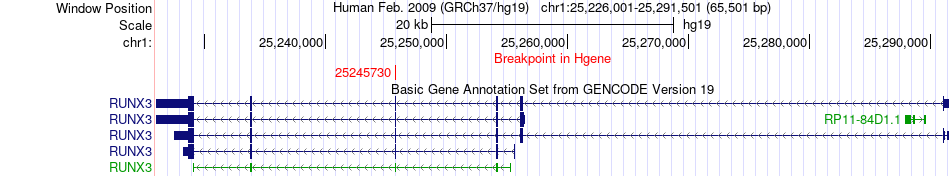 |
 Fusion gene breakpoints across LDLRAP1 (3'-gene) Fusion gene breakpoints across LDLRAP1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
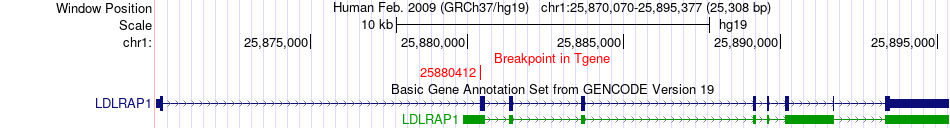 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-1548 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
Top |
Fusion Gene ORF analysis for RUNX3-LDLRAP1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000308873 | ENST00000488127 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| 5CDS-3UTR | ENST00000338888 | ENST00000488127 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| 5CDS-3UTR | ENST00000399916 | ENST00000488127 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| 5CDS-3UTR | ENST00000540420 | ENST00000488127 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| 5UTR-3CDS | ENST00000496967 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| 5UTR-3UTR | ENST00000496967 | ENST00000488127 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| Frame-shift | ENST00000338888 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| In-frame | ENST00000308873 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| In-frame | ENST00000399916 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
| In-frame | ENST00000540420 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000399916 | RUNX3 | chr1 | 25245730 | - | ENST00000374338 | LDLRAP1 | chr1 | 25880412 | + | 3758 | 1025 | 439 | 1863 | 474 |
| ENST00000308873 | RUNX3 | chr1 | 25245730 | - | ENST00000374338 | LDLRAP1 | chr1 | 25880412 | + | 3286 | 553 | 9 | 1391 | 460 |
| ENST00000540420 | RUNX3 | chr1 | 25245730 | - | ENST00000374338 | LDLRAP1 | chr1 | 25880412 | + | 3077 | 344 | 79 | 1182 | 367 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000399916 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + | 0.006667068 | 0.9933329 |
| ENST00000308873 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + | 0.01086403 | 0.989136 |
| ENST00000540420 | ENST00000374338 | RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + | 0.004855441 | 0.99514455 |
Top |
Fusion Genomic Features for RUNX3-LDLRAP1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + | 1.50E-05 | 0.999985 |
| RUNX3 | chr1 | 25245730 | - | LDLRAP1 | chr1 | 25880412 | + | 1.50E-05 | 0.999985 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
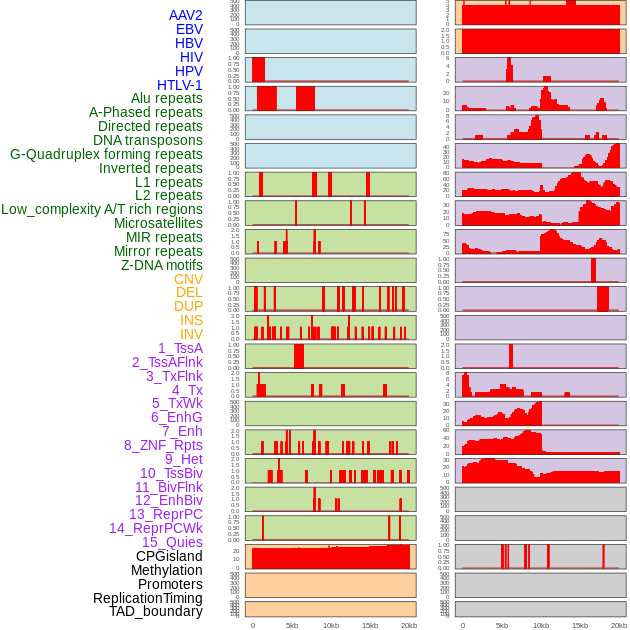 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
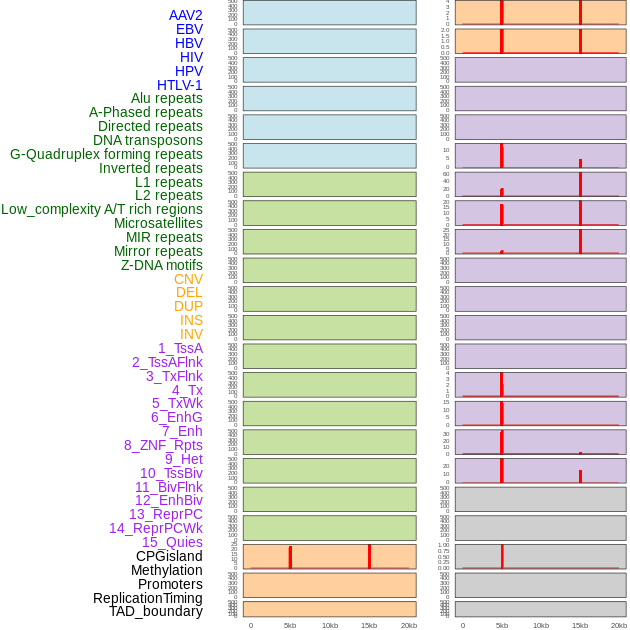 |
Top |
Fusion Protein Features for RUNX3-LDLRAP1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:25245730/chr1:25880412) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LDLRAP1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Adapter protein (clathrin-associated sorting protein (CLASP)) required for efficient endocytosis of the LDL receptor (LDLR) in polarized cells such as hepatocytes and lymphocytes, but not in non-polarized cells (fibroblasts). May be required for LDL binding and internalization but not for receptor clustering in coated pits. May facilitate the endocytocis of LDLR and LDLR-LDL complexes from coated pits by stabilizing the interaction between the receptor and the structural components of the pits. May also be involved in the internalization of other LDLR family members. Binds to phosphoinositides, which regulate clathrin bud assembly at the cell surface. Required for trafficking of LRP2 to the endocytic recycling compartment which is necessary for LRP2 proteolysis, releasing a tail fragment which translocates to the nucleus and mediates transcriptional repression (By similarity). {ECO:0000250|UniProtKB:D3ZAR1, ECO:0000269|PubMed:15728179}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000308873 | - | 3 | 5 | 23_27 | 181 | 416.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000338888 | - | 5 | 7 | 23_27 | 195 | 430.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000399916 | - | 4 | 6 | 23_27 | 195 | 430.0 | Compositional bias | Note=Poly-Gly |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000308873 | - | 3 | 5 | 54_182 | 181 | 416.0 | Domain | Runt |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000338888 | - | 5 | 7 | 54_182 | 195 | 430.0 | Domain | Runt |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000399916 | - | 4 | 6 | 54_182 | 195 | 430.0 | Domain | Runt |
| Tgene | LDLRAP1 | chr1:25245730 | chr1:25880412 | ENST00000374338 | 0 | 9 | 42_196 | 29 | 309.0 | Domain | PID | |
| Tgene | LDLRAP1 | chr1:25245730 | chr1:25880412 | ENST00000374338 | 0 | 9 | 212_216 | 29 | 309.0 | Motif | Clathrin box | |
| Tgene | LDLRAP1 | chr1:25245730 | chr1:25880412 | ENST00000374338 | 0 | 9 | 257_266 | 29 | 309.0 | Motif | [DE]-X(1%2C2)-F-X-X-[FL]-X-X-X-R motif | |
| Tgene | LDLRAP1 | chr1:25245730 | chr1:25880412 | ENST00000374338 | 0 | 9 | 249_276 | 29 | 309.0 | Region | AP-2 complex binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000308873 | - | 3 | 5 | 191_415 | 181 | 416.0 | Compositional bias | Note=Pro/Ser/Thr-rich |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000338888 | - | 5 | 7 | 191_415 | 195 | 430.0 | Compositional bias | Note=Pro/Ser/Thr-rich |
| Hgene | RUNX3 | chr1:25245730 | chr1:25880412 | ENST00000399916 | - | 4 | 6 | 191_415 | 195 | 430.0 | Compositional bias | Note=Pro/Ser/Thr-rich |
| Tgene | LDLRAP1 | chr1:25245730 | chr1:25880412 | ENST00000374338 | 0 | 9 | 23_26 | 29 | 309.0 | Compositional bias | Note=Poly-Gly |
Top |
Fusion Gene Sequence for RUNX3-LDLRAP1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >78714_78714_1_RUNX3-LDLRAP1_RUNX3_chr1_25245730_ENST00000308873_LDLRAP1_chr1_25880412_ENST00000374338_length(transcript)=3286nt_BP=553nt GCCGCTGTTATGCGTATTCCCGTAGACCCAAGCACCAGCCGCCGCTTCACACCTCCCTCCCCGGCCTTCCCCTGCGGCGGCGGCGGCGGC AAGATGGGCGAGAACAGCGGCGCGCTGAGCGCGCAGGCGGCCGTGGGGCCCGGAGGGCGCGCCCGGCCCGAGGTGCGCTCGATGGTGGAC GTGCTGGCGGACCACGCAGGCGAGCTCGTGCGCACCGACAGCCCCAACTTCCTCTGCTCCGTGCTGCCCTCGCACTGGCGCTGCAACAAG ACGCTGCCCGTCGCCTTCAAGGTGGTGGCATTGGGGGACGTGCCGGATGGTACGGTGGTGACTGTGATGGCAGGCAATGACGAGAACTAC TCCGCTGAGCTGCGCAATGCCTCGGCCGTCATGAAGAACCAGGTGGCCAGGTTCAACGACCTTCGCTTCGTGGGCCGCAGTGGGCGAGGG AAGAGTTTCACCCTGACCATCACTGTGTTCACCAACCCCACCCAAGTGGCGACCTACCACCGAGCCATCAAGGTGACCGTGGACGGACCC CGGGAGCCCAGACAGCTGCCTGAGAACTGGACAGACACGCGGGAGACGCTGCTGGAGGGGATGCTGTTCAGCCTCAAGTACCTGGGCATG ACGCTAGTGGAGCAGCCCAAGGGTGAGGAGCTGTCGGCCGCCGCCATCAAGAGGATCGTGGCTACAGCTAAGGCCAGTGGGAAGAAGCTG CAGAAGGTGACTCTGAAGGTGTCGCCACGGGGAATTATCCTGACAGACAACCTCACCAACCAGCTCATTGAGAACGTGTCCATATACAGG ATCTCCTATTGCACAGCAGACAAGATGCACGACAAGGTGTTTGCATACATCGCCCAGAGCCAGCACAACCAGAGCCTCGAGTGCCACGCC TTCCTCTGCACCAAGCGGAAGATGGCACAGGCTGTTACCCTCACCGTAGCCCAGGCCTTCAAAGTCGCCTTTGAGTTTTGGCAGGTGTCC AAGGAAGAGAAAGAGAAGAGGGACAAAGCCAGCCAAGAGGGAGGGGACGTCCTGGGGGCCCGCCAAGACTGCACCCCCTCCTTGAAGAGC TTGGTCGCCACTGGGAACCTGCTGGACTTAGAGGAGACAGCTAAGGCCCCGCTGTCCACGGTCAGCGCCAACACCACCAACATGGACGAG GTGCCGCGGCCACAAGCCTTGAGTGGCAGCAGTGTTGTCTGGGAGCTGGATGATGGCCTGGATGAAGCGTTTTCGAGGCTTGCCCAGTCT CGGACAAACCCTCAGGTCCTGGACACTGGCCTGACAGCCCAGGACATGCATTACGCCCAGTGCCTCTCGCCTGTCGACTGGGACAAGCCT GACAGCAGCGGCACAGAGCAGGATGACCTCTTCAGCTTCTGAGGGCCCGGGGCCAGCCGGACACAAGCGGCCCTGACACGTGATGGACCA AAGCCACCTGCTGCGGGGGAGCCAGTTCTGGGGCCCGCCTGCCACCTCTCCCAGCCCTCAGCATTGTCAGCCTGAAGATCAGAGCTGCAG CCAGTCAGGCAGGGGAGAGATTTTTCTTTTAAGCCCTGCTCTTTCTCTGAGAACCAAAAGATGCCTTGAATATTTATTCAGTGACTTCTG GCTTATGCTCAGAAGCCAGTCTGCGTCAGGCACGTCTCCTGCTGCGTGACATGTGCAGTGCTGTAATCGGCTCCCGCTTGCTCTCCTGGA GCAAGCTCTGCCCTGGCTGTGGGTATCAGGACTGTGACCAAAGCATTTCTAGTCCCTTCTCTCTTTCTAAGGACCCAAATTTCCCTGGGG GCATCCTGCTTCCTGAAAGCTGTTGGATTTCAGTGATTTTTCCCCCCACCCCCCAGCACAGGAGAGCACCCACAGCCGCAGAAGGGGAAT GTGTCCTCCTGCTCTGCTTCCTCAGGGCCCAGCAGGCGGGGGTTTGAGCCCTGGACCCCAGGCTCTTAGAGACTAAGGGGCAGCTCCTGA CCAAAGACGATACAGCTTGGCACTTTAAAGCATTAACAGCAGGTGTGACCCTGAGGGCTCCTCCATGGTGCTGCATTGAGTCCAGCTTTC CTTCTGCCCTTCCTCCAGGAGAAGGGGCCCAAGGTCCCCGTGGATGGTCTCCACCTGTGCTTGGAACCAGTGTAACTGGCTGCTCCCTGC TCCCAGGGACTGACACGGGGATCATCTCTGTGACCGCCCTCCGTCGGGCCCCTGCCTGCCTTCTCCCCTCCACGCAAGGCTGTGCTCTTC CTCTGGTTTCTGTGTGTCCGTTTGAGTGTCTGCGCCCCGCCTCCCCATACTTCCTGGGATGATGTGTGAAACCTGACACCTAGATTTATT TGGAAATATTCTATGACCACTTTACAGATGAGGAAACAGGCCTCAAGCGTGGAGGGGTAGAGTGAAGAGTAGAACCCAGGTCTGATGCCA AAGCTGCTTTCTTCTCTGCCTCCTCCTCACGCAACTCACACCTCCTTTTCTTCTAGCTTTGTTGTCCTCCCAGGAACCAAAAAACCCCAG CTATTTTCTGACCAAAATGTGTTTCATAACAAACCATCTGGTGCCTTTCCACACAGAACTGGCAGGAGCCTCGTGTCCTGCTAGCTGTCT CTCTTGTTGATTTCCGTGAAAATGCAAGTGTTTGAAGTCTGCTCATTCCGAGGGTGAAACAAAATCCAACCCTGTCAGAATCATGCTGTT CTCTTTGCTGACACTGTGACCCTGGGTCGGGACAGACCAGCAGCAATCTGTCTTTAGAATCGCTTTCCTTCCTCCCCTTTTGCCCCCGTG GGGCTCCCGGCATCCTGAAAGCCAGCAAAGCCTCCAGCATCTTTTCCATCCTGAGGTGCCTCCCAGTGGCCTGGCTTGTCGGAGCAAGTT TCATCAGCCCTAGGGAAAACACGGCCCTCCTGGGAACCTCCTTACCTGGAGTAACCGGACACCTTAGACGGAGGTGCCTGAGGGTGGGGT GGGATTTGCAGGGTCATTATCAGAACATGAGGATAACTTCCTTGCCCCTGCTCTGTAGCCACCTCCTTGGCACCGGCCTCTATTTGTCAT AAGGCGGCGTGGGCGAGGCCTGACACAGGCCAGCCTTGGCACGAGGGGGGCCAGGGGTTCTGAGAAGCGCTGCCCTGTGAGAGCCACGCT GGCCTTCGTCTCCATCTCTGGTTGACGGGCTGTCCGTGTGCCTCCTGTGTGTCTGCAGACAAGTCTTGCTGTGCTTTATTTGTGAAACTT >78714_78714_1_RUNX3-LDLRAP1_RUNX3_chr1_25245730_ENST00000308873_LDLRAP1_chr1_25880412_ENST00000374338_length(amino acids)=460AA_BP=182 MRIPVDPSTSRRFTPPSPAFPCGGGGGKMGENSGALSAQAAVGPGGRARPEVRSMVDVLADHAGELVRTDSPNFLCSVLPSHWRCNKTLP VAFKVVALGDVPDGTVVTVMAGNDENYSAELRNASAVMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPTQVATYHRAIKVTVDGPREP RQLPENWTDTRETLLEGMLFSLKYLGMTLVEQPKGEELSAAAIKRIVATAKASGKKLQKVTLKVSPRGIILTDNLTNQLIENVSIYRISY CTADKMHDKVFAYIAQSQHNQSLECHAFLCTKRKMAQAVTLTVAQAFKVAFEFWQVSKEEKEKRDKASQEGGDVLGARQDCTPSLKSLVA TGNLLDLEETAKAPLSTVSANTTNMDEVPRPQALSGSSVVWELDDGLDEAFSRLAQSRTNPQVLDTGLTAQDMHYAQCLSPVDWDKPDSS -------------------------------------------------------------- >78714_78714_2_RUNX3-LDLRAP1_RUNX3_chr1_25245730_ENST00000399916_LDLRAP1_chr1_25880412_ENST00000374338_length(transcript)=3758nt_BP=1025nt ATTCATTCATTCCCCGTGGCACTGGAGGCGGCCCACTCTGCTCTGTCAGCTTCGGAGCTCCTCCACCCTGGCTGCCGAAAGCCCCTTCCC GCCATCTAATGATACACTCTGCATACGCTTCTGTTGAGAATTTGTGGCTAGACATTCCTGTGGGACCGGGAATCCAAATTCTTGGGTACA AACAGAAACTTACTTTCCTTGGGGATTTTTTTCTCTCTCTCACTCACACACACTCTCGCGTTCTTTCCTTTTTTCTTTTTCGTAGCAGCA GGGGGGAAAAAAGAGACAAAAACAAAACAAAAAACAACAAAAAGCAACACCCCCCCCTTTTATTTTCAAAAGTAGCTAGAGGAAAAAAAA ATAAAACAACAGCCAACCAAGTGAATCCCAACCCAACCCCCTGAAGGGCTGAAAATTCTCGCCTTCTTCAGAGCGGGGCATGGCATCGAA CAGCATCTTCGACTCCTTCCCGACCTACTCGCCGACCTTCATCCGCGACCCAAGCACCAGCCGCCGCTTCACACCTCCCTCCCCGGCCTT CCCCTGCGGCGGCGGCGGCGGCAAGATGGGCGAGAACAGCGGCGCGCTGAGCGCGCAGGCGGCCGTGGGGCCCGGAGGGCGCGCCCGGCC CGAGGTGCGCTCGATGGTGGACGTGCTGGCGGACCACGCAGGCGAGCTCGTGCGCACCGACAGCCCCAACTTCCTCTGCTCCGTGCTGCC CTCGCACTGGCGCTGCAACAAGACGCTGCCCGTCGCCTTCAAGGTGGTGGCATTGGGGGACGTGCCGGATGGTACGGTGGTGACTGTGAT GGCAGGCAATGACGAGAACTACTCCGCTGAGCTGCGCAATGCCTCGGCCGTCATGAAGAACCAGGTGGCCAGGTTCAACGACCTTCGCTT CGTGGGCCGCAGTGGGCGAGGGAAGAGTTTCACCCTGACCATCACTGTGTTCACCAACCCCACCCAAGTGGCGACCTACCACCGAGCCAT CAAGGTGACCGTGGACGGACCCCGGGAGCCCAGACAGCTGCCTGAGAACTGGACAGACACGCGGGAGACGCTGCTGGAGGGGATGCTGTT CAGCCTCAAGTACCTGGGCATGACGCTAGTGGAGCAGCCCAAGGGTGAGGAGCTGTCGGCCGCCGCCATCAAGAGGATCGTGGCTACAGC TAAGGCCAGTGGGAAGAAGCTGCAGAAGGTGACTCTGAAGGTGTCGCCACGGGGAATTATCCTGACAGACAACCTCACCAACCAGCTCAT TGAGAACGTGTCCATATACAGGATCTCCTATTGCACAGCAGACAAGATGCACGACAAGGTGTTTGCATACATCGCCCAGAGCCAGCACAA CCAGAGCCTCGAGTGCCACGCCTTCCTCTGCACCAAGCGGAAGATGGCACAGGCTGTTACCCTCACCGTAGCCCAGGCCTTCAAAGTCGC CTTTGAGTTTTGGCAGGTGTCCAAGGAAGAGAAAGAGAAGAGGGACAAAGCCAGCCAAGAGGGAGGGGACGTCCTGGGGGCCCGCCAAGA CTGCACCCCCTCCTTGAAGAGCTTGGTCGCCACTGGGAACCTGCTGGACTTAGAGGAGACAGCTAAGGCCCCGCTGTCCACGGTCAGCGC CAACACCACCAACATGGACGAGGTGCCGCGGCCACAAGCCTTGAGTGGCAGCAGTGTTGTCTGGGAGCTGGATGATGGCCTGGATGAAGC GTTTTCGAGGCTTGCCCAGTCTCGGACAAACCCTCAGGTCCTGGACACTGGCCTGACAGCCCAGGACATGCATTACGCCCAGTGCCTCTC GCCTGTCGACTGGGACAAGCCTGACAGCAGCGGCACAGAGCAGGATGACCTCTTCAGCTTCTGAGGGCCCGGGGCCAGCCGGACACAAGC GGCCCTGACACGTGATGGACCAAAGCCACCTGCTGCGGGGGAGCCAGTTCTGGGGCCCGCCTGCCACCTCTCCCAGCCCTCAGCATTGTC AGCCTGAAGATCAGAGCTGCAGCCAGTCAGGCAGGGGAGAGATTTTTCTTTTAAGCCCTGCTCTTTCTCTGAGAACCAAAAGATGCCTTG AATATTTATTCAGTGACTTCTGGCTTATGCTCAGAAGCCAGTCTGCGTCAGGCACGTCTCCTGCTGCGTGACATGTGCAGTGCTGTAATC GGCTCCCGCTTGCTCTCCTGGAGCAAGCTCTGCCCTGGCTGTGGGTATCAGGACTGTGACCAAAGCATTTCTAGTCCCTTCTCTCTTTCT AAGGACCCAAATTTCCCTGGGGGCATCCTGCTTCCTGAAAGCTGTTGGATTTCAGTGATTTTTCCCCCCACCCCCCAGCACAGGAGAGCA CCCACAGCCGCAGAAGGGGAATGTGTCCTCCTGCTCTGCTTCCTCAGGGCCCAGCAGGCGGGGGTTTGAGCCCTGGACCCCAGGCTCTTA GAGACTAAGGGGCAGCTCCTGACCAAAGACGATACAGCTTGGCACTTTAAAGCATTAACAGCAGGTGTGACCCTGAGGGCTCCTCCATGG TGCTGCATTGAGTCCAGCTTTCCTTCTGCCCTTCCTCCAGGAGAAGGGGCCCAAGGTCCCCGTGGATGGTCTCCACCTGTGCTTGGAACC AGTGTAACTGGCTGCTCCCTGCTCCCAGGGACTGACACGGGGATCATCTCTGTGACCGCCCTCCGTCGGGCCCCTGCCTGCCTTCTCCCC TCCACGCAAGGCTGTGCTCTTCCTCTGGTTTCTGTGTGTCCGTTTGAGTGTCTGCGCCCCGCCTCCCCATACTTCCTGGGATGATGTGTG AAACCTGACACCTAGATTTATTTGGAAATATTCTATGACCACTTTACAGATGAGGAAACAGGCCTCAAGCGTGGAGGGGTAGAGTGAAGA GTAGAACCCAGGTCTGATGCCAAAGCTGCTTTCTTCTCTGCCTCCTCCTCACGCAACTCACACCTCCTTTTCTTCTAGCTTTGTTGTCCT CCCAGGAACCAAAAAACCCCAGCTATTTTCTGACCAAAATGTGTTTCATAACAAACCATCTGGTGCCTTTCCACACAGAACTGGCAGGAG CCTCGTGTCCTGCTAGCTGTCTCTCTTGTTGATTTCCGTGAAAATGCAAGTGTTTGAAGTCTGCTCATTCCGAGGGTGAAACAAAATCCA ACCCTGTCAGAATCATGCTGTTCTCTTTGCTGACACTGTGACCCTGGGTCGGGACAGACCAGCAGCAATCTGTCTTTAGAATCGCTTTCC TTCCTCCCCTTTTGCCCCCGTGGGGCTCCCGGCATCCTGAAAGCCAGCAAAGCCTCCAGCATCTTTTCCATCCTGAGGTGCCTCCCAGTG GCCTGGCTTGTCGGAGCAAGTTTCATCAGCCCTAGGGAAAACACGGCCCTCCTGGGAACCTCCTTACCTGGAGTAACCGGACACCTTAGA CGGAGGTGCCTGAGGGTGGGGTGGGATTTGCAGGGTCATTATCAGAACATGAGGATAACTTCCTTGCCCCTGCTCTGTAGCCACCTCCTT GGCACCGGCCTCTATTTGTCATAAGGCGGCGTGGGCGAGGCCTGACACAGGCCAGCCTTGGCACGAGGGGGGCCAGGGGTTCTGAGAAGC GCTGCCCTGTGAGAGCCACGCTGGCCTTCGTCTCCATCTCTGGTTGACGGGCTGTCCGTGTGCCTCCTGTGTGTCTGCAGACAAGTCTTG >78714_78714_2_RUNX3-LDLRAP1_RUNX3_chr1_25245730_ENST00000399916_LDLRAP1_chr1_25880412_ENST00000374338_length(amino acids)=474AA_BP=196 MASNSIFDSFPTYSPTFIRDPSTSRRFTPPSPAFPCGGGGGKMGENSGALSAQAAVGPGGRARPEVRSMVDVLADHAGELVRTDSPNFLC SVLPSHWRCNKTLPVAFKVVALGDVPDGTVVTVMAGNDENYSAELRNASAVMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPTQVATY HRAIKVTVDGPREPRQLPENWTDTRETLLEGMLFSLKYLGMTLVEQPKGEELSAAAIKRIVATAKASGKKLQKVTLKVSPRGIILTDNLT NQLIENVSIYRISYCTADKMHDKVFAYIAQSQHNQSLECHAFLCTKRKMAQAVTLTVAQAFKVAFEFWQVSKEEKEKRDKASQEGGDVLG ARQDCTPSLKSLVATGNLLDLEETAKAPLSTVSANTTNMDEVPRPQALSGSSVVWELDDGLDEAFSRLAQSRTNPQVLDTGLTAQDMHYA -------------------------------------------------------------- >78714_78714_3_RUNX3-LDLRAP1_RUNX3_chr1_25245730_ENST00000540420_LDLRAP1_chr1_25880412_ENST00000374338_length(transcript)=3077nt_BP=344nt GAAATTTTACCGGACGGCGCTAGGCCGGGATCGGGGCACTGCTCTCCAGACGTTTGCTCGGGGATATTATGCGTCCCGAATGGTGGTGGC ATTGGGGGACGTGCCGGATGGTACGGTGGTGACTGTGATGGCAGGCAATGACGAGAACTACTCCGCTGAGCTGCGCAATGCCTCGGCCGT CATGAAGAACCAGGTGGCCAGGTTCAACGACCTTCGCTTCGTGGGCCGCAGTGGGCGAGGGAAGAGTTTCACCCTGACCATCACTGTGTT CACCAACCCCACCCAAGTGGCGACCTACCACCGAGCCATCAAGGTGACCGTGGACGGACCCCGGGAGCCCAGACAGCTGCCTGAGAACTG GACAGACACGCGGGAGACGCTGCTGGAGGGGATGCTGTTCAGCCTCAAGTACCTGGGCATGACGCTAGTGGAGCAGCCCAAGGGTGAGGA GCTGTCGGCCGCCGCCATCAAGAGGATCGTGGCTACAGCTAAGGCCAGTGGGAAGAAGCTGCAGAAGGTGACTCTGAAGGTGTCGCCACG GGGAATTATCCTGACAGACAACCTCACCAACCAGCTCATTGAGAACGTGTCCATATACAGGATCTCCTATTGCACAGCAGACAAGATGCA CGACAAGGTGTTTGCATACATCGCCCAGAGCCAGCACAACCAGAGCCTCGAGTGCCACGCCTTCCTCTGCACCAAGCGGAAGATGGCACA GGCTGTTACCCTCACCGTAGCCCAGGCCTTCAAAGTCGCCTTTGAGTTTTGGCAGGTGTCCAAGGAAGAGAAAGAGAAGAGGGACAAAGC CAGCCAAGAGGGAGGGGACGTCCTGGGGGCCCGCCAAGACTGCACCCCCTCCTTGAAGAGCTTGGTCGCCACTGGGAACCTGCTGGACTT AGAGGAGACAGCTAAGGCCCCGCTGTCCACGGTCAGCGCCAACACCACCAACATGGACGAGGTGCCGCGGCCACAAGCCTTGAGTGGCAG CAGTGTTGTCTGGGAGCTGGATGATGGCCTGGATGAAGCGTTTTCGAGGCTTGCCCAGTCTCGGACAAACCCTCAGGTCCTGGACACTGG CCTGACAGCCCAGGACATGCATTACGCCCAGTGCCTCTCGCCTGTCGACTGGGACAAGCCTGACAGCAGCGGCACAGAGCAGGATGACCT CTTCAGCTTCTGAGGGCCCGGGGCCAGCCGGACACAAGCGGCCCTGACACGTGATGGACCAAAGCCACCTGCTGCGGGGGAGCCAGTTCT GGGGCCCGCCTGCCACCTCTCCCAGCCCTCAGCATTGTCAGCCTGAAGATCAGAGCTGCAGCCAGTCAGGCAGGGGAGAGATTTTTCTTT TAAGCCCTGCTCTTTCTCTGAGAACCAAAAGATGCCTTGAATATTTATTCAGTGACTTCTGGCTTATGCTCAGAAGCCAGTCTGCGTCAG GCACGTCTCCTGCTGCGTGACATGTGCAGTGCTGTAATCGGCTCCCGCTTGCTCTCCTGGAGCAAGCTCTGCCCTGGCTGTGGGTATCAG GACTGTGACCAAAGCATTTCTAGTCCCTTCTCTCTTTCTAAGGACCCAAATTTCCCTGGGGGCATCCTGCTTCCTGAAAGCTGTTGGATT TCAGTGATTTTTCCCCCCACCCCCCAGCACAGGAGAGCACCCACAGCCGCAGAAGGGGAATGTGTCCTCCTGCTCTGCTTCCTCAGGGCC CAGCAGGCGGGGGTTTGAGCCCTGGACCCCAGGCTCTTAGAGACTAAGGGGCAGCTCCTGACCAAAGACGATACAGCTTGGCACTTTAAA GCATTAACAGCAGGTGTGACCCTGAGGGCTCCTCCATGGTGCTGCATTGAGTCCAGCTTTCCTTCTGCCCTTCCTCCAGGAGAAGGGGCC CAAGGTCCCCGTGGATGGTCTCCACCTGTGCTTGGAACCAGTGTAACTGGCTGCTCCCTGCTCCCAGGGACTGACACGGGGATCATCTCT GTGACCGCCCTCCGTCGGGCCCCTGCCTGCCTTCTCCCCTCCACGCAAGGCTGTGCTCTTCCTCTGGTTTCTGTGTGTCCGTTTGAGTGT CTGCGCCCCGCCTCCCCATACTTCCTGGGATGATGTGTGAAACCTGACACCTAGATTTATTTGGAAATATTCTATGACCACTTTACAGAT GAGGAAACAGGCCTCAAGCGTGGAGGGGTAGAGTGAAGAGTAGAACCCAGGTCTGATGCCAAAGCTGCTTTCTTCTCTGCCTCCTCCTCA CGCAACTCACACCTCCTTTTCTTCTAGCTTTGTTGTCCTCCCAGGAACCAAAAAACCCCAGCTATTTTCTGACCAAAATGTGTTTCATAA CAAACCATCTGGTGCCTTTCCACACAGAACTGGCAGGAGCCTCGTGTCCTGCTAGCTGTCTCTCTTGTTGATTTCCGTGAAAATGCAAGT GTTTGAAGTCTGCTCATTCCGAGGGTGAAACAAAATCCAACCCTGTCAGAATCATGCTGTTCTCTTTGCTGACACTGTGACCCTGGGTCG GGACAGACCAGCAGCAATCTGTCTTTAGAATCGCTTTCCTTCCTCCCCTTTTGCCCCCGTGGGGCTCCCGGCATCCTGAAAGCCAGCAAA GCCTCCAGCATCTTTTCCATCCTGAGGTGCCTCCCAGTGGCCTGGCTTGTCGGAGCAAGTTTCATCAGCCCTAGGGAAAACACGGCCCTC CTGGGAACCTCCTTACCTGGAGTAACCGGACACCTTAGACGGAGGTGCCTGAGGGTGGGGTGGGATTTGCAGGGTCATTATCAGAACATG AGGATAACTTCCTTGCCCCTGCTCTGTAGCCACCTCCTTGGCACCGGCCTCTATTTGTCATAAGGCGGCGTGGGCGAGGCCTGACACAGG CCAGCCTTGGCACGAGGGGGGCCAGGGGTTCTGAGAAGCGCTGCCCTGTGAGAGCCACGCTGGCCTTCGTCTCCATCTCTGGTTGACGGG CTGTCCGTGTGCCTCCTGTGTGTCTGCAGACAAGTCTTGCTGTGCTTTATTTGTGAAACTTTAATGAGGAAAAAACAAATAATAAATGTT >78714_78714_3_RUNX3-LDLRAP1_RUNX3_chr1_25245730_ENST00000540420_LDLRAP1_chr1_25880412_ENST00000374338_length(amino acids)=367AA_BP=89 MVVALGDVPDGTVVTVMAGNDENYSAELRNASAVMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPTQVATYHRAIKVTVDGPREPRQL PENWTDTRETLLEGMLFSLKYLGMTLVEQPKGEELSAAAIKRIVATAKASGKKLQKVTLKVSPRGIILTDNLTNQLIENVSIYRISYCTA DKMHDKVFAYIAQSQHNQSLECHAFLCTKRKMAQAVTLTVAQAFKVAFEFWQVSKEEKEKRDKASQEGGDVLGARQDCTPSLKSLVATGN LLDLEETAKAPLSTVSANTTNMDEVPRPQALSGSSVVWELDDGLDEAFSRLAQSRTNPQVLDTGLTAQDMHYAQCLSPVDWDKPDSSGTE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for RUNX3-LDLRAP1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for RUNX3-LDLRAP1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for RUNX3-LDLRAP1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
