
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SEL1L3-BHLHE41 (FusionGDB2 ID:80202) |
Fusion Gene Summary for SEL1L3-BHLHE41 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SEL1L3-BHLHE41 | Fusion gene ID: 80202 | Hgene | Tgene | Gene symbol | SEL1L3 | BHLHE41 | Gene ID | 23231 | 79365 |
| Gene name | SEL1L family member 3 | basic helix-loop-helix family member e41 | |
| Synonyms | Sel-1L3 | BHLHB3|DEC2|FNSS1|SHARP1|hDEC2 | |
| Cytomap | 4p15.2 | 12p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | protein sel-1 homolog 3sel-1 suppressor of lin-12-like 3suppressor of lin-12-like protein 3 | class E basic helix-loop-helix protein 41basic helix-loop-helix domain containing, class B, 3differentially expressed in chondrocytes protein 2enhancer-of-split and hairy-related protein 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9C0J9 | |
| Ensembl transtripts involved in fusion gene | ENST00000513364, ENST00000502949, ENST00000264868, ENST00000399878, | ENST00000242728, | |
| Fusion gene scores | * DoF score | 11 X 12 X 7=924 | 5 X 2 X 4=40 |
| # samples | 12 | 5 | |
| ** MAII score | log2(12/924*10)=-2.94485844580754 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/40*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: SEL1L3 [Title/Abstract] AND BHLHE41 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SEL1L3(25836818)-BHLHE41(26277515), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | SEL1L3-BHLHE41 seems lost the major protein functional domain in Tgene partner, which is a transcription factor due to the frame-shifted ORF. SEL1L3-BHLHE41 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | BHLHE41 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 14672706|15193144 |
 Fusion gene breakpoints across SEL1L3 (5'-gene) Fusion gene breakpoints across SEL1L3 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
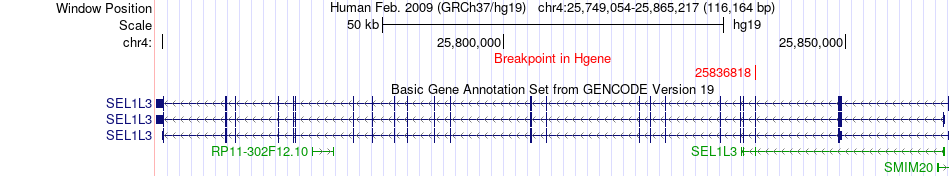 |
 Fusion gene breakpoints across BHLHE41 (3'-gene) Fusion gene breakpoints across BHLHE41 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
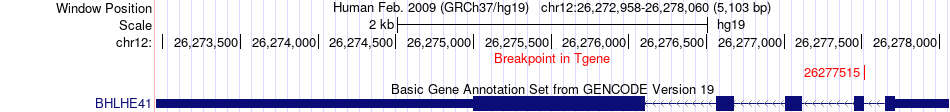 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-25-2042 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - |
Top |
Fusion Gene ORF analysis for SEL1L3-BHLHE41 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000513364 | ENST00000242728 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - |
| Frame-shift | ENST00000502949 | ENST00000242728 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - |
| In-frame | ENST00000264868 | ENST00000242728 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - |
| In-frame | ENST00000399878 | ENST00000242728 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000399878 | SEL1L3 | chr4 | 25836818 | - | ENST00000242728 | BHLHE41 | chr12 | 26277515 | - | 4410 | 983 | 78 | 2369 | 763 |
| ENST00000264868 | SEL1L3 | chr4 | 25836818 | - | ENST00000242728 | BHLHE41 | chr12 | 26277515 | - | 4265 | 838 | 20 | 2224 | 734 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000399878 | ENST00000242728 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - | 0.001162506 | 0.99883753 |
| ENST00000264868 | ENST00000242728 | SEL1L3 | chr4 | 25836818 | - | BHLHE41 | chr12 | 26277515 | - | 0.000594849 | 0.99940515 |
Top |
Fusion Genomic Features for SEL1L3-BHLHE41 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
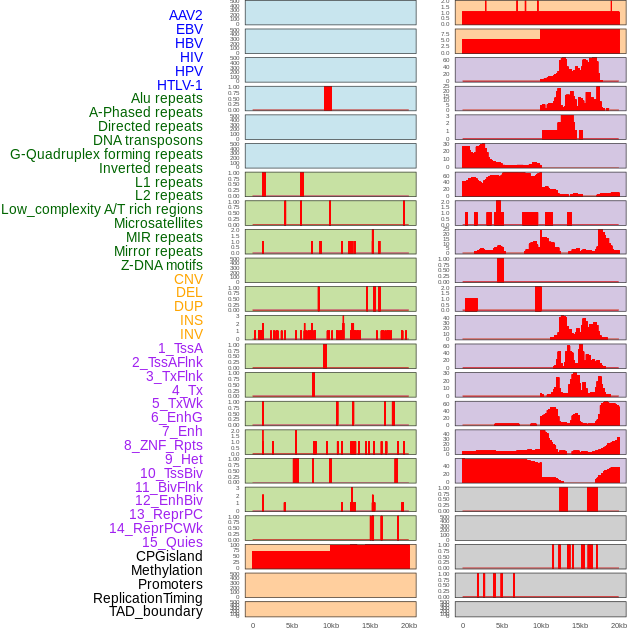 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SEL1L3-BHLHE41 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:25836818/chr12:26277515) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | BHLHE41 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional repressor involved in the regulation of the circadian rhythm by negatively regulating the activity of the clock genes and clock-controlled genes (PubMed:11278948, PubMed:14672706, PubMed:15193144, PubMed:15560782, PubMed:18411297, PubMed:19786558, PubMed:25083013). Acts as the negative limb of a novel autoregulatory feedback loop (DEC loop) which differs from the one formed by the PER and CRY transcriptional repressors (PER/CRY loop). Both these loops are interlocked as it represses the expression of PER1 and in turn is repressed by PER1/2 and CRY1/2. Represses the activity of the circadian transcriptional activator: CLOCK-ARNTL/BMAL1 heterodimer by competing for the binding to E-box elements (5'-CACGTG-3') found within the promoters of its target genes (PubMed:25083013). Negatively regulates its own expression and the expression of DBP and BHLHE41/DEC2. Acts as a corepressor of RXR and the RXR-LXR heterodimers and represses the ligand-induced RXRA/B/G, NR1H3/LXRA, NR1H4 and VDR transactivation activity. Inhibits HNF1A-mediated transactivation of CYP1A2, CYP2E1 AND CYP3A11 (By similarity). {ECO:0000250|UniProtKB:Q99PV5, ECO:0000269|PubMed:11278948, ECO:0000269|PubMed:14672706, ECO:0000269|PubMed:15193144, ECO:0000269|PubMed:15560782, ECO:0000269|PubMed:18411297, ECO:0000269|PubMed:19786558, ECO:0000269|PubMed:25083013}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | BHLHE41 | chr4:25836818 | chr12:26277515 | ENST00000242728 | 0 | 5 | 297_431 | 20 | 483.0 | Compositional bias | Note=Ala/Gly-rich | |
| Tgene | BHLHE41 | chr4:25836818 | chr12:26277515 | ENST00000242728 | 0 | 5 | 131_166 | 20 | 483.0 | Domain | Orange | |
| Tgene | BHLHE41 | chr4:25836818 | chr12:26277515 | ENST00000242728 | 0 | 5 | 44_99 | 20 | 483.0 | Domain | bHLH |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 575_609 | 251 | 1098.0 | Repeat | Note=Sel1-like 1 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 611_647 | 251 | 1098.0 | Repeat | Note=Sel1-like 2 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 694_730 | 251 | 1098.0 | Repeat | Note=Sel1-like 3 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 732_767 | 251 | 1098.0 | Repeat | Note=Sel1-like 4 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 768_800 | 251 | 1098.0 | Repeat | Note=Sel1-like 5 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 801_839 | 251 | 1098.0 | Repeat | Note=Sel1-like 6 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 840_877 | 251 | 1098.0 | Repeat | Note=Sel1-like 7 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 952_988 | 251 | 1098.0 | Repeat | Note=Sel1-like 8 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 575_609 | 286 | 1133.0 | Repeat | Note=Sel1-like 1 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 611_647 | 286 | 1133.0 | Repeat | Note=Sel1-like 2 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 694_730 | 286 | 1133.0 | Repeat | Note=Sel1-like 3 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 732_767 | 286 | 1133.0 | Repeat | Note=Sel1-like 4 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 768_800 | 286 | 1133.0 | Repeat | Note=Sel1-like 5 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 801_839 | 286 | 1133.0 | Repeat | Note=Sel1-like 6 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 840_877 | 286 | 1133.0 | Repeat | Note=Sel1-like 7 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 952_988 | 286 | 1133.0 | Repeat | Note=Sel1-like 8 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 575_609 | 133 | 980.0 | Repeat | Note=Sel1-like 1 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 611_647 | 133 | 980.0 | Repeat | Note=Sel1-like 2 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 694_730 | 133 | 980.0 | Repeat | Note=Sel1-like 3 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 732_767 | 133 | 980.0 | Repeat | Note=Sel1-like 4 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 768_800 | 133 | 980.0 | Repeat | Note=Sel1-like 5 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 801_839 | 133 | 980.0 | Repeat | Note=Sel1-like 6 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 840_877 | 133 | 980.0 | Repeat | Note=Sel1-like 7 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 952_988 | 133 | 980.0 | Repeat | Note=Sel1-like 8 |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000264868 | - | 3 | 24 | 1057_1077 | 251 | 1098.0 | Transmembrane | Helical |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000399878 | - | 3 | 24 | 1057_1077 | 286 | 1133.0 | Transmembrane | Helical |
| Hgene | SEL1L3 | chr4:25836818 | chr12:26277515 | ENST00000502949 | - | 3 | 24 | 1057_1077 | 133 | 980.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for SEL1L3-BHLHE41 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >80202_80202_1_SEL1L3-BHLHE41_SEL1L3_chr4_25836818_ENST00000264868_BHLHE41_chr12_26277515_ENST00000242728_length(transcript)=4265nt_BP=838nt TCAAAATCCATCCTGAAGGCCTGGGAGCCGGGCAGCTGCGCGGCACAACTCCAGGGGGCACACTCGCCCTGCGCCCTGCGGGAATGGCGC CCCGGCCAAAAAAGCAACCAGACAAAAACCCATTGCACGGGCGGGAACTGAATGTTGTACCATCTTTGGGTAGGCAGACTTCCCTGACGA CATCAGTGATACCCAAAGCTGAGCAGAGCGTGGCTTACAAAGACTTTATTTATTTTACTGTCTTTGAAGGAAACGTTCGCAACGTTTCTG AAGTCTCGGTTGAGTATTTATGCTCTCAGCCTTGTGTTGTCAATTTGGAAGCAGTTGTTTCATCTGAGTTCAGAAGTAGCATTCCCGTGT ACAAAAAAAGGTGGAAGAATGAGAAACATCTTCACACCAGCAGGACACAAATAGTACATGTGAAATTTCCAAGCATTATGGTTTACAGAG ATGATTATTTCATCAGACATTCCATCTCTGTATCTGCAGTGATAGTACGCGCCTGGATTACTCACAAATACAGTGGCAGAGACTGGAATG TTAAATGGGAGGAAAACTTGCTCCATGCTGTAGCAAAGAATTATACCCTCCTGCAGACCATCCCGCCTTTTGAACGCCCTTTCAAAGATC ATCAAGTGTGCCTTGAGTGGAACATGGGTTATATTTGGAACCTTCGGGCAAACAGGATTCCACAGTGTCCTCTGGAAAATGATGTGGTTG CCCTGCTTGGCTTTCCTTATGCCTCCAGTGGAGAAAACACAGGCATTGTCAAGAAGTTCCCGAGGTTTCGGAACCGAGAGCTGGAGGCCA CTCGACGCCAGAGGATGGATTACCCAGTACTGGACTATTCCTCTTTGTATATGTGTAAACCCAAAAGGAGCATGAAACGAGACGACACCA AGGATACCTACAAATTACCGCACAGATTAATAGAAAAGAAAAGAAGAGACCGAATTAATGAATGCATTGCTCAGCTGAAAGATTTACTGC CTGAACATCTGAAATTGACAACTCTGGGACATCTGGAGAAAGCTGTAGTCTTGGAATTAACTTTGAAACACTTAAAAGCTTTAACCGCCT TAACCGAGCAACAGCATCAGAAGATAATTGCTTTACAGAATGGGGAGCGATCTCTGAAATCGCCCATTCAGTCCGACTTGGATGCGTTCC ACTCGGGATTTCAAACATGCGCCAAAGAAGTCTTGCAATACCTCTCCCGGTTTGAGAGCTGGACACCCAGGGAGCCGCGGTGTGTCCAGC TGATCAACCACTTGCACGCCGTGGCCACCCAGTTCTTGCCCACCCCGCAGCTGTTGACTCAACAGGTCCCTCTGAGCAAAGGCACCGGCG CTCCCTCGGCCGCCGGGTCCGCGGCCGCCCCCTGCCTGGAGCGCGCGGGGCAGAAGCTGGAGCCCCTCGCCTACTGCGTGCCCGTCATCC AGCGGACTCAGCCCAGCGCCGAGCTCGCCGCCGAGAACGACACGGACACCGACAGCGGCTACGGCGGCGAAGCCGAGGCCCGGCCGGACC GCGAGAAAGGCAAAGGCGCGGGGGCGAGCCGCGTCACCATCAAGCAGGAGCCTCCCGGGGAGGACTCGCCGGCGCCCAAGAGGATGAAGC TGGATTCCCGCGGCGGCGGCAGCGGCGGCGGCCCGGGGGGCGGCGCGGCGGCGGCGGCAGCCGCGCTTCTGGGGCCCGACCCTGCCGCCG CGGCCGCGCTGCTGAGACCCGACGCCGCCCTGCTCAGCTCGCTGGTGGCGTTCGGCGGAGGCGGAGGCGCGCCCTTCCCGCAGCCCGCGG CCGCCGCGGCCCCCTTCTGCCTGCCCTTCTGCTTCCTCTCGCCTTCTGCAGCTGCCGCCTACGTGCAGCCCTTCCTGGACAAGAGCGGCC TGGAGAAGTATCTGTACCCGGCGGCGGCTGCCGCCCCGTTCCCGCTGCTATACCCCGGCATCCCCGCCCCGGCGGCAGCCGCGGCAGCCG CCGCCGCCGCTGCCGCCGCCGCCGCCGCGTTCCCCTGCCTGTCCTCGGTGTTGTCGCCCCCTCCCGAGAAGGCGGGCGCCGCCGCCGCGA CCCTCCTGCCGCACGAGGTGGCGCCCCTTGGGGCGCCGCACCCCCAGCACCCGCACGGCCGCACCCACCTGCCCTTCGCCGGGCCCCGCG AGCCGGGGAACCCGGAGAGCTCTGCTCAGGAAGATCCCTCGCAGCCAGGAAAGGAAGCTCCCTGAATCCTTGCGTCCCGAAGGACGGAGG TTCAAGCAGAGTGAGAAGTTAAAATACCCTTAAGGAGGTTCAAGCAGAGTGAGAAGTTAAAATACCCTTAAGGTCTTTAAGGGAGGAAGT GTAATAGATGCACGACAGGCATAAACAAGAACAACAAAACAGGTGTTATGTGTACATTCGGAGTTCCTGTTTTGCTCATCCCGCACCACC CCACCCTCCACACACTAACATCCCTTTCTTCCCCCCACCAGCTGTAAAAGATCCTATGCGAAAGACACTGGCTCTTTTTTTTAATCCCCC AAATAAATTTTGCCCCCTTTTAGGCCATGTTCCATTATCTCTTAAAATTGGAACCTAATTCGAGAGGAAGTAAGAAGGGTCTGTTCTGTG GCTGAGCTAGGTGAACCCCGGGGTAGGGGAAAGATGTTAACACCTTTGACGTCTTTGGAGTTGACATGGAACAGCAGGTAGTTGTTATGT AGAGCTAGTTCTCAAAGCTGCCCTGCCTGTTTTAGGAGGCGTTCCACAAACAGATTGAGGCTCTTTTAGAATTGAATTTACTCTTCAGTA TTTTCTAATGTTCAGCTTTCTAAAAGGCATATATTTTTCAAAGAAGTGAGGATGCAGTTTCTCACGTTGCAACCTATTCTGAAGTGGTTT AAATGGTATCTCTTAGTAACTTGCACTCGTTAAAGAAACACGGAGCTGGGCCATCGTCAGAACTAAGTCAGGGAAGGAGATGGATGAGAA GGCCAGAATCATTCCTAGTACATTTGCTAACACTTTATTGAGAAATTGACCATGAATTAATGGACTCATCTTAATTTCTTCTAAGTCCAT ATATAGATAGATATCTATCTGTACAGATTTCTATTTATCCATAGATAGGTATCTATACATACACATCTCAAGTGCATCTATTCCCACTCT CATTAATCCATCATGTTCCTAAATTTTTGTAATCTTACTGTAAAAAAAAGTGCACTGAACTTCAAAACAAAACAAAAAACAACAACAACA AAAAACAAGTCCAAACTGATATATCCTATATTCTGTTAAAATTCAAAAGTGAACGAAAGCATTTAACTGGCCAGTTTTGATTGCAAATGC TGTAAAGATATAGAATGAAGTCCTGTGAGGCCTTCCTATCTCCAAGTCTATGTATTTTCTGGAGACCAAACCAGATACCAGATAATCACA AAGAAAGCTTTTTTAATAAGGCTTAAACCAAGACCTTGTCTAGATATTTTTAGTTTGTTGCCAAGGTAGCACTGTGAGAAATCTCACTTG GATGTTATGTAAGGGGTGAGACACAACAGTCTGACTATGAGTGAGGAAAATATCTGGGTCTTTTCGTCAGTTTGGTGCATTTGCTGCTGC TGTTGCTACTGTTTGCCTCAAACGCTGTGTTTAAACAACGTTAAACTCTTAGCCTACAAGGTGGCTCTTATGTACATAGTTGTTAATACA TCCAATTAATGATGTCTGACATGCTATTTTTGTAGGGAGAAAATATGTGCTAATGATATTTTGAGTTAAAATATCTTTTGGGGAGGATTT GCTGAAAAGTTGCACTTTTGTTACAATGCTTATGCTTGGTACAAGCTTATGCTGTCTTAAATTATTTTAAAAAAATAAATACTGTCTGTG AGAAACCAGCTGGTTTAGAAAAGTTTAGTATGTGACGATAAACTAGAAATTACCTTTATATTCTAGTATTTTCAGCACTCCATAAATTCT ATTACCTAAATATTGCCACACTATTTTGTGATTTAAAAATTCTTACTAAGGAATAAAAACTTTAATATACGATATGATATTGTCTAATAA TTAAAAAAGACATAATGGATGCTCAATTAGTTTTAAGATATCTATAACTATAGGGATACAAATCACTACAGTTCTCAGATTTACAGCTTT TTTTTGTCATTGGCTTGATGTCACACATTTCCAATCTCTTGCAAGCCTCCAGGCTCTGGCTTTGTCTACCTGCTCGTTCCCAATGTATCT >80202_80202_1_SEL1L3-BHLHE41_SEL1L3_chr4_25836818_ENST00000264868_BHLHE41_chr12_26277515_ENST00000242728_length(amino acids)=734AA_BP=273 MGAGQLRGTTPGGTLALRPAGMAPRPKKQPDKNPLHGRELNVVPSLGRQTSLTTSVIPKAEQSVAYKDFIYFTVFEGNVRNVSEVSVEYL CSQPCVVNLEAVVSSEFRSSIPVYKKRWKNEKHLHTSRTQIVHVKFPSIMVYRDDYFIRHSISVSAVIVRAWITHKYSGRDWNVKWEENL LHAVAKNYTLLQTIPPFERPFKDHQVCLEWNMGYIWNLRANRIPQCPLENDVVALLGFPYASSGENTGIVKKFPRFRNRELEATRRQRMD YPVLDYSSLYMCKPKRSMKRDDTKDTYKLPHRLIEKKRRDRINECIAQLKDLLPEHLKLTTLGHLEKAVVLELTLKHLKALTALTEQQHQ KIIALQNGERSLKSPIQSDLDAFHSGFQTCAKEVLQYLSRFESWTPREPRCVQLINHLHAVATQFLPTPQLLTQQVPLSKGTGAPSAAGS AAAPCLERAGQKLEPLAYCVPVIQRTQPSAELAAENDTDTDSGYGGEAEARPDREKGKGAGASRVTIKQEPPGEDSPAPKRMKLDSRGGG SGGGPGGGAAAAAAALLGPDPAAAAALLRPDAALLSSLVAFGGGGGAPFPQPAAAAAPFCLPFCFLSPSAAAAYVQPFLDKSGLEKYLYP AAAAAPFPLLYPGIPAPAAAAAAAAAAAAAAAAFPCLSSVLSPPPEKAGAAAATLLPHEVAPLGAPHPQHPHGRTHLPFAGPREPGNPES -------------------------------------------------------------- >80202_80202_2_SEL1L3-BHLHE41_SEL1L3_chr4_25836818_ENST00000399878_BHLHE41_chr12_26277515_ENST00000242728_length(transcript)=4410nt_BP=983nt CGGCAGGTGGCCCCGCGCGCGGCGGCCGGCCCGGCCGGGGGCGGGCGGGAAGGTGGCGCCTCGGGCGGGGGCCGGTCCCTGCACCAGGTG ACCTGTTCCGGCCCGGATCCGGGCGGCCTCGCCATGCAGCGGCGCGGCGCGGGGCTCGGGTGGCCGCGGCAGCAGCAGCAGCAACCCCCG CCGCTCGCGGTCGGCCCCCGGGCCGCAGCCATGGTCCCGAGTGGCGGCGTCCCCCAGGGCCTCGGCGGCCGCTCTGCCTGCGCGCTGCTC CTGCTCTGCTACCTGAATGTTGTACCATCTTTGGGTAGGCAGACTTCCCTGACGACATCAGTGATACCCAAAGCTGAGCAGAGCGTGGCT TACAAAGACTTTATTTATTTTACTGTCTTTGAAGGAAACGTTCGCAACGTTTCTGAAGTCTCGGTTGAGTATTTATGCTCTCAGCCTTGT GTTGTCAATTTGGAAGCAGTTGTTTCATCTGAGTTCAGAAGTAGCATTCCCGTGTACAAAAAAAGGTGGAAGAATGAGAAACATCTTCAC ACCAGCAGGACACAAATAGTACATGTGAAATTTCCAAGCATTATGGTTTACAGAGATGATTATTTCATCAGACATTCCATCTCTGTATCT GCAGTGATAGTACGCGCCTGGATTACTCACAAATACAGTGGCAGAGACTGGAATGTTAAATGGGAGGAAAACTTGCTCCATGCTGTAGCA AAGAATTATACCCTCCTGCAGACCATCCCGCCTTTTGAACGCCCTTTCAAAGATCATCAAGTGTGCCTTGAGTGGAACATGGGTTATATT TGGAACCTTCGGGCAAACAGGATTCCACAGTGTCCTCTGGAAAATGATGTGGTTGCCCTGCTTGGCTTTCCTTATGCCTCCAGTGGAGAA AACACAGGCATTGTCAAGAAGTTCCCGAGGTTTCGGAACCGAGAGCTGGAGGCCACTCGACGCCAGAGGATGGATTACCCAGTACTGGAC TATTCCTCTTTGTATATGTGTAAACCCAAAAGGAGCATGAAACGAGACGACACCAAGGATACCTACAAATTACCGCACAGATTAATAGAA AAGAAAAGAAGAGACCGAATTAATGAATGCATTGCTCAGCTGAAAGATTTACTGCCTGAACATCTGAAATTGACAACTCTGGGACATCTG GAGAAAGCTGTAGTCTTGGAATTAACTTTGAAACACTTAAAAGCTTTAACCGCCTTAACCGAGCAACAGCATCAGAAGATAATTGCTTTA CAGAATGGGGAGCGATCTCTGAAATCGCCCATTCAGTCCGACTTGGATGCGTTCCACTCGGGATTTCAAACATGCGCCAAAGAAGTCTTG CAATACCTCTCCCGGTTTGAGAGCTGGACACCCAGGGAGCCGCGGTGTGTCCAGCTGATCAACCACTTGCACGCCGTGGCCACCCAGTTC TTGCCCACCCCGCAGCTGTTGACTCAACAGGTCCCTCTGAGCAAAGGCACCGGCGCTCCCTCGGCCGCCGGGTCCGCGGCCGCCCCCTGC CTGGAGCGCGCGGGGCAGAAGCTGGAGCCCCTCGCCTACTGCGTGCCCGTCATCCAGCGGACTCAGCCCAGCGCCGAGCTCGCCGCCGAG AACGACACGGACACCGACAGCGGCTACGGCGGCGAAGCCGAGGCCCGGCCGGACCGCGAGAAAGGCAAAGGCGCGGGGGCGAGCCGCGTC ACCATCAAGCAGGAGCCTCCCGGGGAGGACTCGCCGGCGCCCAAGAGGATGAAGCTGGATTCCCGCGGCGGCGGCAGCGGCGGCGGCCCG GGGGGCGGCGCGGCGGCGGCGGCAGCCGCGCTTCTGGGGCCCGACCCTGCCGCCGCGGCCGCGCTGCTGAGACCCGACGCCGCCCTGCTC AGCTCGCTGGTGGCGTTCGGCGGAGGCGGAGGCGCGCCCTTCCCGCAGCCCGCGGCCGCCGCGGCCCCCTTCTGCCTGCCCTTCTGCTTC CTCTCGCCTTCTGCAGCTGCCGCCTACGTGCAGCCCTTCCTGGACAAGAGCGGCCTGGAGAAGTATCTGTACCCGGCGGCGGCTGCCGCC CCGTTCCCGCTGCTATACCCCGGCATCCCCGCCCCGGCGGCAGCCGCGGCAGCCGCCGCCGCCGCTGCCGCCGCCGCCGCCGCGTTCCCC TGCCTGTCCTCGGTGTTGTCGCCCCCTCCCGAGAAGGCGGGCGCCGCCGCCGCGACCCTCCTGCCGCACGAGGTGGCGCCCCTTGGGGCG CCGCACCCCCAGCACCCGCACGGCCGCACCCACCTGCCCTTCGCCGGGCCCCGCGAGCCGGGGAACCCGGAGAGCTCTGCTCAGGAAGAT CCCTCGCAGCCAGGAAAGGAAGCTCCCTGAATCCTTGCGTCCCGAAGGACGGAGGTTCAAGCAGAGTGAGAAGTTAAAATACCCTTAAGG AGGTTCAAGCAGAGTGAGAAGTTAAAATACCCTTAAGGTCTTTAAGGGAGGAAGTGTAATAGATGCACGACAGGCATAAACAAGAACAAC AAAACAGGTGTTATGTGTACATTCGGAGTTCCTGTTTTGCTCATCCCGCACCACCCCACCCTCCACACACTAACATCCCTTTCTTCCCCC CACCAGCTGTAAAAGATCCTATGCGAAAGACACTGGCTCTTTTTTTTAATCCCCCAAATAAATTTTGCCCCCTTTTAGGCCATGTTCCAT TATCTCTTAAAATTGGAACCTAATTCGAGAGGAAGTAAGAAGGGTCTGTTCTGTGGCTGAGCTAGGTGAACCCCGGGGTAGGGGAAAGAT GTTAACACCTTTGACGTCTTTGGAGTTGACATGGAACAGCAGGTAGTTGTTATGTAGAGCTAGTTCTCAAAGCTGCCCTGCCTGTTTTAG GAGGCGTTCCACAAACAGATTGAGGCTCTTTTAGAATTGAATTTACTCTTCAGTATTTTCTAATGTTCAGCTTTCTAAAAGGCATATATT TTTCAAAGAAGTGAGGATGCAGTTTCTCACGTTGCAACCTATTCTGAAGTGGTTTAAATGGTATCTCTTAGTAACTTGCACTCGTTAAAG AAACACGGAGCTGGGCCATCGTCAGAACTAAGTCAGGGAAGGAGATGGATGAGAAGGCCAGAATCATTCCTAGTACATTTGCTAACACTT TATTGAGAAATTGACCATGAATTAATGGACTCATCTTAATTTCTTCTAAGTCCATATATAGATAGATATCTATCTGTACAGATTTCTATT TATCCATAGATAGGTATCTATACATACACATCTCAAGTGCATCTATTCCCACTCTCATTAATCCATCATGTTCCTAAATTTTTGTAATCT TACTGTAAAAAAAAGTGCACTGAACTTCAAAACAAAACAAAAAACAACAACAACAAAAAACAAGTCCAAACTGATATATCCTATATTCTG TTAAAATTCAAAAGTGAACGAAAGCATTTAACTGGCCAGTTTTGATTGCAAATGCTGTAAAGATATAGAATGAAGTCCTGTGAGGCCTTC CTATCTCCAAGTCTATGTATTTTCTGGAGACCAAACCAGATACCAGATAATCACAAAGAAAGCTTTTTTAATAAGGCTTAAACCAAGACC TTGTCTAGATATTTTTAGTTTGTTGCCAAGGTAGCACTGTGAGAAATCTCACTTGGATGTTATGTAAGGGGTGAGACACAACAGTCTGAC TATGAGTGAGGAAAATATCTGGGTCTTTTCGTCAGTTTGGTGCATTTGCTGCTGCTGTTGCTACTGTTTGCCTCAAACGCTGTGTTTAAA CAACGTTAAACTCTTAGCCTACAAGGTGGCTCTTATGTACATAGTTGTTAATACATCCAATTAATGATGTCTGACATGCTATTTTTGTAG GGAGAAAATATGTGCTAATGATATTTTGAGTTAAAATATCTTTTGGGGAGGATTTGCTGAAAAGTTGCACTTTTGTTACAATGCTTATGC TTGGTACAAGCTTATGCTGTCTTAAATTATTTTAAAAAAATAAATACTGTCTGTGAGAAACCAGCTGGTTTAGAAAAGTTTAGTATGTGA CGATAAACTAGAAATTACCTTTATATTCTAGTATTTTCAGCACTCCATAAATTCTATTACCTAAATATTGCCACACTATTTTGTGATTTA AAAATTCTTACTAAGGAATAAAAACTTTAATATACGATATGATATTGTCTAATAATTAAAAAAGACATAATGGATGCTCAATTAGTTTTA AGATATCTATAACTATAGGGATACAAATCACTACAGTTCTCAGATTTACAGCTTTTTTTTGTCATTGGCTTGATGTCACACATTTCCAAT CTCTTGCAAGCCTCCAGGCTCTGGCTTTGTCTACCTGCTCGTTCCCAATGTATCTTAATGAAAAGTGCAAAAGAAAAACCTACCAATTAA >80202_80202_2_SEL1L3-BHLHE41_SEL1L3_chr4_25836818_ENST00000399878_BHLHE41_chr12_26277515_ENST00000242728_length(amino acids)=763AA_BP=302 MHQVTCSGPDPGGLAMQRRGAGLGWPRQQQQQPPPLAVGPRAAAMVPSGGVPQGLGGRSACALLLLCYLNVVPSLGRQTSLTTSVIPKAE QSVAYKDFIYFTVFEGNVRNVSEVSVEYLCSQPCVVNLEAVVSSEFRSSIPVYKKRWKNEKHLHTSRTQIVHVKFPSIMVYRDDYFIRHS ISVSAVIVRAWITHKYSGRDWNVKWEENLLHAVAKNYTLLQTIPPFERPFKDHQVCLEWNMGYIWNLRANRIPQCPLENDVVALLGFPYA SSGENTGIVKKFPRFRNRELEATRRQRMDYPVLDYSSLYMCKPKRSMKRDDTKDTYKLPHRLIEKKRRDRINECIAQLKDLLPEHLKLTT LGHLEKAVVLELTLKHLKALTALTEQQHQKIIALQNGERSLKSPIQSDLDAFHSGFQTCAKEVLQYLSRFESWTPREPRCVQLINHLHAV ATQFLPTPQLLTQQVPLSKGTGAPSAAGSAAAPCLERAGQKLEPLAYCVPVIQRTQPSAELAAENDTDTDSGYGGEAEARPDREKGKGAG ASRVTIKQEPPGEDSPAPKRMKLDSRGGGSGGGPGGGAAAAAAALLGPDPAAAAALLRPDAALLSSLVAFGGGGGAPFPQPAAAAAPFCL PFCFLSPSAAAAYVQPFLDKSGLEKYLYPAAAAAPFPLLYPGIPAPAAAAAAAAAAAAAAAAFPCLSSVLSPPPEKAGAAAATLLPHEVA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SEL1L3-BHLHE41 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SEL1L3-BHLHE41 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SEL1L3-BHLHE41 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
