
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SEMA4B-CERS3 (FusionGDB2 ID:80308) |
Fusion Gene Summary for SEMA4B-CERS3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SEMA4B-CERS3 | Fusion gene ID: 80308 | Hgene | Tgene | Gene symbol | SEMA4B | CERS3 | Gene ID | 10509 | 204219 |
| Gene name | semaphorin 4B | ceramide synthase 3 | |
| Synonyms | SEMAC|SemC | ARCI9|LASS3 | |
| Cytomap | 15q26.1 | 15q26.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | semaphorin-4Bsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4Bsema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, 4Bsemaphorin-C | ceramide synthase 3LAG1 homolog, ceramide synthase 3LAG1 longevity assurance homolog 3dihydroceramide synthase 3sphingosine N-acyltransferase CERS3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q8IU89 | |
| Ensembl transtripts involved in fusion gene | ENST00000332496, ENST00000379122, ENST00000411539, ENST00000560263, | ENST00000560944, ENST00000284382, ENST00000394113, ENST00000538112, | |
| Fusion gene scores | * DoF score | 16 X 9 X 11=1584 | 8 X 7 X 7=392 |
| # samples | 21 | 11 | |
| ** MAII score | log2(21/1584*10)=-2.91511110241349 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/392*10)=-1.83335013059055 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SEMA4B [Title/Abstract] AND CERS3 [Title/Abstract] AND fusion [Title/Abstract] | ||
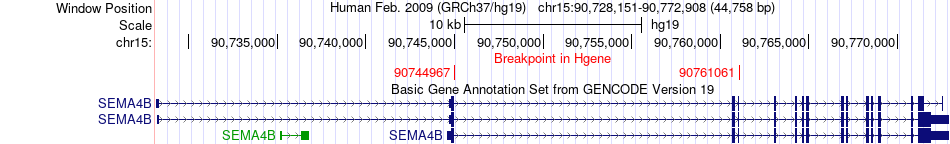
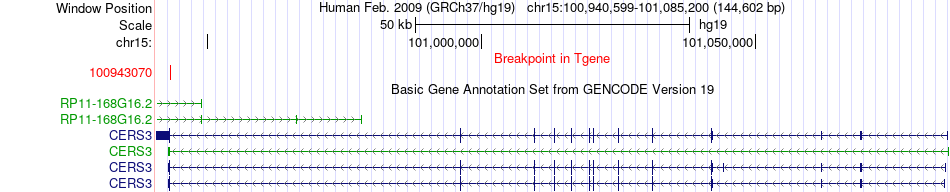
| Most frequent breakpoint | SEMA4B(90744967)-CERS3(100943070), # samples:1 SEMA4B(90761061)-CERS3(100943070), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | SEMA4B-CERS3 seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. SEMA4B-CERS3 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CERS3 | GO:0046513 | ceramide biosynthetic process | 17977534|22038835 |
 Fusion gene breakpoints across SEMA4B (5'-gene) Fusion gene breakpoints across SEMA4B (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CERS3 (3'-gene) Fusion gene breakpoints across CERS3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-J2-A4AD-01A | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| ChimerDB4 | LUAD | TCGA-J2-A4AD-01A | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
Top |
Fusion Gene ORF analysis for SEMA4B-CERS3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000332496 | ENST00000560944 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| 5CDS-5UTR | ENST00000332496 | ENST00000560944 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| 5CDS-5UTR | ENST00000379122 | ENST00000560944 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| 5CDS-5UTR | ENST00000379122 | ENST00000560944 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| 5CDS-5UTR | ENST00000411539 | ENST00000560944 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| 5CDS-5UTR | ENST00000411539 | ENST00000560944 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000332496 | ENST00000284382 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000332496 | ENST00000394113 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000332496 | ENST00000538112 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000332496 | ENST00000538112 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000379122 | ENST00000284382 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000379122 | ENST00000394113 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000379122 | ENST00000538112 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000379122 | ENST00000538112 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000411539 | ENST00000284382 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000411539 | ENST00000394113 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000411539 | ENST00000538112 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| Frame-shift | ENST00000411539 | ENST00000538112 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000332496 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000332496 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000379122 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000379122 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000411539 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| In-frame | ENST00000411539 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000560263 | ENST00000284382 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000560263 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000560263 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000560263 | ENST00000394113 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000560263 | ENST00000538112 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| intron-3CDS | ENST00000560263 | ENST00000538112 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
| intron-5UTR | ENST00000560263 | ENST00000560944 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - |
| intron-5UTR | ENST00000560263 | ENST00000560944 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000379122 | SEMA4B | chr15 | 90744967 | + | ENST00000394113 | CERS3 | chr15 | 100943070 | - | 795 | 440 | 142 | 462 | 106 |
| ENST00000332496 | SEMA4B | chr15 | 90744967 | + | ENST00000394113 | CERS3 | chr15 | 100943070 | - | 752 | 397 | 99 | 419 | 106 |
| ENST00000411539 | SEMA4B | chr15 | 90744967 | + | ENST00000394113 | CERS3 | chr15 | 100943070 | - | 772 | 417 | 405 | 1 | 135 |
| ENST00000379122 | SEMA4B | chr15 | 90761061 | - | ENST00000284382 | CERS3 | chr15 | 100943070 | - | 3138 | 667 | 142 | 819 | 225 |
| ENST00000332496 | SEMA4B | chr15 | 90761061 | - | ENST00000284382 | CERS3 | chr15 | 100943070 | - | 3095 | 624 | 99 | 776 | 225 |
| ENST00000411539 | SEMA4B | chr15 | 90761061 | - | ENST00000284382 | CERS3 | chr15 | 100943070 | - | 3115 | 644 | 191 | 796 | 201 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000379122 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - | 0.59944105 | 0.40055895 |
| ENST00000332496 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - | 0.66969585 | 0.33030415 |
| ENST00000411539 | ENST00000394113 | SEMA4B | chr15 | 90744967 | + | CERS3 | chr15 | 100943070 | - | 0.60230345 | 0.3976965 |
| ENST00000379122 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - | 0.00762498 | 0.9923751 |
| ENST00000332496 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - | 0.007669067 | 0.9923309 |
| ENST00000411539 | ENST00000284382 | SEMA4B | chr15 | 90761061 | - | CERS3 | chr15 | 100943070 | - | 0.007622249 | 0.99237776 |
Top |
Fusion Genomic Features for SEMA4B-CERS3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
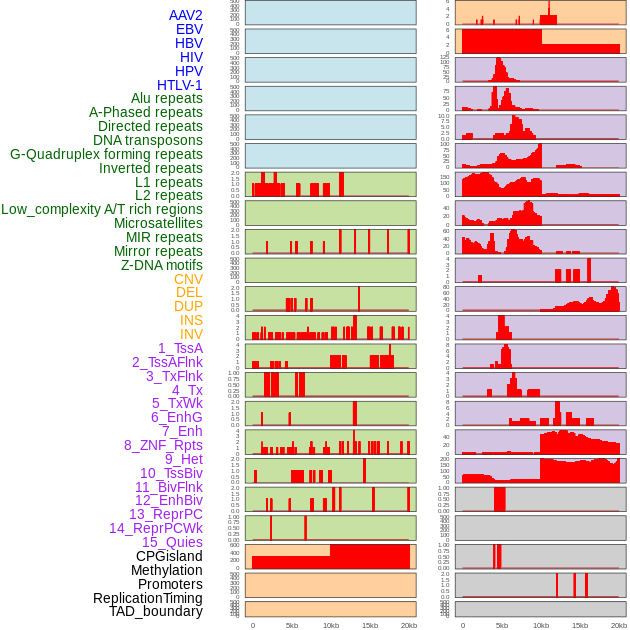
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SEMA4B-CERS3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr15:90744967/chr15:100943070) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CERS3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Ceramide synthase that catalyzes formation of ceramide from sphinganine and acyl-CoA substrates, with high selectivity toward very-long (C22:0-C24:0) and ultra long chain (more than C26:0) as acyl donor (PubMed:17977534, PubMed:22038835, PubMed:26887952). It is crucial for the synthesis of ultra long-chain ceramides in the epidermis, to maintain epidermal lipid homeostasis and terminal differentiation (PubMed:23754960). {ECO:0000269|PubMed:17977534, ECO:0000269|PubMed:22038835, ECO:0000269|PubMed:23754960, ECO:0000269|PubMed:26887952}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 346_355 | 333 | 384.0 | Compositional bias | Note=Poly-Glu |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 758_781 | 47 | 738.0 | Compositional bias | Note=Pro-rich |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 758_781 | 123 | 738.0 | Compositional bias | Note=Pro-rich |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 47_523 | 47 | 738.0 | Domain | Sema |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 525_579 | 47 | 738.0 | Domain | Note=PSI |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 604_663 | 47 | 738.0 | Domain | Note=Ig-like C2-type |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 47_523 | 123 | 738.0 | Domain | Sema |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 525_579 | 123 | 738.0 | Domain | Note=PSI |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 604_663 | 123 | 738.0 | Domain | Note=Ig-like C2-type |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 44_717 | 47 | 738.0 | Topological domain | Extracellular |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 739_837 | 47 | 738.0 | Topological domain | Cytoplasmic |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 44_717 | 123 | 738.0 | Topological domain | Extracellular |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 739_837 | 123 | 738.0 | Topological domain | Cytoplasmic |
| Hgene | SEMA4B | chr15:90744967 | chr15:100943070 | ENST00000379122 | + | 2 | 16 | 718_738 | 47 | 738.0 | Transmembrane | Helical |
| Hgene | SEMA4B | chr15:90761061 | chr15:100943070 | ENST00000379122 | - | 4 | 16 | 718_738 | 123 | 738.0 | Transmembrane | Helical |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 130_331 | 333 | 384.0 | Domain | TLC | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 66_127 | 333 | 384.0 | Region | Homeobox-like | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 319_383 | 333 | 384.0 | Topological domain | Cytoplasmic | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000284382 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000394113 | 12 | 14 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90744967 | chr15:100943070 | ENST00000538112 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000284382 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000394113 | 12 | 14 | 32_52 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 139_159 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 174_194 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 205_225 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 264_284 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 298_318 | 333 | 384.0 | Transmembrane | Helical | |
| Tgene | CERS3 | chr15:90761061 | chr15:100943070 | ENST00000538112 | 11 | 13 | 32_52 | 333 | 384.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for SEMA4B-CERS3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >80308_80308_1_SEMA4B-CERS3_SEMA4B_chr15_90744967_ENST00000332496_CERS3_chr15_100943070_ENST00000394113_length(transcript)=752nt_BP=397nt CTCCGGGTCCCCAGGGGCTGCGCCGGGCCGGCCTGGCAAGGGGGACGAGTCAGTGGACACTCCAGGAAGAGCGGCCCCGCGGGGGGCGAT GACCGTGCGCTGACCCTGACTCACTCCAGGTCCGGAGGCGGGGGCCCCCGGGGCGACTCGGGGGCGGACCGCGGGGCGGAGCTGCCGCCC GTGAGTCCGGCCGAGCCACCTGAGCCCGAGCCGCGGGACACCGTCGCTCCTGCTCTCCGAATGCTGCGCACCGCGATGGGCCTGAGGAGC TGGCTCGCCGCCCCATGGGGCGCGCTGCCGCCTCGGCCACCGCTGCTGCTGCTCCTGCTGCTGCTGCTCCTGCTGCAGCCGCCGCCTCCG ACCTGGGCGCTCAGCCCCCGGATCAGCCTGCCTCTGGAGCATCCAGGATGTGAGGAGTGATGACGAGGATTATGAAGAGGAAGAGGAAGA GGAAGAAGAAGAGGCTACCAAAGGCAAAGAGATGGATTGTTTAAAGAACGGCCTCAGGGCTGAGAGGCACCTCATTCCCAATGGCCAGCA TGGCCATTAGCTGGAAGCCTACAGGACTCCCATGGCACAGCATGCTGCAAGTACTGTTGGCAGCCTGGCTTCCAGGCCCCACACCGACCC CACATTCTGCCCTTCCCTCTTTCTCACCACCGCCTTCCCTCCCACCTAAGATGTGTTTACCAAAATGTTGTTAACTTGTGTTAAAATGTT >80308_80308_1_SEMA4B-CERS3_SEMA4B_chr15_90744967_ENST00000332496_CERS3_chr15_100943070_ENST00000394113_length(amino acids)=106AA_BP= MTLTHSRSGGGGPRGDSGADRGAELPPVSPAEPPEPEPRDTVAPALRMLRTAMGLRSWLAAPWGALPPRPPLLLLLLLLLLLQPPPPTWA -------------------------------------------------------------- >80308_80308_2_SEMA4B-CERS3_SEMA4B_chr15_90744967_ENST00000379122_CERS3_chr15_100943070_ENST00000394113_length(transcript)=795nt_BP=440nt ATTTCCGGGGACGCCGCGCCGGACTGAGGCTGTGCGCCCGAGACTCCGGGTCCCCAGGGGCTGCGCCGGGCCGGCCTGGCAAGGGGGACG AGTCAGTGGACACTCCAGGAAGAGCGGCCCCGCGGGGGGCGATGACCGTGCGCTGACCCTGACTCACTCCAGGTCCGGAGGCGGGGGCCC CCGGGGCGACTCGGGGGCGGACCGCGGGGCGGAGCTGCCGCCCGTGAGTCCGGCCGAGCCACCTGAGCCCGAGCCGCGGGACACCGTCGC TCCTGCTCTCCGAATGCTGCGCACCGCGATGGGCCTGAGGAGCTGGCTCGCCGCCCCATGGGGCGCGCTGCCGCCTCGGCCACCGCTGCT GCTGCTCCTGCTGCTGCTGCTCCTGCTGCAGCCGCCGCCTCCGACCTGGGCGCTCAGCCCCCGGATCAGCCTGCCTCTGGAGCATCCAGG ATGTGAGGAGTGATGACGAGGATTATGAAGAGGAAGAGGAAGAGGAAGAAGAAGAGGCTACCAAAGGCAAAGAGATGGATTGTTTAAAGA ACGGCCTCAGGGCTGAGAGGCACCTCATTCCCAATGGCCAGCATGGCCATTAGCTGGAAGCCTACAGGACTCCCATGGCACAGCATGCTG CAAGTACTGTTGGCAGCCTGGCTTCCAGGCCCCACACCGACCCCACATTCTGCCCTTCCCTCTTTCTCACCACCGCCTTCCCTCCCACCT >80308_80308_2_SEMA4B-CERS3_SEMA4B_chr15_90744967_ENST00000379122_CERS3_chr15_100943070_ENST00000394113_length(amino acids)=106AA_BP= MTLTHSRSGGGGPRGDSGADRGAELPPVSPAEPPEPEPRDTVAPALRMLRTAMGLRSWLAAPWGALPPRPPLLLLLLLLLLLQPPPPTWA -------------------------------------------------------------- >80308_80308_3_SEMA4B-CERS3_SEMA4B_chr15_90744967_ENST00000411539_CERS3_chr15_100943070_ENST00000394113_length(transcript)=772nt_BP=417nt CGCCCTCCGCCGCTTGCGGGTGAGCTCTGCCCAAGCCGAGGCTGCGGGGCCGGCGCCGGCGGGAGGACTGCGGTGCCCCGCGGAGGGGCT GAGTTTGCCAGGGCCCACTTGACCCTGTTTCCCACCTCCCGCCCCCCAGGTCCGGAGGCGGGGGCCCCCGGGGCGACTCGGGGGCGGACC GCGGGGCGGAGCTGCCGCCCGTGAGTCCGGCCGAGCCACCTGAGCCCGAGCCGCGGGACACCGTCGCTCCTGCTCTCCGAATGCTGCGCA CCGCGATGGGCCTGAGGAGCTGGCTCGCCGCCCCATGGGGCGCGCTGCCGCCTCGGCCACCGCTGCTGCTGCTCCTGCTGCTGCTGCTCC TGCTGCAGCCGCCGCCTCCGACCTGGGCGCTCAGCCCCCGGATCAGCCTGCCTCTGGAGCATCCAGGATGTGAGGAGTGATGACGAGGAT TATGAAGAGGAAGAGGAAGAGGAAGAAGAAGAGGCTACCAAAGGCAAAGAGATGGATTGTTTAAAGAACGGCCTCAGGGCTGAGAGGCAC CTCATTCCCAATGGCCAGCATGGCCATTAGCTGGAAGCCTACAGGACTCCCATGGCACAGCATGCTGCAAGTACTGTTGGCAGCCTGGCT TCCAGGCCCCACACCGACCCCACATTCTGCCCTTCCCTCTTTCTCACCACCGCCTTCCCTCCCACCTAAGATGTGTTTACCAAAATGTTG >80308_80308_3_SEMA4B-CERS3_SEMA4B_chr15_90744967_ENST00000411539_CERS3_chr15_100943070_ENST00000394113_length(amino acids)=135AA_BP= MIRGLSAQVGGGGCSRSSSSRSSSSGGRGGSAPHGAASQLLRPIAVRSIRRAGATVSRGSGSGGSAGLTGGSSAPRSAPESPRGPPPPDL -------------------------------------------------------------- >80308_80308_4_SEMA4B-CERS3_SEMA4B_chr15_90761061_ENST00000332496_CERS3_chr15_100943070_ENST00000284382_length(transcript)=3095nt_BP=624nt CTCCGGGTCCCCAGGGGCTGCGCCGGGCCGGCCTGGCAAGGGGGACGAGTCAGTGGACACTCCAGGAAGAGCGGCCCCGCGGGGGGCGAT GACCGTGCGCTGACCCTGACTCACTCCAGGTCCGGAGGCGGGGGCCCCCGGGGCGACTCGGGGGCGGACCGCGGGGCGGAGCTGCCGCCC GTGAGTCCGGCCGAGCCACCTGAGCCCGAGCCGCGGGACACCGTCGCTCCTGCTCTCCGAATGCTGCGCACCGCGATGGGCCTGAGGAGC TGGCTCGCCGCCCCATGGGGCGCGCTGCCGCCTCGGCCACCGCTGCTGCTGCTCCTGCTGCTGCTGCTCCTGCTGCAGCCGCCGCCTCCG ACCTGGGCGCTCAGCCCCCGGATCAGCCTGCCTCTGGGCTCTGAAGAGCGGCCATTCCTCAGATTCGAAGCTGAACACATCTCCAACTAC ACAGCCCTTCTGCTGAGCAGGGATGGCAGGACCCTGTACGTGGGTGCTCGAGAGGCCCTCTTTGCACTCAGTAGCAACCTCAGCTTCCTG CCAGGCGGGGAGTACCAGGAGCTGCTTTGGGGTGCAGACGCAGAGAAGAAACAGCAGTGCAGCTTCAAGGGCAAGGACCCACAGAGCATC CAGGATGTGAGGAGTGATGACGAGGATTATGAAGAGGAAGAGGAAGAGGAAGAAGAAGAGGCTACCAAAGGCAAAGAGATGGATTGTTTA AAGAACGGCCTCAGGGCTGAGAGGCACCTCATTCCCAATGGCCAGCATGGCCATTAGCTGGAAGCCTACAGGACTCCCATGGCACAGCAT GCTGCAAGTACTGTTGGCAGCCTGGCTTCCAGGCCCCACACCGACCCCACATTCTGCCCTTCCCTCTTTCTCACCACCGCCTTCCCTCCC ACCTAAGATGTGTTTACCAAAATGTTGTTAACTTGTGTTAAAATGTTAAATATAAGCATGCCCATGGATTTTTACTGCAGTTAGGACTCA GACTGGTCAAAGATTTCAAAGATTTCTCCACAGAACCGTCTCAGTTCTAATTGCACTCCCTCATGCATGTCACTTTCTCAGGGGCTCGCT TTGTTATAGACCCTTTCGCCTCGCCACCTTGCCTGTCCTCAGGACGCTTTCACAGGTGCTAAGTGATCTCATTTTTCCCAGGTGTGTTTG GCCACAAAGAGCAGCTTCTTTCTCAAAATGAGTTAGAAGTGGCAGTGGGACAGGAGCGGAAGGACCACACCAGGAGACACTTCCATCCTG AAGCTTAGGTGCCTCATCTCCACAGGGCGGTGGCAGTCCCTGCCTGCCACCCCACAGGGTTACAGCAAGGATTAAGTGAGATCACAGAAG TTTAAGTACACACTGTCAACCGTGGGGTCATCGTGAACCAACCCACTTTGCACTGTTTGGGAGACAGAAACCTGGCTAAACATGGCACTG CAATGGGCCTGAACAAAAGGCAGAATCTTTAAAAATTCCTCTTAAATGACTCTGGAGATACCTGGAAATGAAAGTGCCAAGAAAGGTGTT CCATTTTATTTGCTAACTATTTATGCATTAGCTCCCCAAGGGTCATTCTCAGATTCTCCTGGAATTCTTCTCCCTCAGGACCCATGGTTC AAGGAAGGAAGCTTAGGCCCTGTTCCTTCCTGTCCTTTGCTGGTCTTTCCCTTTTTTCCTTTCTAGGAGGGAAGCTTCTGTGCTGCTGCC CTGAGCCCTTCCTTCAGGCCAGCACAGTACCTGGGGACCTCCACGGGGGAATGGGATCCAGGCCAGGTTGCTTGCTGAGCCTCATCACCC AGGAGGCCTGAGCCTCTGGGGAGGGCACGCATGTACACTGCCAGACCCAGGGGAGATCTTGGGAACAGAGGATGCTACGTGATTTCCTCT GGCTTCCAACCCAATCAGCCTGCATCACAGTGAAACACAACACAAAAGGCCATAAAAGGCATACCCCTGAGAAATGTTTAAGGGCAGGTG CTAGAGTCAGTGCCAGCTCCCTGGGAGGAGGGATAGGGACGGGCAGATCAGTAGCCAGCTTGTCCTCACCCTCCGGAAGGGAGCACCGGA GAAGACTTGCACACGTCCCTGCCTCCCTTGCGGCCTTGTCACCAGAGGGACACATCTCCTGGGCACAATGTGCAGGGCTGACCTGGGAGA CTTCTCAGGTGGCTGCTGGCTGAGGAGAGGCCAGGCCTTCCCAAGGAAGACCCTGAGTAAAATCTGCATCTGTCCTCACCACCTGGCACA GTCTCTCATAACGGTCAGATTTTGTATGTTCATCCTATTTATTCAGGGGCTCGTACTAGAAAGCCACAGGAGAGGTCGGCTTGTAGGGAC TGGAAAATCAGCCCCAAGCAGAGCAGCTGCAGGGCCCCTGGAGCCGGAAGGACACTGCACAGAACAGACTGCGTTATTGTTATTTTTAAA TAAAAATATACATTTGAAACCTTAAGGCTAAACAAAAATAAACAAAAAATCCTTTAGAATTCTTTCCACAACAATATCTCTTTCTGAGAA ATTGTTACAAACAAGGTCAGATTTTCTCTGTATAACATTTGCTTTTATGAGGACAATATCATATGCATTATATGCATAATATGATATTAT AAATCAAAATGCCTGCACCCACTTTAGGGTATAGCTATTGACTTATTATTAATATTATATTATTATTATTTTGCTGGAAGAAGGTCACAC TAAGATATAATTTTTTATGTTTTCAGTTAACGGTATGCTTTCTTCTTTGCTTATTTGGTTTTTGTCTCTGTACCAAATATCTTCTTGCTT AAGGTAGAAAAGTATTTGTTTACCTCTATCTCCAGTTTTTTTTCTTATTTGAATGTTGAAGGTAAAATTGATATACCAATTTTAACTATT TCTGATACAGCTGAAAGCACTAAACTACTTCATAAGAAGTAGATACTCATTTTTGTAACACTATTTAGGGCTTTTGTGGTTAATTTTAAA GGAAACCACTCTTTCTACAGGAAACAAGGGCTCAGGATTCTTCAGATGACCTTATAAAAATGCAGTCCACAGTGCTATCAATATTGTAAC >80308_80308_4_SEMA4B-CERS3_SEMA4B_chr15_90761061_ENST00000332496_CERS3_chr15_100943070_ENST00000284382_length(amino acids)=225AA_BP=175 MTLTHSRSGGGGPRGDSGADRGAELPPVSPAEPPEPEPRDTVAPALRMLRTAMGLRSWLAAPWGALPPRPPLLLLLLLLLLLQPPPPTWA LSPRISLPLGSEERPFLRFEAEHISNYTALLLSRDGRTLYVGAREALFALSSNLSFLPGGEYQELLWGADAEKKQQCSFKGKDPQSIQDV -------------------------------------------------------------- >80308_80308_5_SEMA4B-CERS3_SEMA4B_chr15_90761061_ENST00000379122_CERS3_chr15_100943070_ENST00000284382_length(transcript)=3138nt_BP=667nt ATTTCCGGGGACGCCGCGCCGGACTGAGGCTGTGCGCCCGAGACTCCGGGTCCCCAGGGGCTGCGCCGGGCCGGCCTGGCAAGGGGGACG AGTCAGTGGACACTCCAGGAAGAGCGGCCCCGCGGGGGGCGATGACCGTGCGCTGACCCTGACTCACTCCAGGTCCGGAGGCGGGGGCCC CCGGGGCGACTCGGGGGCGGACCGCGGGGCGGAGCTGCCGCCCGTGAGTCCGGCCGAGCCACCTGAGCCCGAGCCGCGGGACACCGTCGC TCCTGCTCTCCGAATGCTGCGCACCGCGATGGGCCTGAGGAGCTGGCTCGCCGCCCCATGGGGCGCGCTGCCGCCTCGGCCACCGCTGCT GCTGCTCCTGCTGCTGCTGCTCCTGCTGCAGCCGCCGCCTCCGACCTGGGCGCTCAGCCCCCGGATCAGCCTGCCTCTGGGCTCTGAAGA GCGGCCATTCCTCAGATTCGAAGCTGAACACATCTCCAACTACACAGCCCTTCTGCTGAGCAGGGATGGCAGGACCCTGTACGTGGGTGC TCGAGAGGCCCTCTTTGCACTCAGTAGCAACCTCAGCTTCCTGCCAGGCGGGGAGTACCAGGAGCTGCTTTGGGGTGCAGACGCAGAGAA GAAACAGCAGTGCAGCTTCAAGGGCAAGGACCCACAGAGCATCCAGGATGTGAGGAGTGATGACGAGGATTATGAAGAGGAAGAGGAAGA GGAAGAAGAAGAGGCTACCAAAGGCAAAGAGATGGATTGTTTAAAGAACGGCCTCAGGGCTGAGAGGCACCTCATTCCCAATGGCCAGCA TGGCCATTAGCTGGAAGCCTACAGGACTCCCATGGCACAGCATGCTGCAAGTACTGTTGGCAGCCTGGCTTCCAGGCCCCACACCGACCC CACATTCTGCCCTTCCCTCTTTCTCACCACCGCCTTCCCTCCCACCTAAGATGTGTTTACCAAAATGTTGTTAACTTGTGTTAAAATGTT AAATATAAGCATGCCCATGGATTTTTACTGCAGTTAGGACTCAGACTGGTCAAAGATTTCAAAGATTTCTCCACAGAACCGTCTCAGTTC TAATTGCACTCCCTCATGCATGTCACTTTCTCAGGGGCTCGCTTTGTTATAGACCCTTTCGCCTCGCCACCTTGCCTGTCCTCAGGACGC TTTCACAGGTGCTAAGTGATCTCATTTTTCCCAGGTGTGTTTGGCCACAAAGAGCAGCTTCTTTCTCAAAATGAGTTAGAAGTGGCAGTG GGACAGGAGCGGAAGGACCACACCAGGAGACACTTCCATCCTGAAGCTTAGGTGCCTCATCTCCACAGGGCGGTGGCAGTCCCTGCCTGC CACCCCACAGGGTTACAGCAAGGATTAAGTGAGATCACAGAAGTTTAAGTACACACTGTCAACCGTGGGGTCATCGTGAACCAACCCACT TTGCACTGTTTGGGAGACAGAAACCTGGCTAAACATGGCACTGCAATGGGCCTGAACAAAAGGCAGAATCTTTAAAAATTCCTCTTAAAT GACTCTGGAGATACCTGGAAATGAAAGTGCCAAGAAAGGTGTTCCATTTTATTTGCTAACTATTTATGCATTAGCTCCCCAAGGGTCATT CTCAGATTCTCCTGGAATTCTTCTCCCTCAGGACCCATGGTTCAAGGAAGGAAGCTTAGGCCCTGTTCCTTCCTGTCCTTTGCTGGTCTT TCCCTTTTTTCCTTTCTAGGAGGGAAGCTTCTGTGCTGCTGCCCTGAGCCCTTCCTTCAGGCCAGCACAGTACCTGGGGACCTCCACGGG GGAATGGGATCCAGGCCAGGTTGCTTGCTGAGCCTCATCACCCAGGAGGCCTGAGCCTCTGGGGAGGGCACGCATGTACACTGCCAGACC CAGGGGAGATCTTGGGAACAGAGGATGCTACGTGATTTCCTCTGGCTTCCAACCCAATCAGCCTGCATCACAGTGAAACACAACACAAAA GGCCATAAAAGGCATACCCCTGAGAAATGTTTAAGGGCAGGTGCTAGAGTCAGTGCCAGCTCCCTGGGAGGAGGGATAGGGACGGGCAGA TCAGTAGCCAGCTTGTCCTCACCCTCCGGAAGGGAGCACCGGAGAAGACTTGCACACGTCCCTGCCTCCCTTGCGGCCTTGTCACCAGAG GGACACATCTCCTGGGCACAATGTGCAGGGCTGACCTGGGAGACTTCTCAGGTGGCTGCTGGCTGAGGAGAGGCCAGGCCTTCCCAAGGA AGACCCTGAGTAAAATCTGCATCTGTCCTCACCACCTGGCACAGTCTCTCATAACGGTCAGATTTTGTATGTTCATCCTATTTATTCAGG GGCTCGTACTAGAAAGCCACAGGAGAGGTCGGCTTGTAGGGACTGGAAAATCAGCCCCAAGCAGAGCAGCTGCAGGGCCCCTGGAGCCGG AAGGACACTGCACAGAACAGACTGCGTTATTGTTATTTTTAAATAAAAATATACATTTGAAACCTTAAGGCTAAACAAAAATAAACAAAA AATCCTTTAGAATTCTTTCCACAACAATATCTCTTTCTGAGAAATTGTTACAAACAAGGTCAGATTTTCTCTGTATAACATTTGCTTTTA TGAGGACAATATCATATGCATTATATGCATAATATGATATTATAAATCAAAATGCCTGCACCCACTTTAGGGTATAGCTATTGACTTATT ATTAATATTATATTATTATTATTTTGCTGGAAGAAGGTCACACTAAGATATAATTTTTTATGTTTTCAGTTAACGGTATGCTTTCTTCTT TGCTTATTTGGTTTTTGTCTCTGTACCAAATATCTTCTTGCTTAAGGTAGAAAAGTATTTGTTTACCTCTATCTCCAGTTTTTTTTCTTA TTTGAATGTTGAAGGTAAAATTGATATACCAATTTTAACTATTTCTGATACAGCTGAAAGCACTAAACTACTTCATAAGAAGTAGATACT CATTTTTGTAACACTATTTAGGGCTTTTGTGGTTAATTTTAAAGGAAACCACTCTTTCTACAGGAAACAAGGGCTCAGGATTCTTCAGAT >80308_80308_5_SEMA4B-CERS3_SEMA4B_chr15_90761061_ENST00000379122_CERS3_chr15_100943070_ENST00000284382_length(amino acids)=225AA_BP=175 MTLTHSRSGGGGPRGDSGADRGAELPPVSPAEPPEPEPRDTVAPALRMLRTAMGLRSWLAAPWGALPPRPPLLLLLLLLLLLQPPPPTWA LSPRISLPLGSEERPFLRFEAEHISNYTALLLSRDGRTLYVGAREALFALSSNLSFLPGGEYQELLWGADAEKKQQCSFKGKDPQSIQDV -------------------------------------------------------------- >80308_80308_6_SEMA4B-CERS3_SEMA4B_chr15_90761061_ENST00000411539_CERS3_chr15_100943070_ENST00000284382_length(transcript)=3115nt_BP=644nt CGCCCTCCGCCGCTTGCGGGTGAGCTCTGCCCAAGCCGAGGCTGCGGGGCCGGCGCCGGCGGGAGGACTGCGGTGCCCCGCGGAGGGGCT GAGTTTGCCAGGGCCCACTTGACCCTGTTTCCCACCTCCCGCCCCCCAGGTCCGGAGGCGGGGGCCCCCGGGGCGACTCGGGGGCGGACC GCGGGGCGGAGCTGCCGCCCGTGAGTCCGGCCGAGCCACCTGAGCCCGAGCCGCGGGACACCGTCGCTCCTGCTCTCCGAATGCTGCGCA CCGCGATGGGCCTGAGGAGCTGGCTCGCCGCCCCATGGGGCGCGCTGCCGCCTCGGCCACCGCTGCTGCTGCTCCTGCTGCTGCTGCTCC TGCTGCAGCCGCCGCCTCCGACCTGGGCGCTCAGCCCCCGGATCAGCCTGCCTCTGGGCTCTGAAGAGCGGCCATTCCTCAGATTCGAAG CTGAACACATCTCCAACTACACAGCCCTTCTGCTGAGCAGGGATGGCAGGACCCTGTACGTGGGTGCTCGAGAGGCCCTCTTTGCACTCA GTAGCAACCTCAGCTTCCTGCCAGGCGGGGAGTACCAGGAGCTGCTTTGGGGTGCAGACGCAGAGAAGAAACAGCAGTGCAGCTTCAAGG GCAAGGACCCACAGAGCATCCAGGATGTGAGGAGTGATGACGAGGATTATGAAGAGGAAGAGGAAGAGGAAGAAGAAGAGGCTACCAAAG GCAAAGAGATGGATTGTTTAAAGAACGGCCTCAGGGCTGAGAGGCACCTCATTCCCAATGGCCAGCATGGCCATTAGCTGGAAGCCTACA GGACTCCCATGGCACAGCATGCTGCAAGTACTGTTGGCAGCCTGGCTTCCAGGCCCCACACCGACCCCACATTCTGCCCTTCCCTCTTTC TCACCACCGCCTTCCCTCCCACCTAAGATGTGTTTACCAAAATGTTGTTAACTTGTGTTAAAATGTTAAATATAAGCATGCCCATGGATT TTTACTGCAGTTAGGACTCAGACTGGTCAAAGATTTCAAAGATTTCTCCACAGAACCGTCTCAGTTCTAATTGCACTCCCTCATGCATGT CACTTTCTCAGGGGCTCGCTTTGTTATAGACCCTTTCGCCTCGCCACCTTGCCTGTCCTCAGGACGCTTTCACAGGTGCTAAGTGATCTC ATTTTTCCCAGGTGTGTTTGGCCACAAAGAGCAGCTTCTTTCTCAAAATGAGTTAGAAGTGGCAGTGGGACAGGAGCGGAAGGACCACAC CAGGAGACACTTCCATCCTGAAGCTTAGGTGCCTCATCTCCACAGGGCGGTGGCAGTCCCTGCCTGCCACCCCACAGGGTTACAGCAAGG ATTAAGTGAGATCACAGAAGTTTAAGTACACACTGTCAACCGTGGGGTCATCGTGAACCAACCCACTTTGCACTGTTTGGGAGACAGAAA CCTGGCTAAACATGGCACTGCAATGGGCCTGAACAAAAGGCAGAATCTTTAAAAATTCCTCTTAAATGACTCTGGAGATACCTGGAAATG AAAGTGCCAAGAAAGGTGTTCCATTTTATTTGCTAACTATTTATGCATTAGCTCCCCAAGGGTCATTCTCAGATTCTCCTGGAATTCTTC TCCCTCAGGACCCATGGTTCAAGGAAGGAAGCTTAGGCCCTGTTCCTTCCTGTCCTTTGCTGGTCTTTCCCTTTTTTCCTTTCTAGGAGG GAAGCTTCTGTGCTGCTGCCCTGAGCCCTTCCTTCAGGCCAGCACAGTACCTGGGGACCTCCACGGGGGAATGGGATCCAGGCCAGGTTG CTTGCTGAGCCTCATCACCCAGGAGGCCTGAGCCTCTGGGGAGGGCACGCATGTACACTGCCAGACCCAGGGGAGATCTTGGGAACAGAG GATGCTACGTGATTTCCTCTGGCTTCCAACCCAATCAGCCTGCATCACAGTGAAACACAACACAAAAGGCCATAAAAGGCATACCCCTGA GAAATGTTTAAGGGCAGGTGCTAGAGTCAGTGCCAGCTCCCTGGGAGGAGGGATAGGGACGGGCAGATCAGTAGCCAGCTTGTCCTCACC CTCCGGAAGGGAGCACCGGAGAAGACTTGCACACGTCCCTGCCTCCCTTGCGGCCTTGTCACCAGAGGGACACATCTCCTGGGCACAATG TGCAGGGCTGACCTGGGAGACTTCTCAGGTGGCTGCTGGCTGAGGAGAGGCCAGGCCTTCCCAAGGAAGACCCTGAGTAAAATCTGCATC TGTCCTCACCACCTGGCACAGTCTCTCATAACGGTCAGATTTTGTATGTTCATCCTATTTATTCAGGGGCTCGTACTAGAAAGCCACAGG AGAGGTCGGCTTGTAGGGACTGGAAAATCAGCCCCAAGCAGAGCAGCTGCAGGGCCCCTGGAGCCGGAAGGACACTGCACAGAACAGACT GCGTTATTGTTATTTTTAAATAAAAATATACATTTGAAACCTTAAGGCTAAACAAAAATAAACAAAAAATCCTTTAGAATTCTTTCCACA ACAATATCTCTTTCTGAGAAATTGTTACAAACAAGGTCAGATTTTCTCTGTATAACATTTGCTTTTATGAGGACAATATCATATGCATTA TATGCATAATATGATATTATAAATCAAAATGCCTGCACCCACTTTAGGGTATAGCTATTGACTTATTATTAATATTATATTATTATTATT TTGCTGGAAGAAGGTCACACTAAGATATAATTTTTTATGTTTTCAGTTAACGGTATGCTTTCTTCTTTGCTTATTTGGTTTTTGTCTCTG TACCAAATATCTTCTTGCTTAAGGTAGAAAAGTATTTGTTTACCTCTATCTCCAGTTTTTTTTCTTATTTGAATGTTGAAGGTAAAATTG ATATACCAATTTTAACTATTTCTGATACAGCTGAAAGCACTAAACTACTTCATAAGAAGTAGATACTCATTTTTGTAACACTATTTAGGG CTTTTGTGGTTAATTTTAAAGGAAACCACTCTTTCTACAGGAAACAAGGGCTCAGGATTCTTCAGATGACCTTATAAAAATGCAGTCCAC >80308_80308_6_SEMA4B-CERS3_SEMA4B_chr15_90761061_ENST00000411539_CERS3_chr15_100943070_ENST00000284382_length(amino acids)=201AA_BP=151 MPPVSPAEPPEPEPRDTVAPALRMLRTAMGLRSWLAAPWGALPPRPPLLLLLLLLLLLQPPPPTWALSPRISLPLGSEERPFLRFEAEHI SNYTALLLSRDGRTLYVGAREALFALSSNLSFLPGGEYQELLWGADAEKKQQCSFKGKDPQSIQDVRSDDEDYEEEEEEEEEEATKGKEM -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SEMA4B-CERS3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SEMA4B-CERS3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SEMA4B-CERS3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
