
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SH3YL1-MTHFD1L (FusionGDB2 ID:81539) |
Fusion Gene Summary for SH3YL1-MTHFD1L |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SH3YL1-MTHFD1L | Fusion gene ID: 81539 | Hgene | Tgene | Gene symbol | SH3YL1 | MTHFD1L | Gene ID | 26751 | 25902 |
| Gene name | SH3 and SYLF domain containing 1 | methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1 like | |
| Synonyms | RAY | FTHFSDC1|MTC1THFS|dJ292B18.2 | |
| Cytomap | 2p25.3 | 6q25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | SH3 domain-containing YSC84-like protein 1SH3 domain containing, Ysc84-like 1 | monofunctional C1-tetrahydrofolate synthase, mitochondrial10-formyl-THF synthetaseformyltetrahydrofolate synthetase domain containing 1 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q6UB35 | |
| Ensembl transtripts involved in fusion gene | ENST00000356150, ENST00000403657, ENST00000403658, ENST00000403712, ENST00000405430, ENST00000415006, ENST00000468321, ENST00000402632, | ENST00000367307, ENST00000478643, ENST00000367321, | |
| Fusion gene scores | * DoF score | 3 X 5 X 3=45 | 14 X 17 X 13=3094 |
| # samples | 4 | 27 | |
| ** MAII score | log2(4/45*10)=-0.169925001442312 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/3094*10)=-3.51844188439821 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SH3YL1 [Title/Abstract] AND MTHFD1L [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SH3YL1(229966)-MTHFD1L(151243341), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | SH3YL1-MTHFD1L seems lost the major protein functional domain in Tgene partner, which is a cell metabolism gene due to the frame-shifted ORF. SH3YL1-MTHFD1L seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MTHFD1L | GO:0006760 | folic acid-containing compound metabolic process | 12937168 |
| Tgene | MTHFD1L | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process | 12937168 |
| Tgene | MTHFD1L | GO:0015942 | formate metabolic process | 16171773 |
 Fusion gene breakpoints across SH3YL1 (5'-gene) Fusion gene breakpoints across SH3YL1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
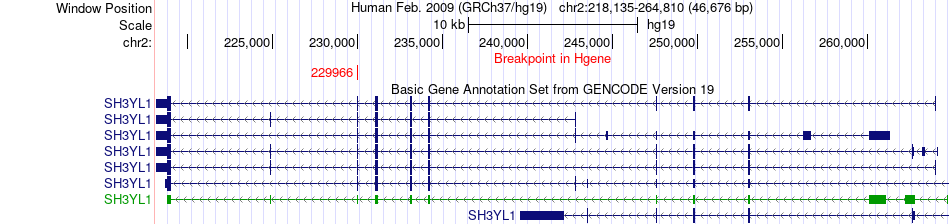 |
 Fusion gene breakpoints across MTHFD1L (3'-gene) Fusion gene breakpoints across MTHFD1L (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
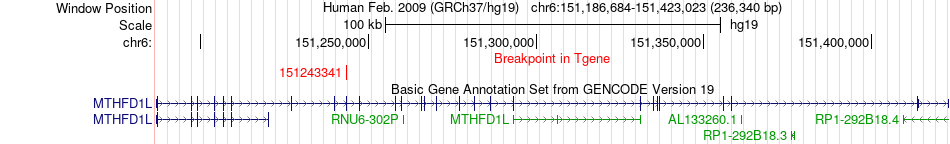 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-X2-A95T-01A | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
Top |
Fusion Gene ORF analysis for SH3YL1-MTHFD1L |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000356150 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000356150 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000403657 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000403657 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000403658 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000403658 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000403712 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000403712 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000405430 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000405430 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000415006 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5CDS-intron | ENST00000415006 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5UTR-3CDS | ENST00000468321 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5UTR-intron | ENST00000468321 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| 5UTR-intron | ENST00000468321 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| Frame-shift | ENST00000356150 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| Frame-shift | ENST00000403658 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| Frame-shift | ENST00000403712 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| Frame-shift | ENST00000415006 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| In-frame | ENST00000403657 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| In-frame | ENST00000405430 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| intron-3CDS | ENST00000402632 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| intron-intron | ENST00000402632 | ENST00000367307 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
| intron-intron | ENST00000402632 | ENST00000478643 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000405430 | SH3YL1 | chr2 | 229966 | - | ENST00000367321 | MTHFD1L | chr6 | 151243341 | + | 3504 | 1158 | 1146 | 3110 | 654 |
| ENST00000403657 | SH3YL1 | chr2 | 229966 | - | ENST00000367321 | MTHFD1L | chr6 | 151243341 | + | 5035 | 2689 | 2677 | 4641 | 654 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000405430 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + | 0.000941255 | 0.9990588 |
| ENST00000403657 | ENST00000367321 | SH3YL1 | chr2 | 229966 | - | MTHFD1L | chr6 | 151243341 | + | 0.000946455 | 0.99905354 |
Top |
Fusion Genomic Features for SH3YL1-MTHFD1L |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| SH3YL1 | chr2 | 229965 | - | MTHFD1L | chr6 | 151243340 | + | 1.04E-08 | 1 |
| SH3YL1 | chr2 | 229965 | - | MTHFD1L | chr6 | 151243340 | + | 1.04E-08 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
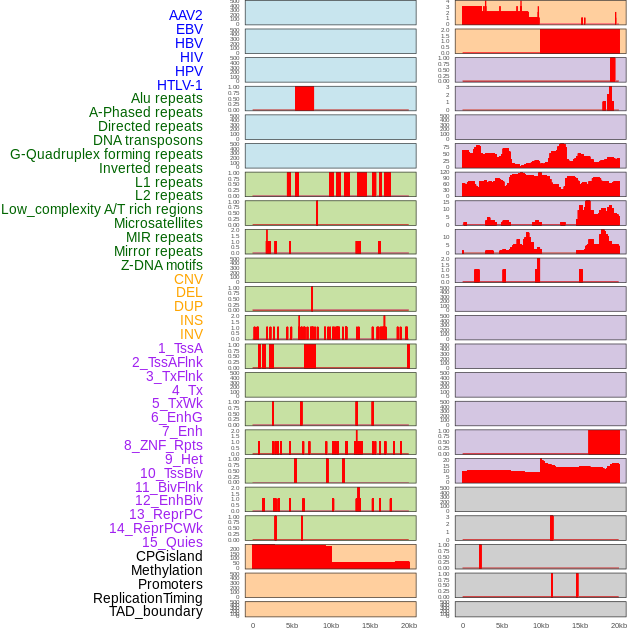 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
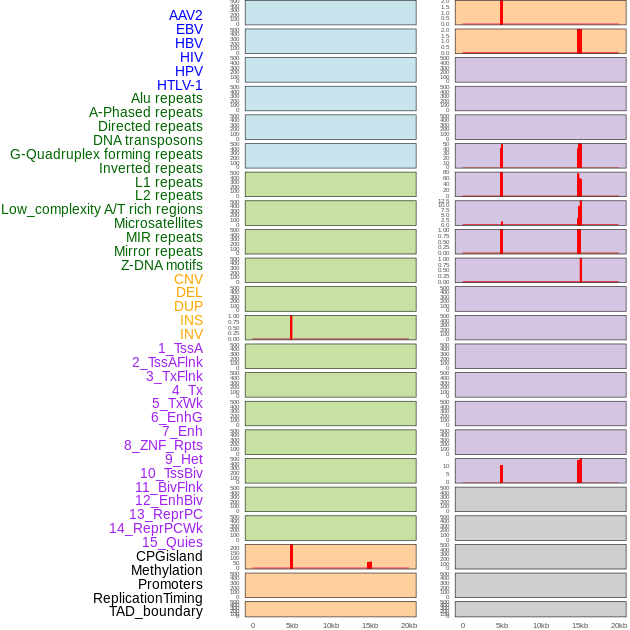 |
Top |
Fusion Protein Features for SH3YL1-MTHFD1L |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:229966/chr6:151243341) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MTHFD1L |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: May provide the missing metabolic reaction required to link the mitochondria and the cytoplasm in the mammalian model of one-carbon folate metabolism in embryonic an transformed cells complementing thus the enzymatic activities of MTHFD2. {ECO:0000250, ECO:0000269|PubMed:16171773}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367307 | 0 | 8 | 34_42 | 0 | 276.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367307 | 0 | 8 | 423_430 | 0 | 276.0 | Nucleotide binding | ATP | |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367321 | 8 | 28 | 423_430 | 328 | 932.6666666666666 | Nucleotide binding | ATP | |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367307 | 0 | 8 | 31_348 | 0 | 276.0 | Region | Note=Methylenetetrahydrofolate dehydrogenase and cyclohydrolase | |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367307 | 0 | 8 | 349_978 | 0 | 276.0 | Region | Note=Formyltetrahydrofolate synthetase | |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367321 | 8 | 28 | 349_978 | 328 | 932.6666666666666 | Region | Note=Formyltetrahydrofolate synthetase |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000356150 | - | 8 | 10 | 283_342 | 260 | 343.0 | Domain | SH3 |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000402632 | - | 1 | 7 | 283_342 | 0 | 945.3333333333334 | Domain | SH3 |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000403657 | - | 11 | 12 | 283_342 | 164 | 228.0 | Domain | SH3 |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000403658 | - | 10 | 11 | 283_342 | 164 | 228.0 | Domain | SH3 |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000403712 | - | 8 | 9 | 283_342 | 260 | 324.0 | Domain | SH3 |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000405430 | - | 10 | 12 | 283_342 | 260 | 343.0 | Domain | SH3 |
| Hgene | SH3YL1 | chr2:229966 | chr6:151243341 | ENST00000415006 | - | 5 | 7 | 283_342 | 164 | 247.0 | Domain | SH3 |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367321 | 8 | 28 | 34_42 | 328 | 932.6666666666666 | Compositional bias | Note=Poly-Gly | |
| Tgene | MTHFD1L | chr2:229966 | chr6:151243341 | ENST00000367321 | 8 | 28 | 31_348 | 328 | 932.6666666666666 | Region | Note=Methylenetetrahydrofolate dehydrogenase and cyclohydrolase |
Top |
Fusion Gene Sequence for SH3YL1-MTHFD1L |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >81539_81539_1_SH3YL1-MTHFD1L_SH3YL1_chr2_229966_ENST00000403657_MTHFD1L_chr6_151243341_ENST00000367321_length(transcript)=5035nt_BP=2689nt AAGAGGCTTCATTAGTAATGACCTGTGTCTTCATCAGTATGATAGAATGGGACATCAGCACTCTGATCAGGTCATGCTCTATATGCCTTA TGGTACAGCAGAGGAAAAATGGGTGAGCCATCATTTGATGCTGAGGATGGGATTTGAGGCTTATGTCTAAATTTTTATTTGCATATTCTT TTTGTTCTGACAGAGAAATTCACTGCATGTAACTATGACCACGCATGATTTTATCAGAAGTTGATTAGAATCTGTAACATTTTCTGTAGT TCAGCTGACACACTGAAGATGTCCTCACTGTGACCAGCCTTGTCAACAGCTGGATCACAGTGACATAAGCTGTGTCTAGGATGAGTCTTG CGGTCTGCAGCCGTGTGAGGCTGCATCTGATTCCCAAGTCCAATGGCTCTCTTCCCGTGTTTACAGATCAGGCCATGACCTTTCCCTACA GCAGGATGTTTGCAAGATTAACTCTCAACTGAGGAATTTGCTTATTTGCTCTAATACTGCTTGGTTGAGAGAGTTCAAAGTAGAAAGTCT TCTGAGGGGATAAGATTCATGTATTATTTACAGTGCTTCAGTTGCATGTATCAGAAAATCTAGTCTAAATTGCTTTAACAGAAAAAGATG TTGCTGGTCCATTTGACGAAGAAGTCCAGAGCGGCTTGTGACTTCAGTCTGGCTTTGCAGAGCCCAGGTGCTCTCTCCACCTTCTGCCAG GTTCCCCTATGAAGGCTTTGCTCTCAGACTCTCCCCTTGTGGAGACAAAAATCCCACAGCAAGTGGAAACAGAAGTGCCTTGGAAAAAGT TATTCTGGCAGCTCCTATACAAGTCCCCCATCAGATGCTGGTATTGGACCAGCTTTGGTTATAAAGCCCTGCTGTGGCACCAAAAGGAAT GTCGTCCTGATACAGCAGCCTCGGGTCACCCAATCCACTCCAGATACTATGGCTGCAGCATCAATGTAAGCAGACAAGTTGTGCTGCCTC TGAAATAAAGTAGATGAAATATTCAGCAGCAGAAAGAACAAGTTTCCACTCCAGTGAACTAATTCAAATGTATCCAGCATCTAGATATAT GTATAAATGTAATTAAAACATATTGAGCATCTAGAGCATCTCTCCCAAGATAATTACCACCTGATAGTAATGGCTATAAAAGCTAAAAGT GGAATCAGTCTACATGTTTCGCAATAGGAATCAATTGAAGCATACATCAAAATGCAGACAGTAGTATTGATGGAGTTTCGTTCTTGCTGC CCAAGCTGGAGTGCAATGGCGCCATCTCGGCTCACTGCAACCTCCACCTTCCAGGTTCAAGTGATTCTCCTGCCTCAGGCTCCTGAGTAG CTGGGATTACAGGCGCACGTCACCACATCTGGCTAATTTTTTGTATTTTTAGTAGAAATGGGGTTTCACCATGTTAGCCAGGCTGGTCTT GAACTCCTGACCTCAGGTGATCCCCCCACCTCGGCCTCCCAAATTGCTGGGATTACAGGTGTGAGCCACCGTGTCCGGCCATAAAACATT TTTTTACAAAAGGCTTACAAAGGAATATTTATAGCAGTTTTATTTGTAATAGCCAAAAGCTGGAAACAACCCAATTGTCTATCAACAGTG ACTGGAAGGACAAATTGTGCTATGTCCACACATTAGAATACCAGTTGGCAGTCAAAAAGGAAGAGCTGATCTGTGCAGCAATTTGAGTCT CAGTAATTTCATGTGAATAACCCTATACCTTCCAATTTGAAATCAGAAGCAAAAAAGGCTGCCAAAATATTAAGAGAATTCACAGAAATA ACTTCCAGAAATGGACCTGATAAGATCATTCCTGCTCACGTAATTGCGAAGGCTAAAGGCCTTGCAATTCTGTCTGTGATCAAAGCCGGG TTCCTGGTGACTGCCAGAGGAGGCAGCGGGATTGTAGTGGCGCGCCTTCCAGATGGAAAATGGTCTGCACCCTCAGCCATTGGGATAGCT GGCCTTGGTGGAGGATTTGAAATAGGAATTGAGAGACTGACGAGACACCCATGAATGCTTCCTTTTCTGTAAAATGGGACAATGACAGGA CCTGTAACCACACAGGGTTTTCCTTTCAAGCACATAGATGGCACACACGTAATCGAGTGCCACTCCAGCTTTCTTAGGTTGCCATCTGCA TGACACATCTTTCTCCATCCTTTTACTGTCAATCTAATGGTATCAGACTTGGTGATAATTCTGAATTATGACCGTGCTGTAGAAGCTTTT GCAAAAGGCGGAAATCTGACCCTCGGAGGGAACTTGACTGTGGCGGTTGGGCCCTTGGGAAGGAACTTGGAAGGAAACGTGGCCCTGAGA AGCTCCGCTGCCGTCTTCACGTACTGCAAGTCAAGGGGACTCTTTGCAGGCGTGTCTTTAGAAGGGAGCTGTTTGATTGAAAGGAAAGAA ACTAATAGAAAATTTTATTGTCAAGATATCCGAGCTTATGACATTTTATTTGGAGATACACCGCGGCCTGCTCAAGCCGAAGATCTTTAT GAAATTCTTGATTCCTTTACTGAAAAGTATGAAAATGAAGGACAACGAATCAATGCAAGAAAAGCAGCAAGGGAGCAGAGGAAGTCTTCT GCTAAAGAATTACCTCCAAAGCCATTGTCAAGACCACAGCAGTCATCTGCACCAGTCCAGCTGAACTCTGGCTCTCAAAAACATGGTCAG TAGTGGAAGGAGATGGCTTCGTGAACAGCAGCACAGGCGGTGGAGACTTCACTGCTTGAAACTTCAGCCTCTCTCCCCTGTGCCAAGTGA CATTGAGATTTCAAGAGGACAAACTCCAAAAGCTGTGGATGTCCTTGCCAAGGAGATTGGATTGCTTGCAGATGAAATTGAAATCTATGG CAAAAGCAAAGCCAAAGTACGTTTGTCCGTGCTAGAAAGGTTAAAGGATCAAGCAGATGGAAAATACGTCTTAGTTGCTGGGATCACACC CACCCCTCTTGGAGAAGGGAAGAGCACAGTCACCATCGGGCTTGTGCAGGCTCTGACCGCACACCTGAATGTCAACTCCTTTGCCTGCTT GAGGCAGCCTTCCCAAGGACCGACGTTTGGAGTGAAAGGAGGAGCCGCGGGTGGTGGATATGCCCAGGTCATCCCCATGGAGGAGTTCAA CCTTCACTTGACTGGAGACATCCACGCCATCACCGCTGCCAATAACTTGCTGGCTGCCGCCATCGACACGAGGATTCTTCATGAAAACAC GCAAACAGATAAGGCTCTGTATAATCGGCTGGTTCCTTTAGTGAATGGTGTCAGAGAATTTTCAGAAATTCAGCTTGCTCGGCTAAAAAA ACTGGGAATAAATAAGACTGATCCGAGCACACTGACAGAAGAGGAAGTGAGTAAATTTGCCCGTCTCGACATCGACCCATCTACCATCAC GTGGCAGAGAGTATTGGATACAAATGACCGATTTCTACGAAAAATAACCATCGGGCAGGGAAACACAGAGAAGGGCCATTACCGGCAGGC GCAGTTTGACATCGCAGTGGCCAGCGAGATCATGGCGGTGCTGGCCCTGACGGACAGCCTCGCAGACATGAAGGCACGGCTGGGAAGGAT GGTGGTGGCCAGTGACAAAAGCGGGCAGCCTGTGACAGCAGATGATTTGGGGGTGACAGGTGCTTTGACAGTTTTGATGAAAGATGCAAT AAAACCAAACCTGATGCAGACCCTGGAAGGGACACCTGTGTTCGTGCATGCGGGCCCTTTTGCTAACATTGCTCACGGCAACTCTTCAGT GTTGGCTGATAAAATTGCCCTGAAACTGGTTGGTGAAGAAGGATTTGTAGTGACCGAAGCTGGCTTTGGTGCTGACATCGGAATGGAGAA ATTCTTCAACATCAAGTGCCGAGCTTCCGGCTTGGTGCCCAACGTGGTTGTGTTAGTGGCAACGGTGCGAGCTCTGAAGATGCATGGAGG CGGGCCAAGTGTAACGGCTGGTGTTCCTCTTAAGAAAGAATATACAGAGGAGAACATCCAGCTGGTGGCAGACGGCTGCTGTAACCTCCA GAAGCAAATTCAGATCACTCAGCTCTTTGGGGTTCCCGTTGTGGTGGCTCTGAATGTCTTCAAGACCGACACCCGCGCTGAGATTGACTT GGTGTGTGAGCTTGCAAAGCGGGCTGGTGCCTTTGATGCAGTCCCCTGCTATCACTGGTCCGTTGGTGGAAAAGGATCGGTGGACTTGGC TCGGGCTGTGAGAGAGGCTGCGAGTAAAAGAAGCCGATTCCAGTTCCTGTATGATGTTCAGGTTCCAATTGTGGACAAGATAAGGACCAT TGCTCAGGCTGTCTATGGAGCCAAAGATATTGAACTCTCTCCTGAGGCACAAGCCAAAATAGATCGTTACACTCAACAGGGTTTTGGAAA TTTGCCCATCTGCATGGCAAAGACCCACCTTTCTCTATCTCACCAACCTGACAAAAAAGGTGTGCCAAGGGACTTCATCTTACCTATCAG TGACGTCCGGGCCAGCATAGGCGCTGGGTTCATTTACCCTTTGGTCGGAACGATGAGCACCATGCCAGGACTGCCCACCCGGCCCTGCTT TTATGACATAGATCTTGATACCGAAACAGAACAAGTTAAAGGCTTGTTCTAAGTGGACAAGGCTCTCACAGGACCCGATGCAGACTCCTG AAACAGACTACTCTTTGCCTTTTTGCTGCAGTTGGAGAAGAAACTGAATTTGAAAAATGTCTGTTATGCAATGCTGGAGACATGGTGAAA TAGGCCAAAGATTTCTTCTTCGTTCAAGATGAATTCTGTTCACAGTGGAGTATGGTGTTCGGCAAAAGGACCTCCACCAAGACTGAAAGA AACTAATTTATTTCTGTTTCTGTGGAGTTTCCATTATTTCTACTGCTTACACTTTAGAATGTTTATTTTATGGGGACTAAGGGATTAGGA >81539_81539_1_SH3YL1-MTHFD1L_SH3YL1_chr2_229966_ENST00000403657_MTHFD1L_chr6_151243341_ENST00000367321_length(amino acids)=654AA_BP=2 MALKNMVSSGRRWLREQQHRRWRLHCLKLQPLSPVPSDIEISRGQTPKAVDVLAKEIGLLADEIEIYGKSKAKVRLSVLERLKDQADGKY VLVAGITPTPLGEGKSTVTIGLVQALTAHLNVNSFACLRQPSQGPTFGVKGGAAGGGYAQVIPMEEFNLHLTGDIHAITAANNLLAAAID TRILHENTQTDKALYNRLVPLVNGVREFSEIQLARLKKLGINKTDPSTLTEEEVSKFARLDIDPSTITWQRVLDTNDRFLRKITIGQGNT EKGHYRQAQFDIAVASEIMAVLALTDSLADMKARLGRMVVASDKSGQPVTADDLGVTGALTVLMKDAIKPNLMQTLEGTPVFVHAGPFAN IAHGNSSVLADKIALKLVGEEGFVVTEAGFGADIGMEKFFNIKCRASGLVPNVVVLVATVRALKMHGGGPSVTAGVPLKKEYTEENIQLV ADGCCNLQKQIQITQLFGVPVVVALNVFKTDTRAEIDLVCELAKRAGAFDAVPCYHWSVGGKGSVDLARAVREAASKRSRFQFLYDVQVP IVDKIRTIAQAVYGAKDIELSPEAQAKIDRYTQQGFGNLPICMAKTHLSLSHQPDKKGVPRDFILPISDVRASIGAGFIYPLVGTMSTMP -------------------------------------------------------------- >81539_81539_2_SH3YL1-MTHFD1L_SH3YL1_chr2_229966_ENST00000405430_MTHFD1L_chr6_151243341_ENST00000367321_length(transcript)=3504nt_BP=1158nt CGAAGGCCTGAGCGCGGCCCGGCGGGCGCGTGGTTTTTGTCCCGCGAAACGTACACGGCGGGTGTTTTGCGCCGCCTCCTTCGTCACCTG CCGCAGGAAGTGTGGAACTGGTCTCAATCGGTTGACCACCTTAAGGAAATAGGATTTACACTGTGGACAGAGATGTGTCCTTTACACTTC ATTTGAACCAATATGCCCATGCTCCTGTGGAGGCTGAGTTGGAAAGTGACACGAAGCTGCCCCCCCGCAAGAGAGCAGAAATAAGGATCT GGCATGTCAACACAAACATAGGAAGTTGAGGCAGTCAAAGGGAAATAACTGCTACCCTGACTCTGCTGAGAGCCTCACAAGTCCCTGTGC ACGCCAGCTTGTTTGCCATGAATAACCCTATACCTTCCAATTTGAAATCAGAAGCAAAAAAGGCTGCCAAAATATTAAGAGAATTCACAG AAATAACTTCCAGAAATGGACCTGATAAGATCATTCCTGCTCACGTAATTGCGAAGGCTAAAGGCCTTGCAATTCTGTCTGTGATCAAAG CCGGGTTCCTGGTGACTGCCAGAGGAGGCAGCGGGATTGTAGTGGCGCGCCTTCCAGATGGAAAATGGTCTGCACCCTCAGCCATTGGGA TAGCTGGCCTTGGTGGAGGATTTGAAATAGGAATTGAGGTATCAGACTTGGTGATAATTCTGAATTATGACCGTGCTGTAGAAGCTTTTG CAAAAGGCGGAAATCTGACCCTCGGAGGGAACTTGACTGTGGCGGTTGGGCCCTTGGGAAGGAACTTGGAAGGAAACGTGGCCCTGAGAA GCTCCGCTGCCGTCTTCACGTACTGCAAGTCAAGGGGACTCTTTGCAGGCGTGTCTTTAGAAGGGAGCTGTTTGATTGAAAGGAAAGAAA CTAATAGAAAATTTTATTGTCAAGATATCCGAGCTTATGACATTTTATTTGGAGATACACCGCGGCCTGCTCAAGCCGAAGATCTTTATG AAATTCTTGATTCCTTTACTGAAAAGTATGAAAATGAAGGACAACGAATCAATGCAAGAAAAGCAGCAAGGGAGCAGAGGAAGTCTTCTG CTAAAGAATTACCTCCAAAGCCATTGTCAAGACCACAGCAGTCATCTGCACCAGTCCAGCTGAACTCTGGCTCTCAAAAACATGGTCAGT AGTGGAAGGAGATGGCTTCGTGAACAGCAGCACAGGCGGTGGAGACTTCACTGCTTGAAACTTCAGCCTCTCTCCCCTGTGCCAAGTGAC ATTGAGATTTCAAGAGGACAAACTCCAAAAGCTGTGGATGTCCTTGCCAAGGAGATTGGATTGCTTGCAGATGAAATTGAAATCTATGGC AAAAGCAAAGCCAAAGTACGTTTGTCCGTGCTAGAAAGGTTAAAGGATCAAGCAGATGGAAAATACGTCTTAGTTGCTGGGATCACACCC ACCCCTCTTGGAGAAGGGAAGAGCACAGTCACCATCGGGCTTGTGCAGGCTCTGACCGCACACCTGAATGTCAACTCCTTTGCCTGCTTG AGGCAGCCTTCCCAAGGACCGACGTTTGGAGTGAAAGGAGGAGCCGCGGGTGGTGGATATGCCCAGGTCATCCCCATGGAGGAGTTCAAC CTTCACTTGACTGGAGACATCCACGCCATCACCGCTGCCAATAACTTGCTGGCTGCCGCCATCGACACGAGGATTCTTCATGAAAACACG CAAACAGATAAGGCTCTGTATAATCGGCTGGTTCCTTTAGTGAATGGTGTCAGAGAATTTTCAGAAATTCAGCTTGCTCGGCTAAAAAAA CTGGGAATAAATAAGACTGATCCGAGCACACTGACAGAAGAGGAAGTGAGTAAATTTGCCCGTCTCGACATCGACCCATCTACCATCACG TGGCAGAGAGTATTGGATACAAATGACCGATTTCTACGAAAAATAACCATCGGGCAGGGAAACACAGAGAAGGGCCATTACCGGCAGGCG CAGTTTGACATCGCAGTGGCCAGCGAGATCATGGCGGTGCTGGCCCTGACGGACAGCCTCGCAGACATGAAGGCACGGCTGGGAAGGATG GTGGTGGCCAGTGACAAAAGCGGGCAGCCTGTGACAGCAGATGATTTGGGGGTGACAGGTGCTTTGACAGTTTTGATGAAAGATGCAATA AAACCAAACCTGATGCAGACCCTGGAAGGGACACCTGTGTTCGTGCATGCGGGCCCTTTTGCTAACATTGCTCACGGCAACTCTTCAGTG TTGGCTGATAAAATTGCCCTGAAACTGGTTGGTGAAGAAGGATTTGTAGTGACCGAAGCTGGCTTTGGTGCTGACATCGGAATGGAGAAA TTCTTCAACATCAAGTGCCGAGCTTCCGGCTTGGTGCCCAACGTGGTTGTGTTAGTGGCAACGGTGCGAGCTCTGAAGATGCATGGAGGC GGGCCAAGTGTAACGGCTGGTGTTCCTCTTAAGAAAGAATATACAGAGGAGAACATCCAGCTGGTGGCAGACGGCTGCTGTAACCTCCAG AAGCAAATTCAGATCACTCAGCTCTTTGGGGTTCCCGTTGTGGTGGCTCTGAATGTCTTCAAGACCGACACCCGCGCTGAGATTGACTTG GTGTGTGAGCTTGCAAAGCGGGCTGGTGCCTTTGATGCAGTCCCCTGCTATCACTGGTCCGTTGGTGGAAAAGGATCGGTGGACTTGGCT CGGGCTGTGAGAGAGGCTGCGAGTAAAAGAAGCCGATTCCAGTTCCTGTATGATGTTCAGGTTCCAATTGTGGACAAGATAAGGACCATT GCTCAGGCTGTCTATGGAGCCAAAGATATTGAACTCTCTCCTGAGGCACAAGCCAAAATAGATCGTTACACTCAACAGGGTTTTGGAAAT TTGCCCATCTGCATGGCAAAGACCCACCTTTCTCTATCTCACCAACCTGACAAAAAAGGTGTGCCAAGGGACTTCATCTTACCTATCAGT GACGTCCGGGCCAGCATAGGCGCTGGGTTCATTTACCCTTTGGTCGGAACGATGAGCACCATGCCAGGACTGCCCACCCGGCCCTGCTTT TATGACATAGATCTTGATACCGAAACAGAACAAGTTAAAGGCTTGTTCTAAGTGGACAAGGCTCTCACAGGACCCGATGCAGACTCCTGA AACAGACTACTCTTTGCCTTTTTGCTGCAGTTGGAGAAGAAACTGAATTTGAAAAATGTCTGTTATGCAATGCTGGAGACATGGTGAAAT AGGCCAAAGATTTCTTCTTCGTTCAAGATGAATTCTGTTCACAGTGGAGTATGGTGTTCGGCAAAAGGACCTCCACCAAGACTGAAAGAA ACTAATTTATTTCTGTTTCTGTGGAGTTTCCATTATTTCTACTGCTTACACTTTAGAATGTTTATTTTATGGGGACTAAGGGATTAGGAG >81539_81539_2_SH3YL1-MTHFD1L_SH3YL1_chr2_229966_ENST00000405430_MTHFD1L_chr6_151243341_ENST00000367321_length(amino acids)=654AA_BP=2 MALKNMVSSGRRWLREQQHRRWRLHCLKLQPLSPVPSDIEISRGQTPKAVDVLAKEIGLLADEIEIYGKSKAKVRLSVLERLKDQADGKY VLVAGITPTPLGEGKSTVTIGLVQALTAHLNVNSFACLRQPSQGPTFGVKGGAAGGGYAQVIPMEEFNLHLTGDIHAITAANNLLAAAID TRILHENTQTDKALYNRLVPLVNGVREFSEIQLARLKKLGINKTDPSTLTEEEVSKFARLDIDPSTITWQRVLDTNDRFLRKITIGQGNT EKGHYRQAQFDIAVASEIMAVLALTDSLADMKARLGRMVVASDKSGQPVTADDLGVTGALTVLMKDAIKPNLMQTLEGTPVFVHAGPFAN IAHGNSSVLADKIALKLVGEEGFVVTEAGFGADIGMEKFFNIKCRASGLVPNVVVLVATVRALKMHGGGPSVTAGVPLKKEYTEENIQLV ADGCCNLQKQIQITQLFGVPVVVALNVFKTDTRAEIDLVCELAKRAGAFDAVPCYHWSVGGKGSVDLARAVREAASKRSRFQFLYDVQVP IVDKIRTIAQAVYGAKDIELSPEAQAKIDRYTQQGFGNLPICMAKTHLSLSHQPDKKGVPRDFILPISDVRASIGAGFIYPLVGTMSTMP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SH3YL1-MTHFD1L |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SH3YL1-MTHFD1L |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SH3YL1-MTHFD1L |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
