
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SHMT2-SLC26A10 (FusionGDB2 ID:81728) |
Fusion Gene Summary for SHMT2-SLC26A10 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SHMT2-SLC26A10 | Fusion gene ID: 81728 | Hgene | Tgene | Gene symbol | SHMT2 | SLC26A10 | Gene ID | 6472 | 65012 |
| Gene name | serine hydroxymethyltransferase 2 | solute carrier family 26 member 10 | |
| Synonyms | GLYA|HEL-S-51e|SHMT | - | |
| Cytomap | 12q13.3 | 12q13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | serine hydroxymethyltransferase, mitochondrialGLY A+epididymis secretory sperm binding protein Li 51eglycine auxotroph A, human complement for hamsterglycine hydroxymethyltransferaseserine aldolaseserine hydroxymethylaseserine hydroxymethyltransfer | solute carrier family 26 member 10 | |
| Modification date | 20200327 | 20200329 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000328923, ENST00000393827, ENST00000414700, ENST00000449049, ENST00000553474, ENST00000557487, ENST00000554600, | ENST00000490243, ENST00000320442, ENST00000379218, | |
| Fusion gene scores | * DoF score | 7 X 6 X 4=168 | 5 X 4 X 3=60 |
| # samples | 8 | 4 | |
| ** MAII score | log2(8/168*10)=-1.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(4/60*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SHMT2 [Title/Abstract] AND SLC26A10 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SHMT2(57627893)-SLC26A10(58018649), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | SHMT2-SLC26A10 seems lost the major protein functional domain in Hgene partner, which is a cell metabolism gene due to the frame-shifted ORF. SHMT2-SLC26A10 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. SHMT2-SLC26A10 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. SHMT2-SLC26A10 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | SHMT2 | GO:0006544 | glycine metabolic process | 25619277|29180469 |
| Hgene | SHMT2 | GO:0006563 | L-serine metabolic process | 25619277|29180469 |
| Hgene | SHMT2 | GO:0006730 | one-carbon metabolic process | 11516159|29180469|29364879|29452640 |
| Hgene | SHMT2 | GO:0034340 | response to type I interferon | 24075985 |
| Hgene | SHMT2 | GO:0046653 | tetrahydrofolate metabolic process | 24075985|25619277 |
| Hgene | SHMT2 | GO:0051262 | protein tetramerization | 25619277 |
| Hgene | SHMT2 | GO:0051289 | protein homotetramerization | 29180469 |
 Fusion gene breakpoints across SHMT2 (5'-gene) Fusion gene breakpoints across SHMT2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
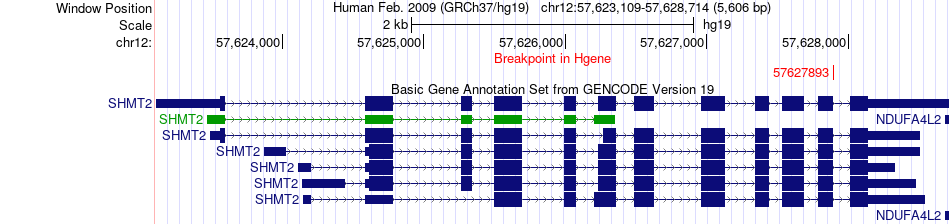 |
 Fusion gene breakpoints across SLC26A10 (3'-gene) Fusion gene breakpoints across SLC26A10 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
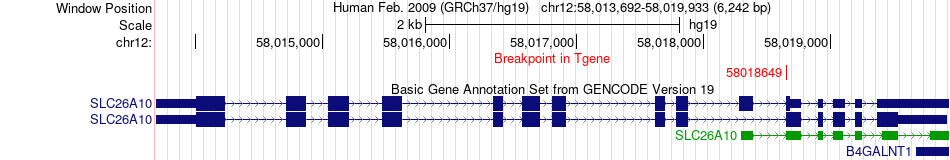 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-28-5207-01A | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
Top |
Fusion Gene ORF analysis for SHMT2-SLC26A10 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000328923 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| 5CDS-3UTR | ENST00000393827 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| 5CDS-3UTR | ENST00000414700 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| 5CDS-3UTR | ENST00000449049 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| 5CDS-3UTR | ENST00000553474 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| 5CDS-3UTR | ENST00000557487 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| Frame-shift | ENST00000328923 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| Frame-shift | ENST00000393827 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| Frame-shift | ENST00000414700 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| Frame-shift | ENST00000449049 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| Frame-shift | ENST00000553474 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| Frame-shift | ENST00000557487 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| In-frame | ENST00000328923 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| In-frame | ENST00000393827 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| In-frame | ENST00000414700 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| In-frame | ENST00000449049 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| In-frame | ENST00000553474 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| In-frame | ENST00000557487 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| intron-3CDS | ENST00000554600 | ENST00000320442 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| intron-3CDS | ENST00000554600 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
| intron-3UTR | ENST00000554600 | ENST00000490243 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000328923 | SHMT2 | chr12 | 57627893 | + | ENST00000379218 | SLC26A10 | chr12 | 58018649 | + | 2709 | 1839 | 452 | 1996 | 514 |
| ENST00000557487 | SHMT2 | chr12 | 57627893 | + | ENST00000379218 | SLC26A10 | chr12 | 58018649 | + | 2295 | 1425 | 68 | 1582 | 504 |
| ENST00000414700 | SHMT2 | chr12 | 57627893 | + | ENST00000379218 | SLC26A10 | chr12 | 58018649 | + | 2383 | 1513 | 102 | 1670 | 522 |
| ENST00000553474 | SHMT2 | chr12 | 57627893 | + | ENST00000379218 | SLC26A10 | chr12 | 58018649 | + | 2317 | 1447 | 33 | 1604 | 523 |
| ENST00000449049 | SHMT2 | chr12 | 57627893 | + | ENST00000379218 | SLC26A10 | chr12 | 58018649 | + | 2525 | 1655 | 295 | 1812 | 505 |
| ENST00000393827 | SHMT2 | chr12 | 57627893 | + | ENST00000379218 | SLC26A10 | chr12 | 58018649 | + | 2225 | 1355 | 256 | 1512 | 418 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000328923 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + | 0.004479228 | 0.9955207 |
| ENST00000557487 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + | 0.006863722 | 0.9931363 |
| ENST00000414700 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + | 0.010356685 | 0.98964334 |
| ENST00000553474 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + | 0.009181701 | 0.9908183 |
| ENST00000449049 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + | 0.010052065 | 0.9899479 |
| ENST00000393827 | ENST00000379218 | SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018649 | + | 0.08328958 | 0.91671044 |
Top |
Fusion Genomic Features for SHMT2-SLC26A10 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018648 | + | 0.001036988 | 0.998963 |
| SHMT2 | chr12 | 57627893 | + | SLC26A10 | chr12 | 58018648 | + | 0.001036988 | 0.998963 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
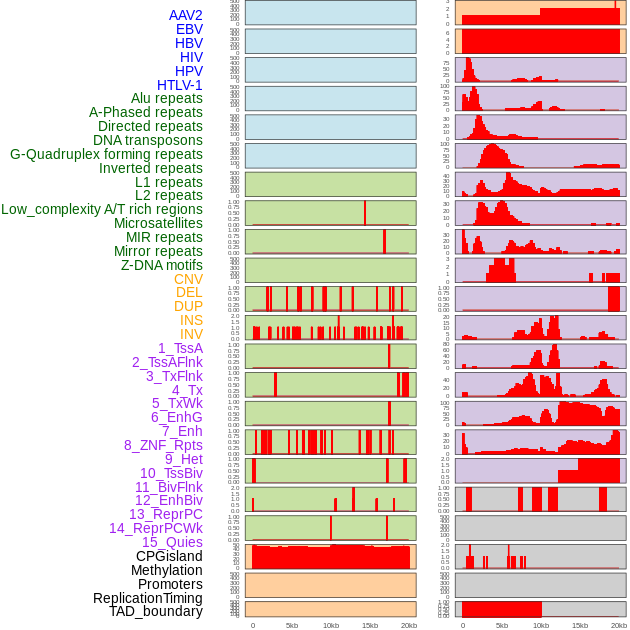 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
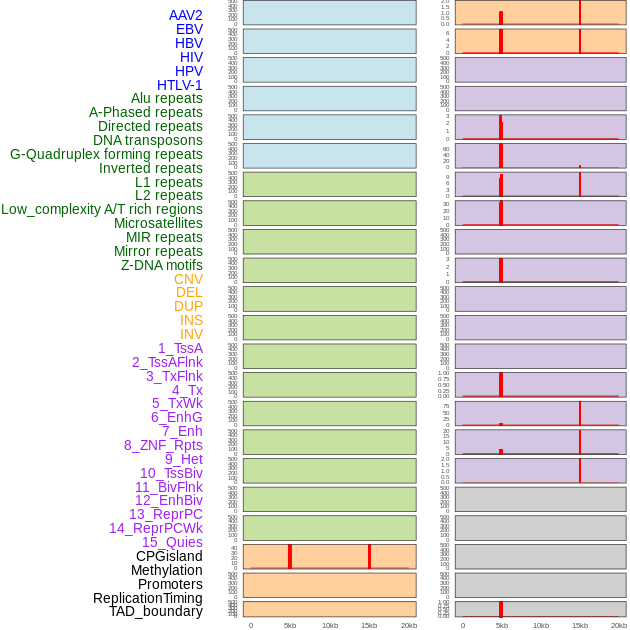 |
Top |
Fusion Protein Features for SHMT2-SLC26A10 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:57627893/chr12:58018649) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000320442 | 8 | 14 | 406_541 | 409 | 564.0 | Domain | STAS |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000379218 | 9 | 15 | 406_541 | 444 | 366.3333333333333 | Domain | STAS | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000320442 | 8 | 14 | 116_136 | 409 | 564.0 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000320442 | 8 | 14 | 152_172 | 409 | 564.0 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000320442 | 8 | 14 | 352_372 | 409 | 564.0 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000320442 | 8 | 14 | 45_65 | 409 | 564.0 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000320442 | 8 | 14 | 75_91 | 409 | 564.0 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000379218 | 9 | 15 | 116_136 | 444 | 366.3333333333333 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000379218 | 9 | 15 | 152_172 | 444 | 366.3333333333333 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000379218 | 9 | 15 | 352_372 | 444 | 366.3333333333333 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000379218 | 9 | 15 | 45_65 | 444 | 366.3333333333333 | Transmembrane | Helical | |
| Tgene | SLC26A10 | chr12:57627893 | chr12:58018649 | ENST00000379218 | 9 | 15 | 75_91 | 444 | 366.3333333333333 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for SHMT2-SLC26A10 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >81728_81728_1_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000328923_SLC26A10_chr12_58018649_ENST00000379218_length(transcript)=2709nt_BP=1839nt ACGGTGCTTTCCAGTGTTCCATAGCTCTTTATCTTGTGTGGGGGTCTAGATCTCCTGGAGTCTAATAATGCTAGCTAGATATTCAGTAGG CACCCTATAAACGTTTGTCTTGACAACAAGCAGAGAGAAAACCCTGAAAAGCTTCAAGATAGTAAGGAGAGGGGGCGGCACTGGACCTCT GTTTTTGTTTTTTGTTTTTTTCCCCCCAGGCAAGGGCCTCTAACCCTGGGGCTACAGACGGGAATCATAAAGAAAAAAGCGGTGAGTGGG CGAACTACAATTCCCAAAAGGCCACAAAGGGGCCACCACTACGCATGCGTAGATCCCTCCCGTTAGCTTTGGCGCCTCAGCGAGCTCTTC TCGCGCATGCGTTCTCCGAACGGTCTTCTTCCGACAGCTTGCTGCCCTAGACCAGAGTTGGTGGCTGGACCTCCTGCGACTTCCGAGTTG CGATGCTGTACTTCTCTTTGTTTTGGGCGGCTCGGCCTCTGCAGAGATGTGGGCAGCTGGTCAGGATGGCCATTCGGGCTCAGCACAGCA ACGCAGCCCAGACTCAGACTGGGGAAGCAAACAGGGGCTGGACAGGCCAGGAGAGCCTGTCGGACAGTGATCCTGAGATGTGGGAGTTGC TGCAGAGGGAGAAGGACAGGCAGTGTCGTGGCCTGGAGCTCATTGCCTCAGAGAACTTCTGCAGCCGAGCTGCGCTGGAGGCCCTGGGGT CCTGTCTGAACAACAAGTACTCGGAGGGTTATCCTGGCAAGAGATACTATGGGGGAGCAGAGGTGGTGGATGAAATTGAGCTGCTGTGCC AGCGCCGGGCCTTGGAAGCCTTTGACCTGGATCCTGCACAGTGGGGAGTCAATGTCCAGCCCTACTCCGGGTCCCCAGCCAACCTGGCCG TCTACACAGCCCTTCTGCAACCTCACGACCGGATCATGGGGCTGGACCTGCCCGATGGGGGCCATCTCACCCACGGCTACATGTCTGACG TCAAGCGGATATCAGCCACGTCCATCTTCTTCGAGTCTATGCCCTATAAGCTCAACCCCAAAACTGGCCTCATTGACTACAACCAGCTGG CACTGACTGCTCGACTTTTCCGGCCACGGCTCATCATAGCTGGCACCAGCGCCTATGCTCGCCTCATTGACTACGCCCGCATGAGAGAGG TGTGTGATGAAGTCAAAGCACACCTGCTGGCAGACATGGCCCACATCAGTGGCCTGGTGGCTGCCAAGGTGATTCCCTCGCCTTTCAAGC ACGCGGACATCGTCACCACCACTACTCACAAGACTCTTCGAGGGGCCAGGTCAGGGCTCATCTTCTACCGGAAAGGGGTGAAGGCTGTGG ACCCCAAGACTGGCCGGGAGATCCCTTACACATTTGAGGACCGAATCAACTTTGCCGTGTTCCCATCCCTGCAGGGGGGCCCCCACAATC ATGCCATTGCTGCAGTAGCTGTGGCCCTAAAGCAGGCCTGCACCCCCATGTTCCGGGAGTACTCCCTGCAGGTTCTGAAGAATGCTCGGG CCATGGCAGATGCCCTGCTAGAGCGAGGCTACTCACTGGTATCAGGTGGTACTGACAACCACCTGGTGCTGGTGGACCTGCGGCCCAAGG GCCTGGATGGAGCTCGGGCTGAGCGGGTGCTAGAGCTTGTATCCATCACTGCCAACAAGAACACCTGTCCTGGAGACCGAAGTGCCATCA CACCGGGCGGCCTGCGGCTTGGGGCCCCAGCCTTAACTTCTCGACAGTTCCGTGAGGATGACTTCCGGAGAGTTGTGGACTTTATAGATG AAGGGGTCAACATTGGCTTAGAGGTGAAGAGCAAGACTGCTCCTCCAGGTCCCGGGGCTCTGCATCCTGAGCTATCCAACACCACTGTAC TTTGGGACCCGTGGGCAGTTTCGCTGCAACCTGGAGTGGCACCTGGGGCTCGGAGAAGGAGAAAAGGAGACTTCAAAGCCAGATGGCCCA ATGGTTGCAGTTGCTGAGCCTGTCAGGGTGGTGGTCCTAGACTTCAGTGGTGTCACCTTTGCAGATGCTGCTGGGGCCAGAGAAGTGGTG CAGGTGAGGGAGAGGCTGGCCAGCCGATGTCGAGATGCTAGGATCCGCCTCCTCCTGGCTCAGTGTAATGCCTTGGTGCAGGGGACACTG ACCCGGGTAGGACTCCTGGACAGGGTGACTCCAGATCAGCTGTTTGTGAGTGTGCAGGATGCAGCTGCTTATGCCCTGGGGAGCCTGTTA AGGGGCAGTAGCACCAGGAGCGGGAGCCAGGAGGCACTGGGCTGCGGCAAGTGAGGCAGGGGTAAGTGGCTGGAGACCCAGGGAGAGGGG TTTGGGAAAGGGTCTGAGGAAGATCAAGAAGAAAAGGGTGGGGACGGAATGAATAGTAGATTGAAGACCAAATCAGGGAGCCCGGGCTGA GCTATGGGTGAGGAACTGAAGAACTCAGATCCCTAAGTAGGTGGGGTACCCCTGAGAGGACTGCCACATTTCATTAAGGAAGAGGGGTGC CTAATCTATTCCTGACATGTCTCCTCTCTTCTCCAGGAGCTCACTGACCCAAAGATTTGCACCGTGTGGGTCTGACCTCATCATGTGGAG TGCAGAGGGCCCTGATGACATGTGTGTGATGAGGACCATGACCCTTGAACCCCCTTACCTAACGTAACTAATAAAATGAAGCTGAGAGCT >81728_81728_1_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000328923_SLC26A10_chr12_58018649_ENST00000379218_length(amino acids)=514AA_BP=0 MLYFSLFWAARPLQRCGQLVRMAIRAQHSNAAQTQTGEANRGWTGQESLSDSDPEMWELLQREKDRQCRGLELIASENFCSRAALEALGS CLNNKYSEGYPGKRYYGGAEVVDEIELLCQRRALEAFDLDPAQWGVNVQPYSGSPANLAVYTALLQPHDRIMGLDLPDGGHLTHGYMSDV KRISATSIFFESMPYKLNPKTGLIDYNQLALTARLFRPRLIIAGTSAYARLIDYARMREVCDEVKAHLLADMAHISGLVAAKVIPSPFKH ADIVTTTTHKTLRGARSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTPMFREYSLQVLKNARA MADALLERGYSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSITANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDE -------------------------------------------------------------- >81728_81728_2_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000393827_SLC26A10_chr12_58018649_ENST00000379218_length(transcript)=2225nt_BP=1355nt ACAATCTGTGGCCTCAGCCCTTGCCAACTCTGCCTCTTCCCCCGCTGACTGGGCCTCTGCAGAGATGTGGGCAGCTGGTCAGGATGGCCA TTCGGGCTCAGCACAGCAACGCAGCCCAGACTCAGACTGGGGAAGCAAACAGGGGCTGGACAGGCCAGGAGAGCCTGTCGGACAGTGATC CTGAGATGTGGGAGTTGCTGCAGAGGGAGAAGGACAGGCAGTGTCGTGGCCTGGAGCTCATTGCCTCAGAGATACTATGGGGGAGCAGAG GTGGTGGATGAAATTGAGCTGCTGTGCCAGCGCCGGGCCTTGGAAGCCTTTGACCTGGATCCTGCACAGTGGGGAGTCAATGTCCAGCCC TACTCCGGGTCCCCAGCCAACCTGGCCGTCTACACAGCCCTTCTGCAACCTCACGACCGGATCATGGGGCTGGACCTGCCCGATGGGGGC CATCTCACCCACGGCTACATGTCTGACGTCAAGCGGATATCAGCCACGTCCATCTTCTTCGAGTCTATGCCCTATAAGCTCAACGGCTTT AGCTGTTTGTGTGTCTGTCCAGCCCAAAACTGGCCTCATTGACTACAACCAGCTGGCACTGACTGCTCGACTTTTCCGGCCACGGCTCAT CATAGCTGGCACCAGCGCCTATGCTCGCCTCATTGACTACGCCCGCATGAGAGAGGTGTGTGATGAAGTCAAAGCACACCTGCTGGCAGA CATGGCCCACATCAGTGGCCTGGTGGCTGCCAAGGTGATTCCCTCGCCTTTCAAGCACGCGGACATCGTCACCACCACTACTCACAAGAC TCTTCGAGGGGCCAGGTCAGGGCTCATCTTCTACCGGAAAGGGGTGAAGGCTGTGGACCCCAAGACTGGCCGGGAGATCCCTTACACATT TGAGGACCGAATCAACTTTGCCGTGTTCCCATCCCTGCAGGGGGGCCCCCACAATCATGCCATTGCTGCAGTAGCTGTGGCCCTAAAGCA GGCCTGCACCCCCATGTTCCGGGAGTACTCCCTGCAGGTTCTGAAGAATGCTCGGGCCATGGCAGATGCCCTGCTAGAGCGAGGCTACTC ACTGGTATCAGGTGGTACTGACAACCACCTGGTGCTGGTGGACCTGCGGCCCAAGGGCCTGGATGGAGCTCGGGCTGAGCGGGTGCTAGA GCTTGTATCCATCACTGCCAACAAGAACACCTGTCCTGGAGACCGAAGTGCCATCACACCGGGCGGCCTGCGGCTTGGGGCCCCAGCCTT AACTTCTCGACAGTTCCGTGAGGATGACTTCCGGAGAGTTGTGGACTTTATAGATGAAGGGGTCAACATTGGCTTAGAGGTGAAGAGCAA GACTGCTCCTCCAGGTCCCGGGGCTCTGCATCCTGAGCTATCCAACACCACTGTACTTTGGGACCCGTGGGCAGTTTCGCTGCAACCTGG AGTGGCACCTGGGGCTCGGAGAAGGAGAAAAGGAGACTTCAAAGCCAGATGGCCCAATGGTTGCAGTTGCTGAGCCTGTCAGGGTGGTGG TCCTAGACTTCAGTGGTGTCACCTTTGCAGATGCTGCTGGGGCCAGAGAAGTGGTGCAGGTGAGGGAGAGGCTGGCCAGCCGATGTCGAG ATGCTAGGATCCGCCTCCTCCTGGCTCAGTGTAATGCCTTGGTGCAGGGGACACTGACCCGGGTAGGACTCCTGGACAGGGTGACTCCAG ATCAGCTGTTTGTGAGTGTGCAGGATGCAGCTGCTTATGCCCTGGGGAGCCTGTTAAGGGGCAGTAGCACCAGGAGCGGGAGCCAGGAGG CACTGGGCTGCGGCAAGTGAGGCAGGGGTAAGTGGCTGGAGACCCAGGGAGAGGGGTTTGGGAAAGGGTCTGAGGAAGATCAAGAAGAAA AGGGTGGGGACGGAATGAATAGTAGATTGAAGACCAAATCAGGGAGCCCGGGCTGAGCTATGGGTGAGGAACTGAAGAACTCAGATCCCT AAGTAGGTGGGGTACCCCTGAGAGGACTGCCACATTTCATTAAGGAAGAGGGGTGCCTAATCTATTCCTGACATGTCTCCTCTCTTCTCC AGGAGCTCACTGACCCAAAGATTTGCACCGTGTGGGTCTGACCTCATCATGTGGAGTGCAGAGGGCCCTGATGACATGTGTGTGATGAGG >81728_81728_2_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000393827_SLC26A10_chr12_58018649_ENST00000379218_length(amino acids)=418AA_BP=0 MGEQRWWMKLSCCASAGPWKPLTWILHSGESMSSPTPGPQPTWPSTQPFCNLTTGSWGWTCPMGAISPTATCLTSSGYQPRPSSSSLCPI SSTALAVCVSVQPKTGLIDYNQLALTARLFRPRLIIAGTSAYARLIDYARMREVCDEVKAHLLADMAHISGLVAAKVIPSPFKHADIVTT TTHKTLRGARSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTPMFREYSLQVLKNARAMADALL ERGYSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSITANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDEGVNIGL -------------------------------------------------------------- >81728_81728_3_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000414700_SLC26A10_chr12_58018649_ENST00000379218_length(transcript)=2383nt_BP=1513nt GCGGACCCGGTGCTGGCAAATGAGCGGAGTTTTCGGCCTGGTCTCACAAGCTGAGCCTTTCCGGCCAGCTCTGAGCCCTGCAGATGATGC AACCGAGCTTCACTGCTTGCATCAGGCAGGGGTCCCGGCGGCAGTGAGGGCTGGAAGGAGGTGGAACGGCCTCTGCAGAGATGTGGGCAG CTGGTCAGGATGGCCATTCGGGCTCAGCACAGCAACGCAGCCCAGACTCAGACTGGGGAAGCAAACAGGGGCTGGACAGGCCAGGAGAGC CTGTCGGACAGTGATCCTGAGATGTGGGAGTTGCTGCAGAGGGAGAAGGACAGGCAGTGTCGTGGCCTGGAGCTCATTGCCTCAGAGAAC TTCTGCAGCCGAGCTGCGCTGGAGGCCCTGGGGTCCTGTCTGAACAACAAGTACTCGGAGGGTTATCCTGGCAAGAGATACTATGGGGGA GCAGAGGTGGTGGATGAAATTGAGCTGCTGTGCCAGCGCCGGGCCTTGGAAGCCTTTGACCTGGATCCTGCACAGTGGGGAGTCAATGTC CAGCCCTACTCCGGGTCCCCAGCCAACCTGGCCGTCTACACAGCCCTTCTGCAACCTCACGACCGGATCATGGGGCTGGACCTGCCCGAT GGGGGCCATCTCACCCACGGCTACATGTCTGACGTCAAGCGGATATCAGCCACGTCCATCTTCTTCGAGTCTATGCCCTATAAGCTCAAC CCCAAAACTGGCCTCATTGACTACAACCAGCTGGCACTGACTGCTCGACTTTTCCGGCCACGGCTCATCATAGCTGGCACCAGCGCCTAT GCTCGCCTCATTGACTACGCCCGCATGAGAGAGGTGTGTGATGAAGTCAAAGCACACCTGCTGGCAGACATGGCCCACATCAGTGGCCTG GTGGCTGCCAAGGTGATTCCCTCGCCTTTCAAGCACGCGGACATCGTCACCACCACTACTCACAAGACTCTTCGAGGGGCCAGGTCAGGG CTCATCTTCTACCGGAAAGGGGTGAAGGCTGTGGACCCCAAGACTGGCCGGGAGATCCCTTACACATTTGAGGACCGAATCAACTTTGCC GTGTTCCCATCCCTGCAGGGGGGCCCCCACAATCATGCCATTGCTGCAGTAGCTGTGGCCCTAAAGCAGGCCTGCACCCCCATGTTCCGG GAGTACTCCCTGCAGGTTCTGAAGAATGCTCGGGCCATGGCAGATGCCCTGCTAGAGCGAGGCTACTCACTGGTATCAGGTGGTACTGAC AACCACCTGGTGCTGGTGGACCTGCGGCCCAAGGGCCTGGATGGAGCTCGGGCTGAGCGGGTGCTAGAGCTTGTATCCATCACTGCCAAC AAGAACACCTGTCCTGGAGACCGAAGTGCCATCACACCGGGCGGCCTGCGGCTTGGGGCCCCAGCCTTAACTTCTCGACAGTTCCGTGAG GATGACTTCCGGAGAGTTGTGGACTTTATAGATGAAGGGGTCAACATTGGCTTAGAGGTGAAGAGCAAGACTGCTCCTCCAGGTCCCGGG GCTCTGCATCCTGAGCTATCCAACACCACTGTACTTTGGGACCCGTGGGCAGTTTCGCTGCAACCTGGAGTGGCACCTGGGGCTCGGAGA AGGAGAAAAGGAGACTTCAAAGCCAGATGGCCCAATGGTTGCAGTTGCTGAGCCTGTCAGGGTGGTGGTCCTAGACTTCAGTGGTGTCAC CTTTGCAGATGCTGCTGGGGCCAGAGAAGTGGTGCAGGTGAGGGAGAGGCTGGCCAGCCGATGTCGAGATGCTAGGATCCGCCTCCTCCT GGCTCAGTGTAATGCCTTGGTGCAGGGGACACTGACCCGGGTAGGACTCCTGGACAGGGTGACTCCAGATCAGCTGTTTGTGAGTGTGCA GGATGCAGCTGCTTATGCCCTGGGGAGCCTGTTAAGGGGCAGTAGCACCAGGAGCGGGAGCCAGGAGGCACTGGGCTGCGGCAAGTGAGG CAGGGGTAAGTGGCTGGAGACCCAGGGAGAGGGGTTTGGGAAAGGGTCTGAGGAAGATCAAGAAGAAAAGGGTGGGGACGGAATGAATAG TAGATTGAAGACCAAATCAGGGAGCCCGGGCTGAGCTATGGGTGAGGAACTGAAGAACTCAGATCCCTAAGTAGGTGGGGTACCCCTGAG AGGACTGCCACATTTCATTAAGGAAGAGGGGTGCCTAATCTATTCCTGACATGTCTCCTCTCTTCTCCAGGAGCTCACTGACCCAAAGAT TTGCACCGTGTGGGTCTGACCTCATCATGTGGAGTGCAGAGGGCCCTGATGACATGTGTGTGATGAGGACCATGACCCTTGAACCCCCTT >81728_81728_3_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000414700_SLC26A10_chr12_58018649_ENST00000379218_length(amino acids)=522AA_BP=1 MLASGRGPGGSEGWKEVERPLQRCGQLVRMAIRAQHSNAAQTQTGEANRGWTGQESLSDSDPEMWELLQREKDRQCRGLELIASENFCSR AALEALGSCLNNKYSEGYPGKRYYGGAEVVDEIELLCQRRALEAFDLDPAQWGVNVQPYSGSPANLAVYTALLQPHDRIMGLDLPDGGHL THGYMSDVKRISATSIFFESMPYKLNPKTGLIDYNQLALTARLFRPRLIIAGTSAYARLIDYARMREVCDEVKAHLLADMAHISGLVAAK VIPSPFKHADIVTTTTHKTLRGARSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTPMFREYSL QVLKNARAMADALLERGYSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSITANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFR -------------------------------------------------------------- >81728_81728_4_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000449049_SLC26A10_chr12_58018649_ENST00000379218_length(transcript)=2525nt_BP=1655nt ATTGCTCTACAATCTGTGGCCTCAGCCCTTGCCAACTCTGCCTCTTCCCCCGCTGACTGGGGTATCAGTTTATGGTGCCCTGCAGGAAGA CCCTCTTGTTCCTGTGGGTGGGAAGGTTCTGGAATCTCAGTTGCCCTCTCTGGAGTTGGGGAGACATAGGCTTTTGGAGGACAAGCTTTG CAGAGTGGTTCATGGAGTGAGGTGGTGGGAGATTTCTGAGGTGCGGTGCGGTGAATGGAGCAAATCAGAACCACCCCTTGGACGTTGATC TCAAGCTTGGCTCCAGAACTGGCTTTTGAAGCCTCTGCAGAGATGTGGGCAGCTGGTCAGGATGGCCATTCGGGCTCAGCACAGCAACGC AGCCCAGACTCAGACTGGGGAAGCAAACAGGGGCTGGACAGGCCAGGAGAGCCTGTCGGACAGTGATCCTGAGATGTGGGAGTTGCTGCA GAGGGAGAAGGACAGGCAGTGTCGTGGCCTGGAGCTCATTGCCTCAGAGAACTTCTGCAGCCGAGCTGCGCTGGAGGCCCTGGGGTCCTG TCTGAACAACAAGTACTCGGAGGGTTATCCTGGCAAGAGATACTATGGGGGAGCAGAGGTGGTGGATGAAATTGAGCTGCTGTGCCAGCG CCGGGCCTTGGAAGCCTTTGACCTGGATCCTGCACAGTGGGGAGTCAATGTCCAGCCCTACTCCGGGTCCCCAGCCAACCTGGCCGTCTA CACAGCCCTTCTGCAACCTCACGACCGGATCATGGGGCTGGACCTGCCCGATGGGGGCCATCTCACCCACGGCTACATGTCTGACGTCAA GCGGATATCAGCCACGTCCATCTTCTTCGAGTCTATGCCCTATAAGCTCAACCCCAAAACTGGCCTCATTGACTACAACCAGCTGGCACT GACTGCTCGACTTTTCCGGCCACGGCTCATCATAGCTGGCACCAGCGCCTATGCTCGCCTCATTGACTACGCCCGCATGAGAGAGGTGTG TGATGAAGTCAAAGCACACCTGCTGGCAGACATGGCCCACATCAGTGGCCTGGTGGCTGCCAAGGTGATTCCCTCGCCTTTCAAGCACGC GGACATCGTCACCACCACTACTCACAAGACTCTTCGAGGGGCCAGGTCAGGGCTCATCTTCTACCGGAAAGGGGTGAAGGCTGTGGACCC CAAGACTGGCCGGGAGATCCCTTACACATTTGAGGACCGAATCAACTTTGCCGTGTTCCCATCCCTGCAGGGGGGCCCCCACAATCATGC CATTGCTGCAGTAGCTGTGGCCCTAAAGCAGGCCTGCACCCCCATGTTCCGGGAGTACTCCCTGCAGGTTCTGAAGAATGCTCGGGCCAT GGCAGATGCCCTGCTAGAGCGAGGCTACTCACTGGTATCAGGTGGTACTGACAACCACCTGGTGCTGGTGGACCTGCGGCCCAAGGGCCT GGATGGAGCTCGGGCTGAGCGGGTGCTAGAGCTTGTATCCATCACTGCCAACAAGAACACCTGTCCTGGAGACCGAAGTGCCATCACACC GGGCGGCCTGCGGCTTGGGGCCCCAGCCTTAACTTCTCGACAGTTCCGTGAGGATGACTTCCGGAGAGTTGTGGACTTTATAGATGAAGG GGTCAACATTGGCTTAGAGGTGAAGAGCAAGACTGCTCCTCCAGGTCCCGGGGCTCTGCATCCTGAGCTATCCAACACCACTGTACTTTG GGACCCGTGGGCAGTTTCGCTGCAACCTGGAGTGGCACCTGGGGCTCGGAGAAGGAGAAAAGGAGACTTCAAAGCCAGATGGCCCAATGG TTGCAGTTGCTGAGCCTGTCAGGGTGGTGGTCCTAGACTTCAGTGGTGTCACCTTTGCAGATGCTGCTGGGGCCAGAGAAGTGGTGCAGG TGAGGGAGAGGCTGGCCAGCCGATGTCGAGATGCTAGGATCCGCCTCCTCCTGGCTCAGTGTAATGCCTTGGTGCAGGGGACACTGACCC GGGTAGGACTCCTGGACAGGGTGACTCCAGATCAGCTGTTTGTGAGTGTGCAGGATGCAGCTGCTTATGCCCTGGGGAGCCTGTTAAGGG GCAGTAGCACCAGGAGCGGGAGCCAGGAGGCACTGGGCTGCGGCAAGTGAGGCAGGGGTAAGTGGCTGGAGACCCAGGGAGAGGGGTTTG GGAAAGGGTCTGAGGAAGATCAAGAAGAAAAGGGTGGGGACGGAATGAATAGTAGATTGAAGACCAAATCAGGGAGCCCGGGCTGAGCTA TGGGTGAGGAACTGAAGAACTCAGATCCCTAAGTAGGTGGGGTACCCCTGAGAGGACTGCCACATTTCATTAAGGAAGAGGGGTGCCTAA TCTATTCCTGACATGTCTCCTCTCTTCTCCAGGAGCTCACTGACCCAAAGATTTGCACCGTGTGGGTCTGACCTCATCATGTGGAGTGCA GAGGGCCCTGATGACATGTGTGTGATGAGGACCATGACCCTTGAACCCCCTTACCTAACGTAACTAATAAAATGAAGCTGAGAGCTTTGG >81728_81728_4_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000449049_SLC26A10_chr12_58018649_ENST00000379218_length(amino acids)=505AA_BP=2 MKPLQRCGQLVRMAIRAQHSNAAQTQTGEANRGWTGQESLSDSDPEMWELLQREKDRQCRGLELIASENFCSRAALEALGSCLNNKYSEG YPGKRYYGGAEVVDEIELLCQRRALEAFDLDPAQWGVNVQPYSGSPANLAVYTALLQPHDRIMGLDLPDGGHLTHGYMSDVKRISATSIF FESMPYKLNPKTGLIDYNQLALTARLFRPRLIIAGTSAYARLIDYARMREVCDEVKAHLLADMAHISGLVAAKVIPSPFKHADIVTTTTH KTLRGARSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTPMFREYSLQVLKNARAMADALLERG YSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSITANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDEGVNIGLEVK -------------------------------------------------------------- >81728_81728_5_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000553474_SLC26A10_chr12_58018649_ENST00000379218_length(transcript)=2317nt_BP=1447nt ACGGACTGCTAAAGGTCTCCCCTCCCACCTGCATTGCTCTACAATCTGTGGCCTCAGCCCTTGCCAACTCTGCCTCTTCCCCCGCTGACT GGGCCTCTGCAGAGATGTGGGCAGCTGGTCAGGATGGCCATTCGGGCTCAGCACAGCAACGCAGCCCAGACTCAGACTGGGGAAGCAAAC AGGGGCTGGACAGGCCAGGAGAGCCTGTCGGACAGTGATCCTGAGATGTGGGAGTTGCTGCAGAGGGAGAAGGACAGGCAGTGTCGTGGC CTGGAGCTCATTGCCTCAGAGAACTTCTGCAGCCGAGCTGCGCTGGAGGCCCTGGGGTCCTGTCTGAACAACAAGTACTCGGAGGGTTAT CCTGGCAAGAGATACTATGGGGGAGCAGAGGTGGTGGATGAAATTGAGCTGCTGTGCCAGCGCCGGGCCTTGGAAGCCTTTGACCTGGAT CCTGCACAGTGGGGAGTCAATGTCCAGCCCTACTCCGGGTCCCCAGCCAACCTGGCCGTCTACACAGCCCTTCTGCAACCTCACGACCGG ATCATGGGGCTGGACCTGCCCGATGGGGGCCATCTCACCCACGGCTACATGTCTGACGTCAAGCGGATATCAGCCACGTCCATCTTCTTC GAGTCTATGCCCTATAAGCTCAACCCCAAAACTGGCCTCATTGACTACAACCAGCTGGCACTGACTGCTCGACTTTTCCGGCCACGGCTC ATCATAGCTGGCACCAGCGCCTATGCTCGCCTCATTGACTACGCCCGCATGAGAGAGGTGTGTGATGAAGTCAAAGCACACCTGCTGGCA GACATGGCCCACATCAGTGGCCTGGTGGCTGCCAAGGTGATTCCCTCGCCTTTCAAGCACGCGGACATCGTCACCACCACTACTCACAAG ACTCTTCGAGGGGCCAGGTCAGGGCTCATCTTCTACCGGAAAGGGGTGAAGGCTGTGGACCCCAAGACTGGCCGGGAGATCCCTTACACA TTTGAGGACCGAATCAACTTTGCCGTGTTCCCATCCCTGCAGGGGGGCCCCCACAATCATGCCATTGCTGCAGTAGCTGTGGCCCTAAAG CAGGCCTGCACCCCCATGTTCCGGGAGTACTCCCTGCAGGTTCTGAAGAATGCTCGGGCCATGGCAGATGCCCTGCTAGAGCGAGGCTAC TCACTGGTATCAGGTGGTACTGACAACCACCTGGTGCTGGTGGACCTGCGGCCCAAGGGCCTGGATGGAGCTCGGGCTGAGCGGGTGCTA GAGCTTGTATCCATCACTGCCAACAAGAACACCTGTCCTGGAGACCGAAGTGCCATCACACCGGGCGGCCTGCGGCTTGGGGCCCCAGCC TTAACTTCTCGACAGTTCCGTGAGGATGACTTCCGGAGAGTTGTGGACTTTATAGATGAAGGGGTCAACATTGGCTTAGAGGTGAAGAGC AAGACTGCTCCTCCAGGTCCCGGGGCTCTGCATCCTGAGCTATCCAACACCACTGTACTTTGGGACCCGTGGGCAGTTTCGCTGCAACCT GGAGTGGCACCTGGGGCTCGGAGAAGGAGAAAAGGAGACTTCAAAGCCAGATGGCCCAATGGTTGCAGTTGCTGAGCCTGTCAGGGTGGT GGTCCTAGACTTCAGTGGTGTCACCTTTGCAGATGCTGCTGGGGCCAGAGAAGTGGTGCAGGTGAGGGAGAGGCTGGCCAGCCGATGTCG AGATGCTAGGATCCGCCTCCTCCTGGCTCAGTGTAATGCCTTGGTGCAGGGGACACTGACCCGGGTAGGACTCCTGGACAGGGTGACTCC AGATCAGCTGTTTGTGAGTGTGCAGGATGCAGCTGCTTATGCCCTGGGGAGCCTGTTAAGGGGCAGTAGCACCAGGAGCGGGAGCCAGGA GGCACTGGGCTGCGGCAAGTGAGGCAGGGGTAAGTGGCTGGAGACCCAGGGAGAGGGGTTTGGGAAAGGGTCTGAGGAAGATCAAGAAGA AAAGGGTGGGGACGGAATGAATAGTAGATTGAAGACCAAATCAGGGAGCCCGGGCTGAGCTATGGGTGAGGAACTGAAGAACTCAGATCC CTAAGTAGGTGGGGTACCCCTGAGAGGACTGCCACATTTCATTAAGGAAGAGGGGTGCCTAATCTATTCCTGACATGTCTCCTCTCTTCT CCAGGAGCTCACTGACCCAAAGATTTGCACCGTGTGGGTCTGACCTCATCATGTGGAGTGCAGAGGGCCCTGATGACATGTGTGTGATGA >81728_81728_5_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000553474_SLC26A10_chr12_58018649_ENST00000379218_length(amino acids)=523AA_BP=1 MLYNLWPQPLPTLPLPPLTGPLQRCGQLVRMAIRAQHSNAAQTQTGEANRGWTGQESLSDSDPEMWELLQREKDRQCRGLELIASENFCS RAALEALGSCLNNKYSEGYPGKRYYGGAEVVDEIELLCQRRALEAFDLDPAQWGVNVQPYSGSPANLAVYTALLQPHDRIMGLDLPDGGH LTHGYMSDVKRISATSIFFESMPYKLNPKTGLIDYNQLALTARLFRPRLIIAGTSAYARLIDYARMREVCDEVKAHLLADMAHISGLVAA KVIPSPFKHADIVTTTTHKTLRGARSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTPMFREYS LQVLKNARAMADALLERGYSLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSITANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDF -------------------------------------------------------------- >81728_81728_6_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000557487_SLC26A10_chr12_58018649_ENST00000379218_length(transcript)=2295nt_BP=1425nt CTTCTTCCGACAGCTTGCTGCCCTAGACCAGAGTTGGTGGCTGGACCTCCTGCGACTTCCGAGTTGCGATGCTGTACTTCTCTTTGTTTT GGGCGGCTCGGCCTCTGCAGAGATGTGGGCAGCTGGTCAGGATGGCCATTCGGGCTCAGCACAGCAACGCAGCCCAGACTCAGACTGGGG AAGCAAACAGGGGCTGGACAGGCCAGGAGAGCCTGTCGGACAGTGATCCTGAGATGTGGGAGTTGCTGCAGAGGGAGAAGGACAGGCAGT GTCGTGGCCTGGAGCTCATTGCCTCAGAGAACTTCTGCAGCCGAGCTGCGCTGGAGGCCCTGGGGTCCTGTCTGAACAACAAGTACTCGG AGGGTTATCCTGGCAAGAGATACTATGGGGGAGCAGAGGTGGTGGATGAAATTGAGCTGCTGTGCCAGCGCCGGGCCTTGGAAGCCTTTG ACCTGGATCCTGCACAGTGGGGAGTCAATGTCCAGCCCTACTCCGGGTCCCCAGCCAACCTGGCCGTCTACACAGCCCTTCTGCAACCTC ACGACCGGATCATGGGGCTGGACCTGCCCGATGGGGGCCATCTCACCCACGGCTACATGTCTGACGTCAAGCGGATATCAGCCACGTCCA TCTTCTTCGAGTCTATGCCCTATAAGCTCAACCTGGCACTGACTGCTCGACTTTTCCGGCCACGGCTCATCATAGCTGGCACCAGCGCCT ATGCTCGCCTCATTGACTACGCCCGCATGAGAGAGGTGTGTGATGAAGTCAAAGCACACCTGCTGGCAGACATGGCCCACATCAGTGGCC TGGTGGCTGCCAAGGTGATTCCCTCGCCTTTCAAGCACGCGGACATCGTCACCACCACTACTCACAAGACTCTTCGAGGGGCCAGGTCAG GGCTCATCTTCTACCGGAAAGGGGTGAAGGCTGTGGACCCCAAGACTGGCCGGGAGATCCCTTACACATTTGAGGACCGAATCAACTTTG CCGTGTTCCCATCCCTGCAGGGGGGCCCCCACAATCATGCCATTGCTGCAGTAGCTGTGGCCCTAAAGCAGGCCTGCACCCCCATGTTCC GGGAGTACTCCCTGCAGGTTCTGAAGAATGCTCGGGCCATGGCAGATGCCCTGCTAGAGCGAGGCTACTCACTGGTATCAGGTGGTACTG ACAACCACCTGGTGCTGGTGGACCTGCGGCCCAAGGGCCTGGATGGAGCTCGGGCTGAGCGGGTGCTAGAGCTTGTATCCATCACTGCCA ACAAGAACACCTGTCCTGGAGACCGAAGTGCCATCACACCGGGCGGCCTGCGGCTTGGGGCCCCAGCCTTAACTTCTCGACAGTTCCGTG AGGATGACTTCCGGAGAGTTGTGGACTTTATAGATGAAGGGGTCAACATTGGCTTAGAGGTGAAGAGCAAGACTGCTCCTCCAGGTCCCG GGGCTCTGCATCCTGAGCTATCCAACACCACTGTACTTTGGGACCCGTGGGCAGTTTCGCTGCAACCTGGAGTGGCACCTGGGGCTCGGA GAAGGAGAAAAGGAGACTTCAAAGCCAGATGGCCCAATGGTTGCAGTTGCTGAGCCTGTCAGGGTGGTGGTCCTAGACTTCAGTGGTGTC ACCTTTGCAGATGCTGCTGGGGCCAGAGAAGTGGTGCAGGTGAGGGAGAGGCTGGCCAGCCGATGTCGAGATGCTAGGATCCGCCTCCTC CTGGCTCAGTGTAATGCCTTGGTGCAGGGGACACTGACCCGGGTAGGACTCCTGGACAGGGTGACTCCAGATCAGCTGTTTGTGAGTGTG CAGGATGCAGCTGCTTATGCCCTGGGGAGCCTGTTAAGGGGCAGTAGCACCAGGAGCGGGAGCCAGGAGGCACTGGGCTGCGGCAAGTGA GGCAGGGGTAAGTGGCTGGAGACCCAGGGAGAGGGGTTTGGGAAAGGGTCTGAGGAAGATCAAGAAGAAAAGGGTGGGGACGGAATGAAT AGTAGATTGAAGACCAAATCAGGGAGCCCGGGCTGAGCTATGGGTGAGGAACTGAAGAACTCAGATCCCTAAGTAGGTGGGGTACCCCTG AGAGGACTGCCACATTTCATTAAGGAAGAGGGGTGCCTAATCTATTCCTGACATGTCTCCTCTCTTCTCCAGGAGCTCACTGACCCAAAG ATTTGCACCGTGTGGGTCTGACCTCATCATGTGGAGTGCAGAGGGCCCTGATGACATGTGTGTGATGAGGACCATGACCCTTGAACCCCC >81728_81728_6_SHMT2-SLC26A10_SHMT2_chr12_57627893_ENST00000557487_SLC26A10_chr12_58018649_ENST00000379218_length(amino acids)=504AA_BP=0 MLYFSLFWAARPLQRCGQLVRMAIRAQHSNAAQTQTGEANRGWTGQESLSDSDPEMWELLQREKDRQCRGLELIASENFCSRAALEALGS CLNNKYSEGYPGKRYYGGAEVVDEIELLCQRRALEAFDLDPAQWGVNVQPYSGSPANLAVYTALLQPHDRIMGLDLPDGGHLTHGYMSDV KRISATSIFFESMPYKLNLALTARLFRPRLIIAGTSAYARLIDYARMREVCDEVKAHLLADMAHISGLVAAKVIPSPFKHADIVTTTTHK TLRGARSGLIFYRKGVKAVDPKTGREIPYTFEDRINFAVFPSLQGGPHNHAIAAVAVALKQACTPMFREYSLQVLKNARAMADALLERGY SLVSGGTDNHLVLVDLRPKGLDGARAERVLELVSITANKNTCPGDRSAITPGGLRLGAPALTSRQFREDDFRRVVDFIDEGVNIGLEVKS -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SHMT2-SLC26A10 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SHMT2-SLC26A10 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SHMT2-SLC26A10 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
