
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SIDT2-CYP2C8 (FusionGDB2 ID:81829) |
Fusion Gene Summary for SIDT2-CYP2C8 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SIDT2-CYP2C8 | Fusion gene ID: 81829 | Hgene | Tgene | Gene symbol | SIDT2 | CYP2C8 | Gene ID | 51092 | 1558 |
| Gene name | SID1 transmembrane family member 2 | cytochrome P450 family 2 subfamily C member 8 | |
| Synonyms | CGI-40 | CPC8|CYP2C8DM|CYPIIC8|MP-12/MP-20 | |
| Cytomap | 11q23.3 | 10q23.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | SID1 transmembrane family member 2 | cytochrome P450 2C8P450 form 1cytochrome P450 IIC2cytochrome P450 MP-12cytochrome P450 MP-20cytochrome P450 form 1cytochrome P450, family 2, subfamily C, polypeptide 8cytochrome P450, subfamily IIC (mephenytoin 4-hydroxylase), polypeptide 8flavopr | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | P10632 | |
| Ensembl transtripts involved in fusion gene | ENST00000324225, ENST00000431081, ENST00000532062, ENST00000530948, | ENST00000539050, ENST00000535898, ENST00000371270, | |
| Fusion gene scores | * DoF score | 4 X 4 X 2=32 | 6 X 2 X 5=60 |
| # samples | 4 | 6 | |
| ** MAII score | log2(4/32*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(6/60*10)=0 | |
| Context | PubMed: SIDT2 [Title/Abstract] AND CYP2C8 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SIDT2(117064679)-CYP2C8(96827448), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | SIDT2 | GO:0050658 | RNA transport | 28277980 |
| Tgene | CYP2C8 | GO:0002933 | lipid hydroxylation | 14559847 |
| Tgene | CYP2C8 | GO:0006082 | organic acid metabolic process | 19651758 |
| Tgene | CYP2C8 | GO:0008202 | steroid metabolic process | 14559847 |
| Tgene | CYP2C8 | GO:0008210 | estrogen metabolic process | 12865317|14559847 |
| Tgene | CYP2C8 | GO:0017144 | drug metabolic process | 19651758 |
| Tgene | CYP2C8 | GO:0019373 | epoxygenase P450 pathway | 7574697 |
| Tgene | CYP2C8 | GO:0042573 | retinoic acid metabolic process | 11093772 |
| Tgene | CYP2C8 | GO:0042738 | exogenous drug catabolic process | 18619574 |
| Tgene | CYP2C8 | GO:0046456 | icosanoid biosynthetic process | 15766564 |
| Tgene | CYP2C8 | GO:0055114 | oxidation-reduction process | 19651758 |
| Tgene | CYP2C8 | GO:0070989 | oxidative demethylation | 18619574 |
 Fusion gene breakpoints across SIDT2 (5'-gene) Fusion gene breakpoints across SIDT2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
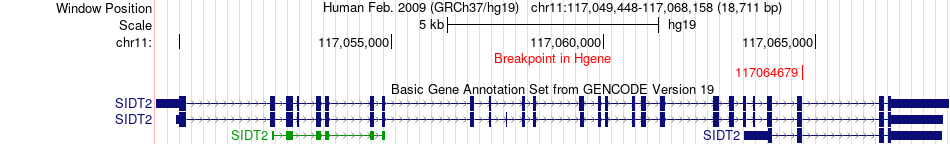 |
 Fusion gene breakpoints across CYP2C8 (3'-gene) Fusion gene breakpoints across CYP2C8 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
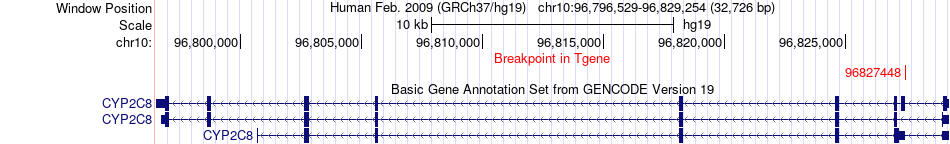 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | THCA | TCGA-EL-A3CP | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
Top |
Fusion Gene ORF analysis for SIDT2-CYP2C8 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000324225 | ENST00000539050 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| 5CDS-5UTR | ENST00000431081 | ENST00000539050 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| 5CDS-5UTR | ENST00000532062 | ENST00000539050 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| 5CDS-intron | ENST00000324225 | ENST00000535898 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| 5CDS-intron | ENST00000431081 | ENST00000535898 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| 5CDS-intron | ENST00000532062 | ENST00000535898 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| In-frame | ENST00000324225 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| In-frame | ENST00000431081 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| In-frame | ENST00000532062 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| intron-3CDS | ENST00000530948 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| intron-5UTR | ENST00000530948 | ENST00000539050 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
| intron-intron | ENST00000530948 | ENST00000535898 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000324225 | SIDT2 | chr11 | 117064679 | + | ENST00000371270 | CYP2C8 | chr10 | 96827448 | - | 4513 | 2853 | 531 | 4157 | 1208 |
| ENST00000431081 | SIDT2 | chr11 | 117064679 | + | ENST00000371270 | CYP2C8 | chr10 | 96827448 | - | 4043 | 2383 | 70 | 3687 | 1205 |
| ENST00000532062 | SIDT2 | chr11 | 117064679 | + | ENST00000371270 | CYP2C8 | chr10 | 96827448 | - | 2413 | 753 | 555 | 2057 | 500 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000324225 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - | 0.00081154 | 0.9991885 |
| ENST00000431081 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - | 0.000265441 | 0.9997346 |
| ENST00000532062 | ENST00000371270 | SIDT2 | chr11 | 117064679 | + | CYP2C8 | chr10 | 96827448 | - | 0.001072549 | 0.9989274 |
Top |
Fusion Genomic Features for SIDT2-CYP2C8 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
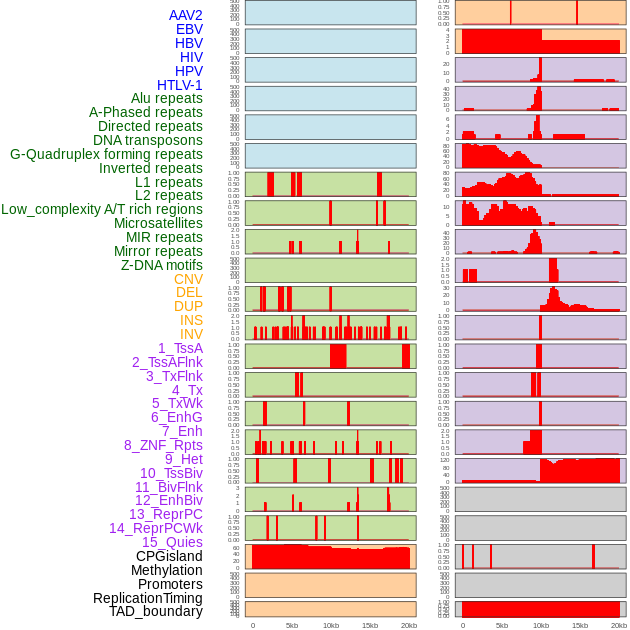 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SIDT2-CYP2C8 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:117064679/chr10:96827448) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CYP2C8 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: A cytochrome P450 monooxygenase involved in the metabolism of various endogenous substrates, including fatty acids, steroid hormones and vitamins (PubMed:7574697, PubMed:11093772, PubMed:14559847, PubMed:15766564, PubMed:19965576). Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase) (PubMed:7574697, PubMed:11093772, PubMed:14559847, PubMed:15766564, PubMed:19965576). Primarily catalyzes the epoxidation of double bonds of polyunsaturated fatty acids (PUFA) with a preference for the last double bond (PubMed:7574697, PubMed:15766564, PubMed:19965576). Catalyzes the hydroxylation of carbon-hydrogen bonds. Metabolizes all trans-retinoic acid toward its 4-hydroxylated form (PubMed:11093772). Displays 16-alpha hydroxylase activity toward estrogen steroid hormones, 17beta-estradiol (E2) and estrone (E1) (PubMed:14559847). Plays a role in the oxidative metabolism of xenobiotics. It is the principal enzyme responsible for the metabolism of the anti-cancer drug paclitaxel (taxol) (PubMed:26427316). {ECO:0000269|PubMed:11093772, ECO:0000269|PubMed:14559847, ECO:0000269|PubMed:15766564, ECO:0000269|PubMed:19965576, ECO:0000269|PubMed:26427316, ECO:0000269|PubMed:7574697}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 764_767 | 774 | 833.0 | Compositional bias | Note=Poly-Phe |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 19_296 | 774 | 833.0 | Topological domain | Extracellular |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 318_447 | 774 | 833.0 | Topological domain | Cytoplasmic |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 469_499 | 774 | 833.0 | Topological domain | Extracellular |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 521_546 | 774 | 833.0 | Topological domain | Cytoplasmic |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 568_605 | 774 | 833.0 | Topological domain | Extracellular |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 627_631 | 774 | 833.0 | Topological domain | Cytoplasmic |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 653_688 | 774 | 833.0 | Topological domain | Extracellular |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 710_715 | 774 | 833.0 | Topological domain | Cytoplasmic |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 737_746 | 774 | 833.0 | Topological domain | Extracellular |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 297_317 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 448_468 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 500_520 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 547_567 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 606_626 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 632_652 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 689_709 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 716_736 | 774 | 833.0 | Transmembrane | Helical |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 747_767 | 774 | 833.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 768_796 | 774 | 833.0 | Topological domain | Cytoplasmic |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 818_832 | 774 | 833.0 | Topological domain | Extracellular |
| Hgene | SIDT2 | chr11:117064679 | chr10:96827448 | ENST00000324225 | + | 24 | 26 | 797_817 | 774 | 833.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for SIDT2-CYP2C8 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >81829_81829_1_SIDT2-CYP2C8_SIDT2_chr11_117064679_ENST00000324225_CYP2C8_chr10_96827448_ENST00000371270_length(transcript)=4513nt_BP=2853nt CTCACTTTTCAGTGTTCGTGCGTCGGGACGGCGCGTCCGCCCGCCCTTAAAGGGCCCGCACCTCGGAAAACTCGCAGCCCAGCACGGCGT CGGGTAGCTACCACCTATCACGCCCCTCACTCTGCGACTCGCCTTCCTCCGCGGCCAGCCCCCGCCGCCGGCTCTTCCTCCCTCCCCTTT CCCGGCGTCTCCAGGCTCCCGGCAACCAGCCGCCCTCCTTCTCACGGCCCTGGCCCGGGGCTCCAGGGTCTCAGGTCGTACTCAGCCCCT CGCGAGCTGGGACCCCTTCCCGTAGCCAGCATCCCCTCCCTCCCCGCCCCCTCCCGGCCGCCCCCTCCCCTGCGGGGACCCTGGGAGCCT CTTCGGGGCTCTTGGGAGGGGACATTTCCTAATCTCAGGCCGGGGTTAAAGTTCTGGTCCTGGTGAGATGCTGGAAGCTGCGGCCGCAGC CGCAACCCGTCCCGGAGGTGTCCTGTCTCCTGTCGCCGCCGCCGCCGCCACCACCGCTGCCACTGCCGCCCTGCCGGGGCCATGTTCGCT CTGGGCTTGCCCTTCTTGGTGCTCTTGGTGGCCTCGGTCGAGAGCCATCTGGGGGTTCTGGGGCCCAAGAACGTCTCGCAGAAAGACGCC GAGTTTGAGCGCACCTACGTGGACGAGGTCAACAGCGAGCTGGTCAACATCTACACCTTCAACCATACTGTGACCCGCAACAGGACAGAG GGCGTGCGTGTGTCTGTGAACGTCCTGAACAAGCAGAAGGGGGCGCCGTTGCTGTTTGTGGTCCGCCAGAAGGAGGCTGTGGTGTCCTTC CAGGTGCCCCTAATCCTGCGAGGGATGTTTCAGCGCAAGTACCTCTACCAAAAAGTGGAACGAACCCTGTGTCAGCCCCCCACCAAGAAT GAGTCGGAGATTCAGTTCTTCTACGTGGATGTGTCCACCCTGTCACCAGTCAACACCACATACCAGCTCCGGGTCAGCCGCATGGACGAT TTTGTGCTCAGGACTGGGGAGCAGTTCAGCTTCAATACCACAGCAGCACAGCCCCAGTACTTCAAGTATGAGTTCCCTGAAGGCGTGGAC TCGGTAATTGTCAAGGTGACCTCCAACAAGGCCTTCCCCTGCTCAGTCATCTCCATTCAGGATGTGCTGTGTCCTGTCTATGACCTGGAC AACAACGTAGCCTTCATCGGCATGTACCAGACGATGACCAAGAAGGCGGCCATCACCGTACAGCGCAAAGACTTCCCCAGCAACAGCTTT TATGTGGTGGTGGTGGTGAAGACCGAAGACCAAGCCTGCGGGGGCTCCCTGCCTTTCTACCCCTTCGCAGAAGATGAACCGGTCGATCAA GGGCACCGCCAGAAAACCCTGTCAGTGCTGGTGTCTCAAGCAGTCACGTCTGAGGCATACGTCAGTGGGATGCTCTTTTGCCTGGGTATA TTTCTCTCCTTTTACCTGCTGACCGTCCTCCTGGCCTGCTGGGAGAACTGGAGGCAGAAGAAGAAGACCCTGCTGGTGGCCATTGACCGA GCCTGCCCAGAAAGCGGTCACCCTCGAGTCCTGGCTGATTCTTTTCCTGGCAGTTCCCCTTATGAGGGTTACAACTATGGCTCCTTTGAG AATGTTTCTGGATCTACCGATGGTCTGGTTGACAGCGCTGGCACTGGGGACCTCTCTTACGGTTACCAGGGCCGCTCCTTTGAACCTGTA GGTACTCGGCCCCGAGTGGACTCCATGAGCTCTGTGGAGGAGGATGACTACGACACATTGACCGACATCGATTCCGACAAGAATGTCATT CGCACCAAGCAATACCTCTATGTGGCTGACCTGGCACGGAAGGACAAGCGTGTTCTGCGGAAAAAGTACCAGATCTACTTCTGGAACATT GCCACCATTGCTGTCTTCTATGCCCTTCCTGTGGTGCAGCTGGTGATCACCTACCAGACGGTGGTGAATGTCACAGGGAATCAGGACATC TGCTACTACAACTTCCTCTGCGCCCACCCACTGGGCAATCTCAGCGCCTTCAACAACATCCTCAGCAACCTGGGGTACATCCTGCTGGGG CTGCTTTTCCTGCTCATCATCCTGCAACGGGAGATCAACCACAACCGGGCCCTGCTGCGCAATGACCTCTGTGCCCTGGAATGTGGGATC CCCAAACACTTTGGGCTTTTCTACGCCATGGGCACAGCCCTGATGATGGAGGGGCTGCTCAGTGCTTGCTATCATGTGTGCCCCAACTAT ACCAATTTCCAGTTTGACACATCGTTCATGTACATGATCGCCGGACTCTGCATGCTGAAGCTCTACCAGAAGCGGCACCCGGACATCAAC GCCAGCGCCTACAGTGCCTACGCCTGCCTGGCCATTGTCATCTTCTTCTCTGTGCTGGGCGTGGTCTTTGGCAAAGGGAACACGGCGTTC TGGATCGTCTTCTCCATCATTCACATCATCGCCACCCTGCTCCTCAGCACGCAGCTCTATTACATGGGCCGGTGGAAACTGGACTCGGGG ATCTTCCGCCGCATCCTCCACGTGCTCTACACAGACTGCATCCGGCAGTGCAGCGGGCCGCTCTACGTGGACCGCATGGTGCTGCTGGTC ATGGGCAACGTCATCAACTGGTCGCTGGCTGCCTATGGGCTTATCATGCGCCCCAATGATTTCGCTTCCTACTTGTTGGCCATTGGCATC TGCAACCTGCTCCTTTACTTCGCCTTCTACATCATCATGAAGCTCCGGAGTGGGGAGAGGATCAAGCTCATCCCCCTGCTCTGCATCGTT TGCACCTCCGTGGTCTGGGGCTTCGCGCTCTTCTTCTTCTTCCAGGGACTCAGCACCTGGCAGTTCTCAAAAGTCTATGGTCCTGTGTTC ACCGTGTATTTTGGCATGAATCCCATAGTGGTGTTTCATGGATATGAGGCAGTGAAGGAAGCCCTGATTGATAATGGAGAGGAGTTTTCT GGAAGAGGCAATTCCCCAATATCTCAAAGAATTACTAAAGGACTTGGAATCATTTCCAGCAATGGAAAGAGATGGAAGGAGATCCGGCGT TTCTCCCTCACAACCTTGCGGAATTTTGGGATGGGGAAGAGGAGCATTGAGGACCGTGTTCAAGAGGAAGCTCACTGCCTTGTGGAGGAG TTGAGAAAAACCAAGGCTTCACCCTGTGATCCCACTTTCATCCTGGGCTGTGCTCCCTGCAATGTGATCTGCTCCGTTGTTTTCCAGAAA CGATTTGATTATAAAGATCAGAATTTTCTCACCCTGATGAAAAGATTCAATGAAAACTTCAGGATTCTGAACTCCCCATGGATCCAGGTC TGCAATAATTTCCCTCTACTCATTGATTGTTTCCCAGGAACTCACAACAAAGTGCTTAAAAATGTTGCTCTTACACGAAGTTACATTAGG GAGAAAGTAAAAGAACACCAAGCATCACTGGATGTTAACAATCCTCGGGACTTTATCGATTGCTTCCTGATCAAAATGGAGCAGGAAAAG GACAACCAAAAGTCAGAATTCAATATTGAAAACTTGGTTGGCACTGTAGCTGATCTATTTGTTGCTGGAACAGAGACAACAAGCACCACT CTGAGATATGGACTCCTGCTCCTGCTGAAGCACCCAGAGGTCACAGCTAAAGTCCAGGAAGAGATTGATCATGTAATTGGCAGACACAGG AGCCCCTGCATGCAGGATAGGAGCCACATGCCTTACACTGATGCTGTAGTGCACGAGATCCAGAGATACAGTGACCTTGTCCCCACCGGT GTGCCCCATGCAGTGACCACTGATACTAAGTTCAGAAACTACCTCATCCCCAAGGGCACAACCATAATGGCATTACTGACTTCCGTGCTA CATGATGACAAAGAATTTCCTAATCCAAATATCTTTGACCCTGGCCACTTTCTAGATAAGAATGGCAACTTTAAGAAAAGTGACTACTTC ATGCCTTTCTCAGCAGGAAAACGAATTTGTGCAGGAGAAGGACTTGCCCGCATGGAGCTATTTTTATTTCTAACCACAATTTTACAGAAC TTTAACCTGAAATCTGTTGATGATTTAAAGAACCTCAATACTACTGCAGTTACCAAAGGGATTGTTTCTCTGCCACCCTCATACCAGATC TGCTTCATCCCTGTCTGAAGAATGCTAGCCCATCTGGCTGCCGATCTGCTATCACCTGCAACTCTTTTTTTATCAAGGACATTCCCACTA TTATGTCTTCTCTGACCTCTCATCAAATCTTCCCATTCACTCAATATCCCATAAGCATCCAAACTCCATTAAGGAGAGTTGTTCAGGTCA CTGCACAAATATATCTGCAATTATTCATACTCTGTAACACTTGTATTAATTGCTGCATATGCTAATACTTTTCTAATGCTGACTTTTTAA TATGTTATCACTGTAAAACACAGAAAAGTGATTAATGAATGATAATTTAGATCCATTTCTTTTGTGAATGTGCTAAATAAAAAGTGTTAT >81829_81829_1_SIDT2-CYP2C8_SIDT2_chr11_117064679_ENST00000324225_CYP2C8_chr10_96827448_ENST00000371270_length(amino acids)=1208AA_BP=762 MFALGLPFLVLLVASVESHLGVLGPKNVSQKDAEFERTYVDEVNSELVNIYTFNHTVTRNRTEGVRVSVNVLNKQKGAPLLFVVRQKEAV VSFQVPLILRGMFQRKYLYQKVERTLCQPPTKNESEIQFFYVDVSTLSPVNTTYQLRVSRMDDFVLRTGEQFSFNTTAAQPQYFKYEFPE GVDSVIVKVTSNKAFPCSVISIQDVLCPVYDLDNNVAFIGMYQTMTKKAAITVQRKDFPSNSFYVVVVVKTEDQACGGSLPFYPFAEDEP VDQGHRQKTLSVLVSQAVTSEAYVSGMLFCLGIFLSFYLLTVLLACWENWRQKKKTLLVAIDRACPESGHPRVLADSFPGSSPYEGYNYG SFENVSGSTDGLVDSAGTGDLSYGYQGRSFEPVGTRPRVDSMSSVEEDDYDTLTDIDSDKNVIRTKQYLYVADLARKDKRVLRKKYQIYF WNIATIAVFYALPVVQLVITYQTVVNVTGNQDICYYNFLCAHPLGNLSAFNNILSNLGYILLGLLFLLIILQREINHNRALLRNDLCALE CGIPKHFGLFYAMGTALMMEGLLSACYHVCPNYTNFQFDTSFMYMIAGLCMLKLYQKRHPDINASAYSAYACLAIVIFFSVLGVVFGKGN TAFWIVFSIIHIIATLLLSTQLYYMGRWKLDSGIFRRILHVLYTDCIRQCSGPLYVDRMVLLVMGNVINWSLAAYGLIMRPNDFASYLLA IGICNLLLYFAFYIIMKLRSGERIKLIPLLCIVCTSVVWGFALFFFFQGLSTWQFSKVYGPVFTVYFGMNPIVVFHGYEAVKEALIDNGE EFSGRGNSPISQRITKGLGIISSNGKRWKEIRRFSLTTLRNFGMGKRSIEDRVQEEAHCLVEELRKTKASPCDPTFILGCAPCNVICSVV FQKRFDYKDQNFLTLMKRFNENFRILNSPWIQVCNNFPLLIDCFPGTHNKVLKNVALTRSYIREKVKEHQASLDVNNPRDFIDCFLIKME QEKDNQKSEFNIENLVGTVADLFVAGTETTSTTLRYGLLLLLKHPEVTAKVQEEIDHVIGRHRSPCMQDRSHMPYTDAVVHEIQRYSDLV PTGVPHAVTTDTKFRNYLIPKGTTIMALLTSVLHDDKEFPNPNIFDPGHFLDKNGNFKKSDYFMPFSAGKRICAGEGLARMELFLFLTTI -------------------------------------------------------------- >81829_81829_2_SIDT2-CYP2C8_SIDT2_chr11_117064679_ENST00000431081_CYP2C8_chr10_96827448_ENST00000371270_length(transcript)=4043nt_BP=2383nt CCGGAGGTGTCCTGTCTCCTGTCGCCGCCGCCGCCGCCACCACCGCTGCCACTGCCGCCCTGCCGGGGCCATGTTCGCTCTGGGCTTGCC CTTCTTGGTGCTCTTGGTGGCCTCGGTCGAGAGCCATCTGGGGGTTCTGGGGCCCAAGAACGTCTCGCAGAAAGACGCCGAGTTTGAGCG CACCTACGTGGACGAGGTCAACAGCGAGCTGGTCAACATCTACACCTTCAACCATACTGTGACCCGCAACAGGACAGAGGGCGTGCGTGT GTCTGTGAACGTCCTGAACAAGCAGAAGGGGGCGCCGTTGCTGTTTGTGGTCCGCCAGAAGGAGGCTGTGGTGTCCTTCCAGGTGCCCCT AATCCTGCGAGGGATGTTTCAGCGCAAGTACCTCTACCAAAAAGTGGAACGAACCCTGTGTCAGCCCCCCACCAAGAATGAGTCGGAGAT TCAGTTCTTCTACGTGGATGTGTCCACCCTGTCACCAGTCAACACCACATACCAGCTCCGGGTCAGCCGCATGGACGATTTTGTGCTCAG GACTGGGGAGCAGTTCAGCTTCAATACCACAGCAGCACAGCCCCAGTACTTCAAGTATGAGTTCCCTGAAGGCGTGGACTCGGTAATTGT CAAGGTGACCTCCAACAAGGCCTTCCCCTGCTCAGTCATCTCCATTCAGGATGTGCTGTGTCCTGTCTATGACCTGGACAACAACGTAGC CTTCATCGGCATGTACCAGACGATGACCAAGAAGGCGGCCATCACCGTACAGCGCAAAGACTTCCCCAGCAACAGCTTTTATGTGGTGGT GGTGGTGAAGACCGAAGACCAAGCCTGCGGGGGCTCCCTGCCTTTCTACCCCTTCGCAGAAGATGAACCGGTCGATCAAGGGCACCGCCA GAAAACCCTGTCAGTGCTGGTGTCTCAAGCAGTCACGTCTGAGGCATACGTCAGTGGGATGCTCTTTTGCCTGGGTATATTTCTCTCCTT TTACCTGCTGACCGTCCTCCTGGCCTGCTGGGAGAACTGGAGGCAGAAGAAGAAGACCCTGCTGGTGGCCATTGACCGAGCCTGCCCAGA AAGCGCTTCTCTCCTTGGTCACCCTCGAGTCCTGGCTGATTCTTTTCCTGGCAGTTCCCCTTATGAGGGTTACAACTATGGCTCCTTTGA GAATGTTTCTGGATCTACCGATGGTCTGGTTGACAGCGCTGGCACTGGGGACCTCTCTTACGGTTACCAGGGTACTCGGCCCCGAGTGGA CTCCATGAGCTCTGTGGAGGAGGATGACTACGACACATTGACCGACATCGATTCCGACAAGAATGTCATTCGCACCAAGCAATACCTCTA TGTGGCTGACCTGGCACGGAAGGACAAGCGTGTTCTGCGGAAAAAGTACCAGATCTACTTCTGGAACATTGCCACCATTGCTGTCTTCTA TGCCCTTCCTGTGGTGCAGCTGGTGATCACCTACCAGACGGTGGTGAATGTCACAGGGAATCAGGACATCTGCTACTACAACTTCCTCTG CGCCCACCCACTGGGCAATCTCAGCGCCTTCAACAACATCCTCAGCAACCTGGGGTACATCCTGCTGGGGCTGCTTTTCCTGCTCATCAT CCTGCAACGGGAGATCAACCACAACCGGGCCCTGCTGCGCAATGACCTCTGTGCCCTGGAATGTGGGATCCCCAAACACTTTGGGCTTTT CTACGCCATGGGCACAGCCCTGATGATGGAGGGGCTGCTCAGTGCTTGCTATCATGTGTGCCCCAACTATACCAATTTCCAGTTTGACAC ATCGTTCATGTACATGATCGCCGGACTCTGCATGCTGAAGCTCTACCAGAAGCGGCACCCGGACATCAACGCCAGCGCCTACAGTGCCTA CGCCTGCCTGGCCATTGTCATCTTCTTCTCTGTGCTGGGCGTGGTCTTTGGCAAAGGGAACACGGCGTTCTGGATCGTCTTCTCCATCAT TCACATCATCGCCACCCTGCTCCTCAGCACGCAGCTCTATTACATGGGCCGGTGGAAACTGGACTCGGGGATCTTCCGCCGCATCCTCCA CGTGCTCTACACAGACTGCATCCGGCAGTGCAGCGGGCCGCTCTACGTGGACCGCATGGTGCTGCTGGTCATGGGCAACGTCATCAACTG GTCGCTGGCTGCCTATGGGCTTATCATGCGCCCCAATGATTTCGCTTCCTACTTGTTGGCCATTGGCATCTGCAACCTGCTCCTTTACTT CGCCTTCTACATCATCATGAAGCTCCGGAGTGGGGAGAGGATCAAGCTCATCCCCCTGCTCTGCATCGTTTGCACCTCCGTGGTCTGGGG CTTCGCGCTCTTCTTCTTCTTCCAGGGACTCAGCACCTGGCAGTTCTCAAAAGTCTATGGTCCTGTGTTCACCGTGTATTTTGGCATGAA TCCCATAGTGGTGTTTCATGGATATGAGGCAGTGAAGGAAGCCCTGATTGATAATGGAGAGGAGTTTTCTGGAAGAGGCAATTCCCCAAT ATCTCAAAGAATTACTAAAGGACTTGGAATCATTTCCAGCAATGGAAAGAGATGGAAGGAGATCCGGCGTTTCTCCCTCACAACCTTGCG GAATTTTGGGATGGGGAAGAGGAGCATTGAGGACCGTGTTCAAGAGGAAGCTCACTGCCTTGTGGAGGAGTTGAGAAAAACCAAGGCTTC ACCCTGTGATCCCACTTTCATCCTGGGCTGTGCTCCCTGCAATGTGATCTGCTCCGTTGTTTTCCAGAAACGATTTGATTATAAAGATCA GAATTTTCTCACCCTGATGAAAAGATTCAATGAAAACTTCAGGATTCTGAACTCCCCATGGATCCAGGTCTGCAATAATTTCCCTCTACT CATTGATTGTTTCCCAGGAACTCACAACAAAGTGCTTAAAAATGTTGCTCTTACACGAAGTTACATTAGGGAGAAAGTAAAAGAACACCA AGCATCACTGGATGTTAACAATCCTCGGGACTTTATCGATTGCTTCCTGATCAAAATGGAGCAGGAAAAGGACAACCAAAAGTCAGAATT CAATATTGAAAACTTGGTTGGCACTGTAGCTGATCTATTTGTTGCTGGAACAGAGACAACAAGCACCACTCTGAGATATGGACTCCTGCT CCTGCTGAAGCACCCAGAGGTCACAGCTAAAGTCCAGGAAGAGATTGATCATGTAATTGGCAGACACAGGAGCCCCTGCATGCAGGATAG GAGCCACATGCCTTACACTGATGCTGTAGTGCACGAGATCCAGAGATACAGTGACCTTGTCCCCACCGGTGTGCCCCATGCAGTGACCAC TGATACTAAGTTCAGAAACTACCTCATCCCCAAGGGCACAACCATAATGGCATTACTGACTTCCGTGCTACATGATGACAAAGAATTTCC TAATCCAAATATCTTTGACCCTGGCCACTTTCTAGATAAGAATGGCAACTTTAAGAAAAGTGACTACTTCATGCCTTTCTCAGCAGGAAA ACGAATTTGTGCAGGAGAAGGACTTGCCCGCATGGAGCTATTTTTATTTCTAACCACAATTTTACAGAACTTTAACCTGAAATCTGTTGA TGATTTAAAGAACCTCAATACTACTGCAGTTACCAAAGGGATTGTTTCTCTGCCACCCTCATACCAGATCTGCTTCATCCCTGTCTGAAG AATGCTAGCCCATCTGGCTGCCGATCTGCTATCACCTGCAACTCTTTTTTTATCAAGGACATTCCCACTATTATGTCTTCTCTGACCTCT CATCAAATCTTCCCATTCACTCAATATCCCATAAGCATCCAAACTCCATTAAGGAGAGTTGTTCAGGTCACTGCACAAATATATCTGCAA TTATTCATACTCTGTAACACTTGTATTAATTGCTGCATATGCTAATACTTTTCTAATGCTGACTTTTTAATATGTTATCACTGTAAAACA >81829_81829_2_SIDT2-CYP2C8_SIDT2_chr11_117064679_ENST00000431081_CYP2C8_chr10_96827448_ENST00000371270_length(amino acids)=1205AA_BP=759 MFALGLPFLVLLVASVESHLGVLGPKNVSQKDAEFERTYVDEVNSELVNIYTFNHTVTRNRTEGVRVSVNVLNKQKGAPLLFVVRQKEAV VSFQVPLILRGMFQRKYLYQKVERTLCQPPTKNESEIQFFYVDVSTLSPVNTTYQLRVSRMDDFVLRTGEQFSFNTTAAQPQYFKYEFPE GVDSVIVKVTSNKAFPCSVISIQDVLCPVYDLDNNVAFIGMYQTMTKKAAITVQRKDFPSNSFYVVVVVKTEDQACGGSLPFYPFAEDEP VDQGHRQKTLSVLVSQAVTSEAYVSGMLFCLGIFLSFYLLTVLLACWENWRQKKKTLLVAIDRACPESASLLGHPRVLADSFPGSSPYEG YNYGSFENVSGSTDGLVDSAGTGDLSYGYQGTRPRVDSMSSVEEDDYDTLTDIDSDKNVIRTKQYLYVADLARKDKRVLRKKYQIYFWNI ATIAVFYALPVVQLVITYQTVVNVTGNQDICYYNFLCAHPLGNLSAFNNILSNLGYILLGLLFLLIILQREINHNRALLRNDLCALECGI PKHFGLFYAMGTALMMEGLLSACYHVCPNYTNFQFDTSFMYMIAGLCMLKLYQKRHPDINASAYSAYACLAIVIFFSVLGVVFGKGNTAF WIVFSIIHIIATLLLSTQLYYMGRWKLDSGIFRRILHVLYTDCIRQCSGPLYVDRMVLLVMGNVINWSLAAYGLIMRPNDFASYLLAIGI CNLLLYFAFYIIMKLRSGERIKLIPLLCIVCTSVVWGFALFFFFQGLSTWQFSKVYGPVFTVYFGMNPIVVFHGYEAVKEALIDNGEEFS GRGNSPISQRITKGLGIISSNGKRWKEIRRFSLTTLRNFGMGKRSIEDRVQEEAHCLVEELRKTKASPCDPTFILGCAPCNVICSVVFQK RFDYKDQNFLTLMKRFNENFRILNSPWIQVCNNFPLLIDCFPGTHNKVLKNVALTRSYIREKVKEHQASLDVNNPRDFIDCFLIKMEQEK DNQKSEFNIENLVGTVADLFVAGTETTSTTLRYGLLLLLKHPEVTAKVQEEIDHVIGRHRSPCMQDRSHMPYTDAVVHEIQRYSDLVPTG VPHAVTTDTKFRNYLIPKGTTIMALLTSVLHDDKEFPNPNIFDPGHFLDKNGNFKKSDYFMPFSAGKRICAGEGLARMELFLFLTTILQN -------------------------------------------------------------- >81829_81829_3_SIDT2-CYP2C8_SIDT2_chr11_117064679_ENST00000532062_CYP2C8_chr10_96827448_ENST00000371270_length(transcript)=2413nt_BP=753nt CTGCATCCGGCAGTGCAGCGGGCCGCTCTACGTGGTACCTGCCTGGTTCCCCTGCTCCATTCTCCACATCTCCTTTCTTCCCTCTGATGG GGGAGTGGGGCTGGGCTGAGGACCCAGGGGAGAGTGGGGACCAGCTGGCTGGGCCTTCTTCCACCACCCTCCCTGCCCCTGCCCTCAGTC CCTGTGTCCCTGCTTCCCTCCAGGACCGCATGGTGCTGCTGGTCATGGGCAACGTCATCAACTGGTCGCTGTGAGTATGGTCAGCAGTAG TGGCCTGGTTGGGTGGACAGCTGGGGACTCGGTCAGCCACTGGCTGCCTTGGGGGCTAAGGACAACTTCCAAATGTTGGGCATAAGACAT GGGGGCTAAGAGAGATCTTGGCCTCTCTTCTCTCTCTTGGCATCTGGGCTTCTAGAATCTTCTGTGGGCTTCTGAACACAGCCCAACATC ATAATAAAACATGCCTAGGCAGGGCAGGAAGAGCACTTCCTTGCCCTTGGGCTTCCTGCTTCACCACCCTTCATCCCTCTTGCCAGGGCT GCCTATGGGCTTATCATGCGCCCCAATGATTTCGCTTCCTACTTGTTGGCCATTGGCATCTGCAACCTGCTCCTTTACTTCGCCTTCTAC ATCATCATGAAGCTCCGGAGTGGGGAGAGGATCAAGCTCATCCCCCTGCTCTGCATCGTTTGCACCTCCGTGGTCTGGGGCTTCGCGCTC TTCTTCTTCTTCCAGGGACTCAGCACCTGGCAGTTCTCAAAAGTCTATGGTCCTGTGTTCACCGTGTATTTTGGCATGAATCCCATAGTG GTGTTTCATGGATATGAGGCAGTGAAGGAAGCCCTGATTGATAATGGAGAGGAGTTTTCTGGAAGAGGCAATTCCCCAATATCTCAAAGA ATTACTAAAGGACTTGGAATCATTTCCAGCAATGGAAAGAGATGGAAGGAGATCCGGCGTTTCTCCCTCACAACCTTGCGGAATTTTGGG ATGGGGAAGAGGAGCATTGAGGACCGTGTTCAAGAGGAAGCTCACTGCCTTGTGGAGGAGTTGAGAAAAACCAAGGCTTCACCCTGTGAT CCCACTTTCATCCTGGGCTGTGCTCCCTGCAATGTGATCTGCTCCGTTGTTTTCCAGAAACGATTTGATTATAAAGATCAGAATTTTCTC ACCCTGATGAAAAGATTCAATGAAAACTTCAGGATTCTGAACTCCCCATGGATCCAGGTCTGCAATAATTTCCCTCTACTCATTGATTGT TTCCCAGGAACTCACAACAAAGTGCTTAAAAATGTTGCTCTTACACGAAGTTACATTAGGGAGAAAGTAAAAGAACACCAAGCATCACTG GATGTTAACAATCCTCGGGACTTTATCGATTGCTTCCTGATCAAAATGGAGCAGGAAAAGGACAACCAAAAGTCAGAATTCAATATTGAA AACTTGGTTGGCACTGTAGCTGATCTATTTGTTGCTGGAACAGAGACAACAAGCACCACTCTGAGATATGGACTCCTGCTCCTGCTGAAG CACCCAGAGGTCACAGCTAAAGTCCAGGAAGAGATTGATCATGTAATTGGCAGACACAGGAGCCCCTGCATGCAGGATAGGAGCCACATG CCTTACACTGATGCTGTAGTGCACGAGATCCAGAGATACAGTGACCTTGTCCCCACCGGTGTGCCCCATGCAGTGACCACTGATACTAAG TTCAGAAACTACCTCATCCCCAAGGGCACAACCATAATGGCATTACTGACTTCCGTGCTACATGATGACAAAGAATTTCCTAATCCAAAT ATCTTTGACCCTGGCCACTTTCTAGATAAGAATGGCAACTTTAAGAAAAGTGACTACTTCATGCCTTTCTCAGCAGGAAAACGAATTTGT GCAGGAGAAGGACTTGCCCGCATGGAGCTATTTTTATTTCTAACCACAATTTTACAGAACTTTAACCTGAAATCTGTTGATGATTTAAAG AACCTCAATACTACTGCAGTTACCAAAGGGATTGTTTCTCTGCCACCCTCATACCAGATCTGCTTCATCCCTGTCTGAAGAATGCTAGCC CATCTGGCTGCCGATCTGCTATCACCTGCAACTCTTTTTTTATCAAGGACATTCCCACTATTATGTCTTCTCTGACCTCTCATCAAATCT TCCCATTCACTCAATATCCCATAAGCATCCAAACTCCATTAAGGAGAGTTGTTCAGGTCACTGCACAAATATATCTGCAATTATTCATAC TCTGTAACACTTGTATTAATTGCTGCATATGCTAATACTTTTCTAATGCTGACTTTTTAATATGTTATCACTGTAAAACACAGAAAAGTG >81829_81829_3_SIDT2-CYP2C8_SIDT2_chr11_117064679_ENST00000532062_CYP2C8_chr10_96827448_ENST00000371270_length(amino acids)=500AA_BP=54 MRPNDFASYLLAIGICNLLLYFAFYIIMKLRSGERIKLIPLLCIVCTSVVWGFALFFFFQGLSTWQFSKVYGPVFTVYFGMNPIVVFHGY EAVKEALIDNGEEFSGRGNSPISQRITKGLGIISSNGKRWKEIRRFSLTTLRNFGMGKRSIEDRVQEEAHCLVEELRKTKASPCDPTFIL GCAPCNVICSVVFQKRFDYKDQNFLTLMKRFNENFRILNSPWIQVCNNFPLLIDCFPGTHNKVLKNVALTRSYIREKVKEHQASLDVNNP RDFIDCFLIKMEQEKDNQKSEFNIENLVGTVADLFVAGTETTSTTLRYGLLLLLKHPEVTAKVQEEIDHVIGRHRSPCMQDRSHMPYTDA VVHEIQRYSDLVPTGVPHAVTTDTKFRNYLIPKGTTIMALLTSVLHDDKEFPNPNIFDPGHFLDKNGNFKKSDYFMPFSAGKRICAGEGL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SIDT2-CYP2C8 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SIDT2-CYP2C8 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SIDT2-CYP2C8 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
