
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SLC25A25-WDR34 (FusionGDB2 ID:82622) |
Fusion Gene Summary for SLC25A25-WDR34 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SLC25A25-WDR34 | Fusion gene ID: 82622 | Hgene | Tgene | Gene symbol | SLC25A25 | WDR34 | Gene ID | 114789 | 89891 |
| Gene name | solute carrier family 25 member 25 | WD repeat domain 34 | |
| Synonyms | MCSC|PCSCL|SCAMC-2 | CFAP133|DIC5|FAP133|SRTD11|bA216B9.3 | |
| Cytomap | 9q34.11 | 9q34.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | calcium-binding mitochondrial carrier protein SCaMC-2mitochondrial ATP-Mg/Pi carrier protein 3mitochondrial Ca(2+)-dependent solute carrier protein 3short calcium-binding mitochondrial carrier 2small calcium-binding mitochondrial carrier 2solute carr | WD repeat-containing protein 34 | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000373064, ENST00000373066, ENST00000373068, ENST00000373069, ENST00000432073, ENST00000433501, | ENST00000483181, ENST00000372715, | |
| Fusion gene scores | * DoF score | 8 X 5 X 6=240 | 7 X 4 X 6=168 |
| # samples | 8 | 8 | |
| ** MAII score | log2(8/240*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(8/168*10)=-1.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SLC25A25 [Title/Abstract] AND WDR34 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SLC25A25(130864760)-WDR34(131403218), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across SLC25A25 (5'-gene) Fusion gene breakpoints across SLC25A25 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
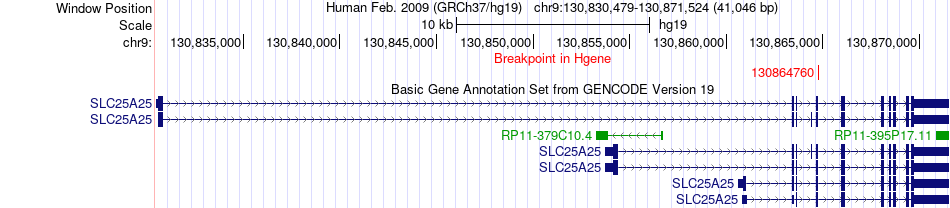 |
 Fusion gene breakpoints across WDR34 (3'-gene) Fusion gene breakpoints across WDR34 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
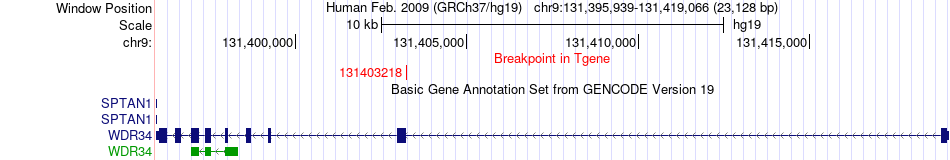 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4H2 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
Top |
Fusion Gene ORF analysis for SLC25A25-WDR34 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000373064 | ENST00000483181 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| 5CDS-intron | ENST00000373066 | ENST00000483181 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| 5CDS-intron | ENST00000373068 | ENST00000483181 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| 5CDS-intron | ENST00000373069 | ENST00000483181 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| 5CDS-intron | ENST00000432073 | ENST00000483181 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| 5CDS-intron | ENST00000433501 | ENST00000483181 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| In-frame | ENST00000373064 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| In-frame | ENST00000373066 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| In-frame | ENST00000373068 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| In-frame | ENST00000373069 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| In-frame | ENST00000432073 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
| In-frame | ENST00000433501 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000373068 | SLC25A25 | chr9 | 130864760 | + | ENST00000372715 | WDR34 | chr9 | 131403218 | - | 2215 | 707 | 29 | 2131 | 700 |
| ENST00000373069 | SLC25A25 | chr9 | 130864760 | + | ENST00000372715 | WDR34 | chr9 | 131403218 | - | 2159 | 651 | 27 | 2075 | 682 |
| ENST00000432073 | SLC25A25 | chr9 | 130864760 | + | ENST00000372715 | WDR34 | chr9 | 131403218 | - | 2461 | 953 | 251 | 2377 | 708 |
| ENST00000373066 | SLC25A25 | chr9 | 130864760 | + | ENST00000372715 | WDR34 | chr9 | 131403218 | - | 2497 | 989 | 251 | 2413 | 720 |
| ENST00000373064 | SLC25A25 | chr9 | 130864760 | + | ENST00000372715 | WDR34 | chr9 | 131403218 | - | 2257 | 749 | 263 | 2173 | 636 |
| ENST00000433501 | SLC25A25 | chr9 | 130864760 | + | ENST00000372715 | WDR34 | chr9 | 131403218 | - | 2047 | 539 | 299 | 1963 | 554 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000373068 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - | 0.016532354 | 0.98346764 |
| ENST00000373069 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - | 0.011867473 | 0.98813254 |
| ENST00000432073 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - | 0.009513347 | 0.9904867 |
| ENST00000373066 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - | 0.009453072 | 0.99054694 |
| ENST00000373064 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - | 0.021229457 | 0.97877055 |
| ENST00000433501 | ENST00000372715 | SLC25A25 | chr9 | 130864760 | + | WDR34 | chr9 | 131403218 | - | 0.02727 | 0.97273004 |
Top |
Fusion Genomic Features for SLC25A25-WDR34 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
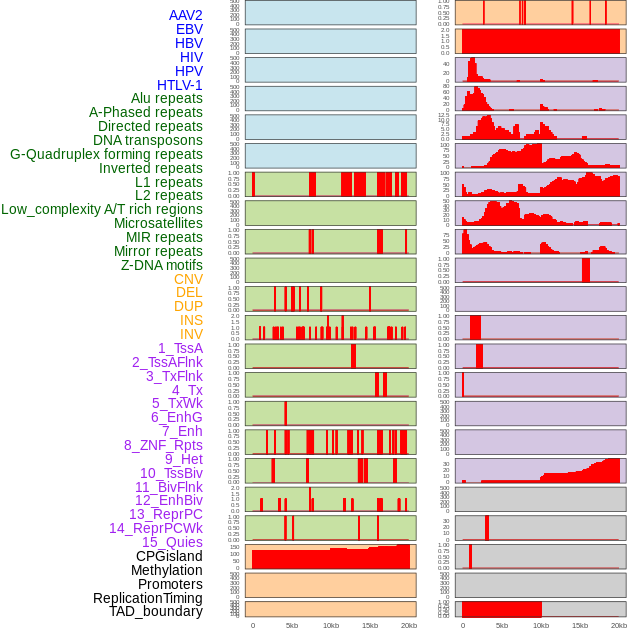 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SLC25A25-WDR34 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:130864760/chr9:131403218) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 60_71 | 162 | 470.0 | Calcium binding | 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 60_71 | 194 | 502.0 | Calcium binding | 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 60_71 | 196 | 504.0 | Calcium binding | 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 60_71 | 208 | 516.0 | Calcium binding | 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 60_71 | 182 | 490.0 | Calcium binding | 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 114_149 | 162 | 470.0 | Domain | EF-hand 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 47_80 | 162 | 470.0 | Domain | EF-hand 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 78_113 | 162 | 470.0 | Domain | EF-hand 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 114_149 | 194 | 502.0 | Domain | EF-hand 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 47_80 | 194 | 502.0 | Domain | EF-hand 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 78_113 | 194 | 502.0 | Domain | EF-hand 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 114_149 | 196 | 504.0 | Domain | EF-hand 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 47_80 | 196 | 504.0 | Domain | EF-hand 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 78_113 | 196 | 504.0 | Domain | EF-hand 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 114_149 | 208 | 516.0 | Domain | EF-hand 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 47_80 | 208 | 516.0 | Domain | EF-hand 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 78_113 | 208 | 516.0 | Domain | EF-hand 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 114_149 | 182 | 490.0 | Domain | EF-hand 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 47_80 | 182 | 490.0 | Domain | EF-hand 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 78_113 | 182 | 490.0 | Domain | EF-hand 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 1_189 | 194 | 502.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 1_189 | 196 | 504.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 1_189 | 208 | 516.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 190_207 | 208 | 516.0 | Transmembrane | Helical%3B Name%3D1 |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 106_131 | 62 | 537.0 | Region | DYNLRB1 binding | |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 80_93 | 62 | 537.0 | Region | DYNLL2 binding | |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 215_255 | 62 | 537.0 | Repeat | WD 1 | |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 264_308 | 62 | 537.0 | Repeat | WD 2 | |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 390_430 | 62 | 537.0 | Repeat | WD 3 | |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 433_473 | 62 | 537.0 | Repeat | WD 4 | |
| Tgene | WDR34 | chr9:130864760 | chr9:131403218 | ENST00000372715 | 0 | 9 | 480_520 | 62 | 537.0 | Repeat | WD 5 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 60_71 | 59 | 367.0 | Calcium binding | 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 114_149 | 59 | 367.0 | Domain | EF-hand 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 47_80 | 59 | 367.0 | Domain | EF-hand 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 78_113 | 59 | 367.0 | Domain | EF-hand 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 184_270 | 162 | 470.0 | Repeat | Note=Solcar 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 278_363 | 162 | 470.0 | Repeat | Note=Solcar 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 375_463 | 162 | 470.0 | Repeat | Note=Solcar 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 184_270 | 194 | 502.0 | Repeat | Note=Solcar 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 278_363 | 194 | 502.0 | Repeat | Note=Solcar 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 375_463 | 194 | 502.0 | Repeat | Note=Solcar 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 184_270 | 196 | 504.0 | Repeat | Note=Solcar 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 278_363 | 196 | 504.0 | Repeat | Note=Solcar 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 375_463 | 196 | 504.0 | Repeat | Note=Solcar 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 184_270 | 208 | 516.0 | Repeat | Note=Solcar 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 278_363 | 208 | 516.0 | Repeat | Note=Solcar 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 375_463 | 208 | 516.0 | Repeat | Note=Solcar 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 184_270 | 182 | 490.0 | Repeat | Note=Solcar 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 278_363 | 182 | 490.0 | Repeat | Note=Solcar 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 375_463 | 182 | 490.0 | Repeat | Note=Solcar 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 184_270 | 59 | 367.0 | Repeat | Note=Solcar 1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 278_363 | 59 | 367.0 | Repeat | Note=Solcar 2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 375_463 | 59 | 367.0 | Repeat | Note=Solcar 3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 1_189 | 162 | 470.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 208_244 | 162 | 470.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 265_287 | 162 | 470.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 302_337 | 162 | 470.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 358_380 | 162 | 470.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 399_437 | 162 | 470.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 458_469 | 162 | 470.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 208_244 | 194 | 502.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 265_287 | 194 | 502.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 302_337 | 194 | 502.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 358_380 | 194 | 502.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 399_437 | 194 | 502.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 458_469 | 194 | 502.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 208_244 | 196 | 504.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 265_287 | 196 | 504.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 302_337 | 196 | 504.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 358_380 | 196 | 504.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 399_437 | 196 | 504.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 458_469 | 196 | 504.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 208_244 | 208 | 516.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 265_287 | 208 | 516.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 302_337 | 208 | 516.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 358_380 | 208 | 516.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 399_437 | 208 | 516.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 458_469 | 208 | 516.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 1_189 | 182 | 490.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 208_244 | 182 | 490.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 265_287 | 182 | 490.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 302_337 | 182 | 490.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 358_380 | 182 | 490.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 399_437 | 182 | 490.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 458_469 | 182 | 490.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 1_189 | 59 | 367.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 208_244 | 59 | 367.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 265_287 | 59 | 367.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 302_337 | 59 | 367.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 358_380 | 59 | 367.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 399_437 | 59 | 367.0 | Topological domain | Mitochondrial matrix |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 458_469 | 59 | 367.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 190_207 | 162 | 470.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 245_264 | 162 | 470.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 288_301 | 162 | 470.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 338_357 | 162 | 470.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 381_398 | 162 | 470.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373064 | + | 4 | 10 | 438_457 | 162 | 470.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 190_207 | 194 | 502.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 245_264 | 194 | 502.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 288_301 | 194 | 502.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 338_357 | 194 | 502.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 381_398 | 194 | 502.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373066 | + | 5 | 11 | 438_457 | 194 | 502.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 190_207 | 196 | 504.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 245_264 | 196 | 504.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 288_301 | 196 | 504.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 338_357 | 196 | 504.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 381_398 | 196 | 504.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373068 | + | 4 | 10 | 438_457 | 196 | 504.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 245_264 | 208 | 516.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 288_301 | 208 | 516.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 338_357 | 208 | 516.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 381_398 | 208 | 516.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000373069 | + | 5 | 11 | 438_457 | 208 | 516.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 190_207 | 182 | 490.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 245_264 | 182 | 490.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 288_301 | 182 | 490.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 338_357 | 182 | 490.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 381_398 | 182 | 490.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000432073 | + | 4 | 10 | 438_457 | 182 | 490.0 | Transmembrane | Helical%3B Name%3D6 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 190_207 | 59 | 367.0 | Transmembrane | Helical%3B Name%3D1 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 245_264 | 59 | 367.0 | Transmembrane | Helical%3B Name%3D2 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 288_301 | 59 | 367.0 | Transmembrane | Helical%3B Name%3D3 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 338_357 | 59 | 367.0 | Transmembrane | Helical%3B Name%3D4 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 381_398 | 59 | 367.0 | Transmembrane | Helical%3B Name%3D5 |
| Hgene | SLC25A25 | chr9:130864760 | chr9:131403218 | ENST00000433501 | + | 4 | 10 | 438_457 | 59 | 367.0 | Transmembrane | Helical%3B Name%3D6 |
Top |
Fusion Gene Sequence for SLC25A25-WDR34 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >82622_82622_1_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373064_WDR34_chr9_131403218_ENST00000372715_length(transcript)=2257nt_BP=749nt CGGTGACGTCAGCCCTGATCTTTGTGCCTCTCACTTGAGCAACCTGATGCCACCTTCTCCCCCGGGCTCTGTGTTCATTGGCTGTTGGCA GGACTTGCCGTGGGAGGGCTTTTTTCTTTTTTCAATTAAGTAAAAGGGCAGGGCTTAAGCAGCCAATGACAGCTGAGGCCAGGCTTGTCT TATGCAAGTTTCCCGGGACCGGCAGCATTTAGGGAACTTGCTCCCGGTGGTGAGGGCAGTGGAGCACCCAGCAGGCCGCCAACATGCTCT GTCTGTGCCTGTATGTGCCGGTCATCGGGGAAGCCCAGACCGAGTTCCAGTACTTTGAGTCGAAGGGGCTCCCTGCCGAGCTGAAGTCCA TTTTCAAGCTCAGTGTCTTCATCCCCTCCCAGGAATTCTCCACCTACCGCCAGTGGAAGCAGAAAATTGTACAAGCTGGAGATAAGGACC TTGATGGGCAGCTAGACTTTGAAGAATTTGTCCATTATCTCCAAGATCATGAGAAGAAGCTGAGGCTGGTGTTTAAGAGTTTGGACAAAA AGAATGATGGACGCATTGACGCGCAGGAGATCATGCAGTCCCTGCGGGACTTGGGAGTCAAGATATCTGAACAGCAGGCAGAAAAAATTC TCAAGAGCATGGATAAAAACGGCACGATGACCATTGACTGGAACGAGTGGAGAGACTACCACCTCCTCCACCCCGTGGAAAACATCCCCG AGATCATCCTCTACTGGAAGCATTCCACGAAAAGTTGCCAGACGGCCAGCATTGCCACTGCCAGTGCATCCGCCCAGGCCAGGAATCATG TGGACGCCCAGGTGCAGACGGAGGCCCCCGTGCCTGTCAGCGTGCAGCCCCCGTCCCAGTATGACATACCCAGGCTCGCAGCCTTTCTTC GGAGAGTGGAGGCCATGGTCATCCGAGAGCTGAACAAGAATTGGCAGAGCCACGCGTTTGATGGCTTCGAGGTGAACTGGACCGAGCAGC AGCAGATGGTGTCTTGTCTGTATACCCTGGGCTACCCGCCAGCCCAAGCGCAGGGTCTGCATGTGACCAGCATCTCCTGGAACTCCACTG GCTCTGTGGTGGCCTGTGCCTACGGCCGGCTGGACCATGGGGACTGGAGCACGCTTAAGTCCTTCGTGTGTGCCTGGAACCTGGACCGGC GAGACCTGCGTCCCCAGCAGCCGTCGGCCGTGGTGGAGGTCCCCAGCGCTGTCCTGTGTCTGGCCTTCCACCCCACGCAGCCCTCCCACG TCGCAGGAGGGCTGTACAGTGGTGAGGTGTTGGTGTGGGACCTGAGCCGTCTTGAGGACCCGCTGCTGTGGCGCACAGGCCTGACGGATG ACACCCACACAGACCCTGTGTCCCAGGTGGTGTGGCTGCCCGAGCCTGGGCACAGCCACCGCTTCCAGGTGCTGAGTGTGGCCACCGACG GGAAGGTGCTACTCTGGCAGGGCATCGGGGTAGGCCAGCTGCAGCTCACAGAGGGCTTCGCCCTGGTCATGCAGCAGCTGCCACGGAGCA CCAAGCTCAAGAAGCATCCCCGCGGGGAGACCGAGGTGGGCGCCACGGCAGTGGCCTTCTCCAGCTTTGACCCTAGGCTGTTCATTCTGG GCACGGAAGGCGGCTTCCCGCTCAAGTGTTCCCTGGCAGCTGGAGAGGCAGCCCTCACGCGGATGCCCAGCTCCGTGCCCCTGCGGGCCC CAGCACAGTTTACCTTCTCCCCCCACGGCGGTCCCATCTACTCTGTGAGCTGTTCCCCCTTCCACAGGAATCTCTTCCTGAGCGCTGGGA CTGACGGGCATGTCCACCTGTACTCCATGCTGCAGGCCCCTCCCTTGACTTCGCTGCAGCTCTCCCTCAAGTATCTGTTTGCTGTGCGCT GGTCCCCAGTGCGGCCCTTGGTTTTTGCAGCTGCCTCTGGGAAAGGTGACGTGCAGCTGTTTGATCTCCAGAAAAGCTCCCAGAAACCCA CAGTTTTGATCAAGCAAACCCAGGATGAAAGCCCTGTCTACTGTCTGGAGTTCAACAGCCAGCAGACTCAGCTCTTGGCTGCGGGCGATG CCCAGGGCACAGTGAAGGTGTGGCAGCTGAGCACAGAGTTCACGGAACAAGGGCCCCGGGAAGCTGAGGACCTGGACTGCCTGGCAGCAG AGGTGGCGGCCTGAGGGGTCCCGGGAGGCGGGTGCAAGCCTTCGCTGTGCCGAGCCTTGTGTTTCTGACGCAAGCCAAATGAAGAAAAGC >82622_82622_1_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373064_WDR34_chr9_131403218_ENST00000372715_length(amino acids)=636AA_BP=162 MLCLCLYVPVIGEAQTEFQYFESKGLPAELKSIFKLSVFIPSQEFSTYRQWKQKIVQAGDKDLDGQLDFEEFVHYLQDHEKKLRLVFKSL DKKNDGRIDAQEIMQSLRDLGVKISEQQAEKILKSMDKNGTMTIDWNEWRDYHLLHPVENIPEIILYWKHSTKSCQTASIATASASAQAR NHVDAQVQTEAPVPVSVQPPSQYDIPRLAAFLRRVEAMVIRELNKNWQSHAFDGFEVNWTEQQQMVSCLYTLGYPPAQAQGLHVTSISWN STGSVVACAYGRLDHGDWSTLKSFVCAWNLDRRDLRPQQPSAVVEVPSAVLCLAFHPTQPSHVAGGLYSGEVLVWDLSRLEDPLLWRTGL TDDTHTDPVSQVVWLPEPGHSHRFQVLSVATDGKVLLWQGIGVGQLQLTEGFALVMQQLPRSTKLKKHPRGETEVGATAVAFSSFDPRLF ILGTEGGFPLKCSLAAGEAALTRMPSSVPLRAPAQFTFSPHGGPIYSVSCSPFHRNLFLSAGTDGHVHLYSMLQAPPLTSLQLSLKYLFA VRWSPVRPLVFAAASGKGDVQLFDLQKSSQKPTVLIKQTQDESPVYCLEFNSQQTQLLAAGDAQGTVKVWQLSTEFTEQGPREAEDLDCL -------------------------------------------------------------- >82622_82622_2_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373066_WDR34_chr9_131403218_ENST00000372715_length(transcript)=2497nt_BP=989nt GCGAGGACAGCTGAATAACTGGGAGCTCGAAGTTGCTTGCTGCCTCCGGGAGTGGTCTGCTTGGCTGTCGCGTTTTCCTTTTCTTTTTTT TTTTTTTTAAAAAAAAGCCAAACGTTTGCTCACCATCACCTGCAGAAGTGAGTTTTTATTGAAGAAGCTCCAGCTGCTTTCTCCTTATTG CCGGCAGCCTGGCAAGCCTCCCTTAGAGTGAGCTACTTCTCAGGCCGGGCACGACAGCAGTGCTAGCACATTTGAAAAAGCAGGAAACCG CCCCTGCATGCAGAGCTTCCAGCCTGAGAACCCCAGGCCCGTCCTCCCAGCAGCTGTGTAGCCAGAGAGCGTTGGTACTGTGGTCACATC CCTCAAAAGTGAACAGTCGCCATCAGAGGCGTTTGGAGGAGACCGTGATGTTGCAGATGCTGTGGCATTTCCTAGCTAGCTTTTTCCCCA GGGCTGGGTGCCACGGCTCCAGAGAGGGGGACGATCGTGAAGTCAGAGGCACCCCAGCCCCTGCCTGGAGAGACCAGATGGCAAGCTTTT TGGGGAAACAGGACGGAAGGGCTGAGGCCACGGAAAAAAGACCCACCATTTTGCTGGTGGTTGGACCTGCAGAGCAGTTTCCTAAGAAAA TTGTACAAGCTGGAGATAAGGACCTTGATGGGCAGCTAGACTTTGAAGAATTTGTCCATTATCTCCAAGATCATGAGAAGAAGCTGAGGC TGGTGTTTAAGAGTTTGGACAAAAAGAATGATGGACGCATTGACGCGCAGGAGATCATGCAGTCCCTGCGGGACTTGGGAGTCAAGATAT CTGAACAGCAGGCAGAAAAAATTCTCAAGAGAATACGAACGGGCCATTTCTGGGGCCCTGTCACCTACATGGATAAAAACGGCACGATGA CCATTGACTGGAACGAGTGGAGAGACTACCACCTCCTCCACCCCGTGGAAAACATCCCCGAGATCATCCTCTACTGGAAGCATTCCACGA AAAGTTGCCAGACGGCCAGCATTGCCACTGCCAGTGCATCCGCCCAGGCCAGGAATCATGTGGACGCCCAGGTGCAGACGGAGGCCCCCG TGCCTGTCAGCGTGCAGCCCCCGTCCCAGTATGACATACCCAGGCTCGCAGCCTTTCTTCGGAGAGTGGAGGCCATGGTCATCCGAGAGC TGAACAAGAATTGGCAGAGCCACGCGTTTGATGGCTTCGAGGTGAACTGGACCGAGCAGCAGCAGATGGTGTCTTGTCTGTATACCCTGG GCTACCCGCCAGCCCAAGCGCAGGGTCTGCATGTGACCAGCATCTCCTGGAACTCCACTGGCTCTGTGGTGGCCTGTGCCTACGGCCGGC TGGACCATGGGGACTGGAGCACGCTTAAGTCCTTCGTGTGTGCCTGGAACCTGGACCGGCGAGACCTGCGTCCCCAGCAGCCGTCGGCCG TGGTGGAGGTCCCCAGCGCTGTCCTGTGTCTGGCCTTCCACCCCACGCAGCCCTCCCACGTCGCAGGAGGGCTGTACAGTGGTGAGGTGT TGGTGTGGGACCTGAGCCGTCTTGAGGACCCGCTGCTGTGGCGCACAGGCCTGACGGATGACACCCACACAGACCCTGTGTCCCAGGTGG TGTGGCTGCCCGAGCCTGGGCACAGCCACCGCTTCCAGGTGCTGAGTGTGGCCACCGACGGGAAGGTGCTACTCTGGCAGGGCATCGGGG TAGGCCAGCTGCAGCTCACAGAGGGCTTCGCCCTGGTCATGCAGCAGCTGCCACGGAGCACCAAGCTCAAGAAGCATCCCCGCGGGGAGA CCGAGGTGGGCGCCACGGCAGTGGCCTTCTCCAGCTTTGACCCTAGGCTGTTCATTCTGGGCACGGAAGGCGGCTTCCCGCTCAAGTGTT CCCTGGCAGCTGGAGAGGCAGCCCTCACGCGGATGCCCAGCTCCGTGCCCCTGCGGGCCCCAGCACAGTTTACCTTCTCCCCCCACGGCG GTCCCATCTACTCTGTGAGCTGTTCCCCCTTCCACAGGAATCTCTTCCTGAGCGCTGGGACTGACGGGCATGTCCACCTGTACTCCATGC TGCAGGCCCCTCCCTTGACTTCGCTGCAGCTCTCCCTCAAGTATCTGTTTGCTGTGCGCTGGTCCCCAGTGCGGCCCTTGGTTTTTGCAG CTGCCTCTGGGAAAGGTGACGTGCAGCTGTTTGATCTCCAGAAAAGCTCCCAGAAACCCACAGTTTTGATCAAGCAAACCCAGGATGAAA GCCCTGTCTACTGTCTGGAGTTCAACAGCCAGCAGACTCAGCTCTTGGCTGCGGGCGATGCCCAGGGCACAGTGAAGGTGTGGCAGCTGA GCACAGAGTTCACGGAACAAGGGCCCCGGGAAGCTGAGGACCTGGACTGCCTGGCAGCAGAGGTGGCGGCCTGAGGGGTCCCGGGAGGCG >82622_82622_2_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373066_WDR34_chr9_131403218_ENST00000372715_length(amino acids)=720AA_BP=246 MKKQETAPACRASSLRTPGPSSQQLCSQRALVLWSHPSKVNSRHQRRLEETVMLQMLWHFLASFFPRAGCHGSREGDDREVRGTPAPAWR DQMASFLGKQDGRAEATEKRPTILLVVGPAEQFPKKIVQAGDKDLDGQLDFEEFVHYLQDHEKKLRLVFKSLDKKNDGRIDAQEIMQSLR DLGVKISEQQAEKILKRIRTGHFWGPVTYMDKNGTMTIDWNEWRDYHLLHPVENIPEIILYWKHSTKSCQTASIATASASAQARNHVDAQ VQTEAPVPVSVQPPSQYDIPRLAAFLRRVEAMVIRELNKNWQSHAFDGFEVNWTEQQQMVSCLYTLGYPPAQAQGLHVTSISWNSTGSVV ACAYGRLDHGDWSTLKSFVCAWNLDRRDLRPQQPSAVVEVPSAVLCLAFHPTQPSHVAGGLYSGEVLVWDLSRLEDPLLWRTGLTDDTHT DPVSQVVWLPEPGHSHRFQVLSVATDGKVLLWQGIGVGQLQLTEGFALVMQQLPRSTKLKKHPRGETEVGATAVAFSSFDPRLFILGTEG GFPLKCSLAAGEAALTRMPSSVPLRAPAQFTFSPHGGPIYSVSCSPFHRNLFLSAGTDGHVHLYSMLQAPPLTSLQLSLKYLFAVRWSPV RPLVFAAASGKGDVQLFDLQKSSQKPTVLIKQTQDESPVYCLEFNSQQTQLLAAGDAQGTVKVWQLSTEFTEQGPREAEDLDCLAAEVAA -------------------------------------------------------------- >82622_82622_3_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373068_WDR34_chr9_131403218_ENST00000372715_length(transcript)=2215nt_BP=707nt TGCCGGCGTCTCCGAGCCCGCGCTCCCAGCTGCAGAGCGCCTGGCTTGCCTCCCGCGCGGTCACCGCCGGCCCGCCGCCCCCGCTCCCGC CCGCGCCCGGAGCCCCTGCCTCGCGCCCGATGGTGAGCAGTGTGTTGTGCCGCTGTGTGGCCTCCCCGCCGCCGGACGCCGCCGCCACCG CCGCCTCTTCGTCTGCCTCATCGCCGGCGTCCGTGGGGGACCCCTGCGGCGGCGCTATCTGCGGGGGCCCGGACCACCGGCTGCGCCTGT GGAGACTCTTTCAGACGCTCGACGTCAACCGGGACGGCGGCCTGTGTGTCAACGACCTGGCGGTGGGGCTGCGGCGCCTGGGACTGCACC GCACCGAGGGCGAGCTCCAGAAAATTGTACAAGCTGGAGATAAGGACCTTGATGGGCAGCTAGACTTTGAAGAATTTGTCCATTATCTCC AAGATCATGAGAAGAAGCTGAGGCTGGTGTTTAAGAGTTTGGACAAAAAGAATGATGGACGCATTGACGCGCAGGAGATCATGCAGTCCC TGCGGGACTTGGGAGTCAAGATATCTGAACAGCAGGCAGAAAAAATTCTCAAGAGCATGGATAAAAACGGCACGATGACCATTGACTGGA ACGAGTGGAGAGACTACCACCTCCTCCACCCCGTGGAAAACATCCCCGAGATCATCCTCTACTGGAAGCATTCCACGAAAAGTTGCCAGA CGGCCAGCATTGCCACTGCCAGTGCATCCGCCCAGGCCAGGAATCATGTGGACGCCCAGGTGCAGACGGAGGCCCCCGTGCCTGTCAGCG TGCAGCCCCCGTCCCAGTATGACATACCCAGGCTCGCAGCCTTTCTTCGGAGAGTGGAGGCCATGGTCATCCGAGAGCTGAACAAGAATT GGCAGAGCCACGCGTTTGATGGCTTCGAGGTGAACTGGACCGAGCAGCAGCAGATGGTGTCTTGTCTGTATACCCTGGGCTACCCGCCAG CCCAAGCGCAGGGTCTGCATGTGACCAGCATCTCCTGGAACTCCACTGGCTCTGTGGTGGCCTGTGCCTACGGCCGGCTGGACCATGGGG ACTGGAGCACGCTTAAGTCCTTCGTGTGTGCCTGGAACCTGGACCGGCGAGACCTGCGTCCCCAGCAGCCGTCGGCCGTGGTGGAGGTCC CCAGCGCTGTCCTGTGTCTGGCCTTCCACCCCACGCAGCCCTCCCACGTCGCAGGAGGGCTGTACAGTGGTGAGGTGTTGGTGTGGGACC TGAGCCGTCTTGAGGACCCGCTGCTGTGGCGCACAGGCCTGACGGATGACACCCACACAGACCCTGTGTCCCAGGTGGTGTGGCTGCCCG AGCCTGGGCACAGCCACCGCTTCCAGGTGCTGAGTGTGGCCACCGACGGGAAGGTGCTACTCTGGCAGGGCATCGGGGTAGGCCAGCTGC AGCTCACAGAGGGCTTCGCCCTGGTCATGCAGCAGCTGCCACGGAGCACCAAGCTCAAGAAGCATCCCCGCGGGGAGACCGAGGTGGGCG CCACGGCAGTGGCCTTCTCCAGCTTTGACCCTAGGCTGTTCATTCTGGGCACGGAAGGCGGCTTCCCGCTCAAGTGTTCCCTGGCAGCTG GAGAGGCAGCCCTCACGCGGATGCCCAGCTCCGTGCCCCTGCGGGCCCCAGCACAGTTTACCTTCTCCCCCCACGGCGGTCCCATCTACT CTGTGAGCTGTTCCCCCTTCCACAGGAATCTCTTCCTGAGCGCTGGGACTGACGGGCATGTCCACCTGTACTCCATGCTGCAGGCCCCTC CCTTGACTTCGCTGCAGCTCTCCCTCAAGTATCTGTTTGCTGTGCGCTGGTCCCCAGTGCGGCCCTTGGTTTTTGCAGCTGCCTCTGGGA AAGGTGACGTGCAGCTGTTTGATCTCCAGAAAAGCTCCCAGAAACCCACAGTTTTGATCAAGCAAACCCAGGATGAAAGCCCTGTCTACT GTCTGGAGTTCAACAGCCAGCAGACTCAGCTCTTGGCTGCGGGCGATGCCCAGGGCACAGTGAAGGTGTGGCAGCTGAGCACAGAGTTCA CGGAACAAGGGCCCCGGGAAGCTGAGGACCTGGACTGCCTGGCAGCAGAGGTGGCGGCCTGAGGGGTCCCGGGAGGCGGGTGCAAGCCTT >82622_82622_3_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373068_WDR34_chr9_131403218_ENST00000372715_length(amino acids)=700AA_BP=11 MQSAWLASRAVTAGPPPPLPPAPGAPASRPMVSSVLCRCVASPPPDAAATAASSSASSPASVGDPCGGAICGGPDHRLRLWRLFQTLDVN RDGGLCVNDLAVGLRRLGLHRTEGELQKIVQAGDKDLDGQLDFEEFVHYLQDHEKKLRLVFKSLDKKNDGRIDAQEIMQSLRDLGVKISE QQAEKILKSMDKNGTMTIDWNEWRDYHLLHPVENIPEIILYWKHSTKSCQTASIATASASAQARNHVDAQVQTEAPVPVSVQPPSQYDIP RLAAFLRRVEAMVIRELNKNWQSHAFDGFEVNWTEQQQMVSCLYTLGYPPAQAQGLHVTSISWNSTGSVVACAYGRLDHGDWSTLKSFVC AWNLDRRDLRPQQPSAVVEVPSAVLCLAFHPTQPSHVAGGLYSGEVLVWDLSRLEDPLLWRTGLTDDTHTDPVSQVVWLPEPGHSHRFQV LSVATDGKVLLWQGIGVGQLQLTEGFALVMQQLPRSTKLKKHPRGETEVGATAVAFSSFDPRLFILGTEGGFPLKCSLAAGEAALTRMPS SVPLRAPAQFTFSPHGGPIYSVSCSPFHRNLFLSAGTDGHVHLYSMLQAPPLTSLQLSLKYLFAVRWSPVRPLVFAAASGKGDVQLFDLQ -------------------------------------------------------------- >82622_82622_4_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373069_WDR34_chr9_131403218_ENST00000372715_length(transcript)=2159nt_BP=651nt GCGCCCGGAGCCCCTGCCTCGCGCCCGATGGTGAGCAGTGTGTTGTGCCGCTGTGTGGCCTCCCCGCCGCCGGACGCCGCCGCCACCGCC GCCTCTTCGTCTGCCTCATCGCCGGCGTCCGTGGGGGACCCCTGCGGCGGCGCTATCTGCGGGGGCCCGGACCACCGGCTGCGCCTGTGG AGACTCTTTCAGACGCTCGACGTCAACCGGGACGGCGGCCTGTGTGTCAACGACCTGGCGGTGGGGCTGCGGCGCCTGGGACTGCACCGC ACCGAGGGCGAGCTCCAGAAAATTGTACAAGCTGGAGATAAGGACCTTGATGGGCAGCTAGACTTTGAAGAATTTGTCCATTATCTCCAA GATCATGAGAAGAAGCTGAGGCTGGTGTTTAAGAGTTTGGACAAAAAGAATGATGGACGCATTGACGCGCAGGAGATCATGCAGTCCCTG CGGGACTTGGGAGTCAAGATATCTGAACAGCAGGCAGAAAAAATTCTCAAGAGAATACGAACGGGCCATTTCTGGGGCCCTGTCACCTAC ATGGATAAAAACGGCACGATGACCATTGACTGGAACGAGTGGAGAGACTACCACCTCCTCCACCCCGTGGAAAACATCCCCGAGATCATC CTCTACTGGAAGCATTCCACGAAAAGTTGCCAGACGGCCAGCATTGCCACTGCCAGTGCATCCGCCCAGGCCAGGAATCATGTGGACGCC CAGGTGCAGACGGAGGCCCCCGTGCCTGTCAGCGTGCAGCCCCCGTCCCAGTATGACATACCCAGGCTCGCAGCCTTTCTTCGGAGAGTG GAGGCCATGGTCATCCGAGAGCTGAACAAGAATTGGCAGAGCCACGCGTTTGATGGCTTCGAGGTGAACTGGACCGAGCAGCAGCAGATG GTGTCTTGTCTGTATACCCTGGGCTACCCGCCAGCCCAAGCGCAGGGTCTGCATGTGACCAGCATCTCCTGGAACTCCACTGGCTCTGTG GTGGCCTGTGCCTACGGCCGGCTGGACCATGGGGACTGGAGCACGCTTAAGTCCTTCGTGTGTGCCTGGAACCTGGACCGGCGAGACCTG CGTCCCCAGCAGCCGTCGGCCGTGGTGGAGGTCCCCAGCGCTGTCCTGTGTCTGGCCTTCCACCCCACGCAGCCCTCCCACGTCGCAGGA GGGCTGTACAGTGGTGAGGTGTTGGTGTGGGACCTGAGCCGTCTTGAGGACCCGCTGCTGTGGCGCACAGGCCTGACGGATGACACCCAC ACAGACCCTGTGTCCCAGGTGGTGTGGCTGCCCGAGCCTGGGCACAGCCACCGCTTCCAGGTGCTGAGTGTGGCCACCGACGGGAAGGTG CTACTCTGGCAGGGCATCGGGGTAGGCCAGCTGCAGCTCACAGAGGGCTTCGCCCTGGTCATGCAGCAGCTGCCACGGAGCACCAAGCTC AAGAAGCATCCCCGCGGGGAGACCGAGGTGGGCGCCACGGCAGTGGCCTTCTCCAGCTTTGACCCTAGGCTGTTCATTCTGGGCACGGAA GGCGGCTTCCCGCTCAAGTGTTCCCTGGCAGCTGGAGAGGCAGCCCTCACGCGGATGCCCAGCTCCGTGCCCCTGCGGGCCCCAGCACAG TTTACCTTCTCCCCCCACGGCGGTCCCATCTACTCTGTGAGCTGTTCCCCCTTCCACAGGAATCTCTTCCTGAGCGCTGGGACTGACGGG CATGTCCACCTGTACTCCATGCTGCAGGCCCCTCCCTTGACTTCGCTGCAGCTCTCCCTCAAGTATCTGTTTGCTGTGCGCTGGTCCCCA GTGCGGCCCTTGGTTTTTGCAGCTGCCTCTGGGAAAGGTGACGTGCAGCTGTTTGATCTCCAGAAAAGCTCCCAGAAACCCACAGTTTTG ATCAAGCAAACCCAGGATGAAAGCCCTGTCTACTGTCTGGAGTTCAACAGCCAGCAGACTCAGCTCTTGGCTGCGGGCGATGCCCAGGGC ACAGTGAAGGTGTGGCAGCTGAGCACAGAGTTCACGGAACAAGGGCCCCGGGAAGCTGAGGACCTGGACTGCCTGGCAGCAGAGGTGGCG >82622_82622_4_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000373069_WDR34_chr9_131403218_ENST00000372715_length(amino acids)=682AA_BP=208 MVSSVLCRCVASPPPDAAATAASSSASSPASVGDPCGGAICGGPDHRLRLWRLFQTLDVNRDGGLCVNDLAVGLRRLGLHRTEGELQKIV QAGDKDLDGQLDFEEFVHYLQDHEKKLRLVFKSLDKKNDGRIDAQEIMQSLRDLGVKISEQQAEKILKRIRTGHFWGPVTYMDKNGTMTI DWNEWRDYHLLHPVENIPEIILYWKHSTKSCQTASIATASASAQARNHVDAQVQTEAPVPVSVQPPSQYDIPRLAAFLRRVEAMVIRELN KNWQSHAFDGFEVNWTEQQQMVSCLYTLGYPPAQAQGLHVTSISWNSTGSVVACAYGRLDHGDWSTLKSFVCAWNLDRRDLRPQQPSAVV EVPSAVLCLAFHPTQPSHVAGGLYSGEVLVWDLSRLEDPLLWRTGLTDDTHTDPVSQVVWLPEPGHSHRFQVLSVATDGKVLLWQGIGVG QLQLTEGFALVMQQLPRSTKLKKHPRGETEVGATAVAFSSFDPRLFILGTEGGFPLKCSLAAGEAALTRMPSSVPLRAPAQFTFSPHGGP IYSVSCSPFHRNLFLSAGTDGHVHLYSMLQAPPLTSLQLSLKYLFAVRWSPVRPLVFAAASGKGDVQLFDLQKSSQKPTVLIKQTQDESP -------------------------------------------------------------- >82622_82622_5_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000432073_WDR34_chr9_131403218_ENST00000372715_length(transcript)=2461nt_BP=953nt GCGAGGACAGCTGAATAACTGGGAGCTCGAAGTTGCTTGCTGCCTCCGGGAGTGGTCTGCTTGGCTGTCGCGTTTTCCTTTTCTTTTTTT TTTTTTTTAAAAAAAAGCCAAACGTTTGCTCACCATCACCTGCAGAAGTGAGTTTTTATTGAAGAAGCTCCAGCTGCTTTCTCCTTATTG CCGGCAGCCTGGCAAGCCTCCCTTAGAGTGAGCTACTTCTCAGGCCGGGCACGACAGCAGTGCTAGCACATTTGAAAAAGCAGGAAACCG CCCCTGCATGCAGAGCTTCCAGCCTGAGAACCCCAGGCCCGTCCTCCCAGCAGCTGTGTAGCCAGAGAGCGTTGGTACTGTGGTCACATC CCTCAAAAGTGAACAGTCGCCATCAGAGGCGTTTGGAGGAGACCGTGATGTTGCAGATGCTGTGGCATTTCCTAGCTAGCTTTTTCCCCA GGGCTGGGTGCCACGGCTCCAGAGAGGGGGACGATCGTGAAGTCAGAGGCACCCCAGCCCCTGCCTGGAGAGACCAGATGGCAAGCTTTT TGGGGAAACAGGACGGAAGGGCTGAGGCCACGGAAAAAAGACCCACCATTTTGCTGGTGGTTGGACCTGCAGAGCAGTTTCCTAAGAAAA TTGTACAAGCTGGAGATAAGGACCTTGATGGGCAGCTAGACTTTGAAGAATTTGTCCATTATCTCCAAGATCATGAGAAGAAGCTGAGGC TGGTGTTTAAGAGTTTGGACAAAAAGAATGATGGACGCATTGACGCGCAGGAGATCATGCAGTCCCTGCGGGACTTGGGAGTCAAGATAT CTGAACAGCAGGCAGAAAAAATTCTCAAGAGCATGGATAAAAACGGCACGATGACCATTGACTGGAACGAGTGGAGAGACTACCACCTCC TCCACCCCGTGGAAAACATCCCCGAGATCATCCTCTACTGGAAGCATTCCACGAAAAGTTGCCAGACGGCCAGCATTGCCACTGCCAGTG CATCCGCCCAGGCCAGGAATCATGTGGACGCCCAGGTGCAGACGGAGGCCCCCGTGCCTGTCAGCGTGCAGCCCCCGTCCCAGTATGACA TACCCAGGCTCGCAGCCTTTCTTCGGAGAGTGGAGGCCATGGTCATCCGAGAGCTGAACAAGAATTGGCAGAGCCACGCGTTTGATGGCT TCGAGGTGAACTGGACCGAGCAGCAGCAGATGGTGTCTTGTCTGTATACCCTGGGCTACCCGCCAGCCCAAGCGCAGGGTCTGCATGTGA CCAGCATCTCCTGGAACTCCACTGGCTCTGTGGTGGCCTGTGCCTACGGCCGGCTGGACCATGGGGACTGGAGCACGCTTAAGTCCTTCG TGTGTGCCTGGAACCTGGACCGGCGAGACCTGCGTCCCCAGCAGCCGTCGGCCGTGGTGGAGGTCCCCAGCGCTGTCCTGTGTCTGGCCT TCCACCCCACGCAGCCCTCCCACGTCGCAGGAGGGCTGTACAGTGGTGAGGTGTTGGTGTGGGACCTGAGCCGTCTTGAGGACCCGCTGC TGTGGCGCACAGGCCTGACGGATGACACCCACACAGACCCTGTGTCCCAGGTGGTGTGGCTGCCCGAGCCTGGGCACAGCCACCGCTTCC AGGTGCTGAGTGTGGCCACCGACGGGAAGGTGCTACTCTGGCAGGGCATCGGGGTAGGCCAGCTGCAGCTCACAGAGGGCTTCGCCCTGG TCATGCAGCAGCTGCCACGGAGCACCAAGCTCAAGAAGCATCCCCGCGGGGAGACCGAGGTGGGCGCCACGGCAGTGGCCTTCTCCAGCT TTGACCCTAGGCTGTTCATTCTGGGCACGGAAGGCGGCTTCCCGCTCAAGTGTTCCCTGGCAGCTGGAGAGGCAGCCCTCACGCGGATGC CCAGCTCCGTGCCCCTGCGGGCCCCAGCACAGTTTACCTTCTCCCCCCACGGCGGTCCCATCTACTCTGTGAGCTGTTCCCCCTTCCACA GGAATCTCTTCCTGAGCGCTGGGACTGACGGGCATGTCCACCTGTACTCCATGCTGCAGGCCCCTCCCTTGACTTCGCTGCAGCTCTCCC TCAAGTATCTGTTTGCTGTGCGCTGGTCCCCAGTGCGGCCCTTGGTTTTTGCAGCTGCCTCTGGGAAAGGTGACGTGCAGCTGTTTGATC TCCAGAAAAGCTCCCAGAAACCCACAGTTTTGATCAAGCAAACCCAGGATGAAAGCCCTGTCTACTGTCTGGAGTTCAACAGCCAGCAGA CTCAGCTCTTGGCTGCGGGCGATGCCCAGGGCACAGTGAAGGTGTGGCAGCTGAGCACAGAGTTCACGGAACAAGGGCCCCGGGAAGCTG AGGACCTGGACTGCCTGGCAGCAGAGGTGGCGGCCTGAGGGGTCCCGGGAGGCGGGTGCAAGCCTTCGCTGTGCCGAGCCTTGTGTTTCT >82622_82622_5_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000432073_WDR34_chr9_131403218_ENST00000372715_length(amino acids)=708AA_BP=234 MKKQETAPACRASSLRTPGPSSQQLCSQRALVLWSHPSKVNSRHQRRLEETVMLQMLWHFLASFFPRAGCHGSREGDDREVRGTPAPAWR DQMASFLGKQDGRAEATEKRPTILLVVGPAEQFPKKIVQAGDKDLDGQLDFEEFVHYLQDHEKKLRLVFKSLDKKNDGRIDAQEIMQSLR DLGVKISEQQAEKILKSMDKNGTMTIDWNEWRDYHLLHPVENIPEIILYWKHSTKSCQTASIATASASAQARNHVDAQVQTEAPVPVSVQ PPSQYDIPRLAAFLRRVEAMVIRELNKNWQSHAFDGFEVNWTEQQQMVSCLYTLGYPPAQAQGLHVTSISWNSTGSVVACAYGRLDHGDW STLKSFVCAWNLDRRDLRPQQPSAVVEVPSAVLCLAFHPTQPSHVAGGLYSGEVLVWDLSRLEDPLLWRTGLTDDTHTDPVSQVVWLPEP GHSHRFQVLSVATDGKVLLWQGIGVGQLQLTEGFALVMQQLPRSTKLKKHPRGETEVGATAVAFSSFDPRLFILGTEGGFPLKCSLAAGE AALTRMPSSVPLRAPAQFTFSPHGGPIYSVSCSPFHRNLFLSAGTDGHVHLYSMLQAPPLTSLQLSLKYLFAVRWSPVRPLVFAAASGKG -------------------------------------------------------------- >82622_82622_6_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000433501_WDR34_chr9_131403218_ENST00000372715_length(transcript)=2047nt_BP=539nt GGCCGCCAACATGCTCTGTCTGTGCCTGTATGTGCCGGTCATCGGGGAAGCCCAGACCGAGTTCCAGTACTTTGAGTCGAAGGGGCTCCC TGCCGAGCTGAAGTCCATTTTCAAGCTCAGTGTCTTCATCCCCTCCCAGGAATTCTCCACCTACCGCCAGTGGAAGCAGGTCTGTGGGGT GACCGCGCGGAAGGGAGAGGCGCATTCAACCGAAAATTGTACAAGCTGGAGATAAGGACCTTGATGGGCAGCTAGACTTTGAAGAATTTG TCCATTATCTCCAAGATCATGAGAAGAAGCTGAGGCTGGTGTTTAAGAGTTTGGACAAAAAGAATGATGGACGCATTGACGCGCAGGAGA TCATGCAGTCCCTGCGGGACTTGGGAGTCAAGATATCTGAACAGCAGGCAGAAAAAATTCTCAAGAGCATGGATAAAAACGGCACGATGA CCATTGACTGGAACGAGTGGAGAGACTACCACCTCCTCCACCCCGTGGAAAACATCCCCGAGATCATCCTCTACTGGAAGCATTCCACGA AAAGTTGCCAGACGGCCAGCATTGCCACTGCCAGTGCATCCGCCCAGGCCAGGAATCATGTGGACGCCCAGGTGCAGACGGAGGCCCCCG TGCCTGTCAGCGTGCAGCCCCCGTCCCAGTATGACATACCCAGGCTCGCAGCCTTTCTTCGGAGAGTGGAGGCCATGGTCATCCGAGAGC TGAACAAGAATTGGCAGAGCCACGCGTTTGATGGCTTCGAGGTGAACTGGACCGAGCAGCAGCAGATGGTGTCTTGTCTGTATACCCTGG GCTACCCGCCAGCCCAAGCGCAGGGTCTGCATGTGACCAGCATCTCCTGGAACTCCACTGGCTCTGTGGTGGCCTGTGCCTACGGCCGGC TGGACCATGGGGACTGGAGCACGCTTAAGTCCTTCGTGTGTGCCTGGAACCTGGACCGGCGAGACCTGCGTCCCCAGCAGCCGTCGGCCG TGGTGGAGGTCCCCAGCGCTGTCCTGTGTCTGGCCTTCCACCCCACGCAGCCCTCCCACGTCGCAGGAGGGCTGTACAGTGGTGAGGTGT TGGTGTGGGACCTGAGCCGTCTTGAGGACCCGCTGCTGTGGCGCACAGGCCTGACGGATGACACCCACACAGACCCTGTGTCCCAGGTGG TGTGGCTGCCCGAGCCTGGGCACAGCCACCGCTTCCAGGTGCTGAGTGTGGCCACCGACGGGAAGGTGCTACTCTGGCAGGGCATCGGGG TAGGCCAGCTGCAGCTCACAGAGGGCTTCGCCCTGGTCATGCAGCAGCTGCCACGGAGCACCAAGCTCAAGAAGCATCCCCGCGGGGAGA CCGAGGTGGGCGCCACGGCAGTGGCCTTCTCCAGCTTTGACCCTAGGCTGTTCATTCTGGGCACGGAAGGCGGCTTCCCGCTCAAGTGTT CCCTGGCAGCTGGAGAGGCAGCCCTCACGCGGATGCCCAGCTCCGTGCCCCTGCGGGCCCCAGCACAGTTTACCTTCTCCCCCCACGGCG GTCCCATCTACTCTGTGAGCTGTTCCCCCTTCCACAGGAATCTCTTCCTGAGCGCTGGGACTGACGGGCATGTCCACCTGTACTCCATGC TGCAGGCCCCTCCCTTGACTTCGCTGCAGCTCTCCCTCAAGTATCTGTTTGCTGTGCGCTGGTCCCCAGTGCGGCCCTTGGTTTTTGCAG CTGCCTCTGGGAAAGGTGACGTGCAGCTGTTTGATCTCCAGAAAAGCTCCCAGAAACCCACAGTTTTGATCAAGCAAACCCAGGATGAAA GCCCTGTCTACTGTCTGGAGTTCAACAGCCAGCAGACTCAGCTCTTGGCTGCGGGCGATGCCCAGGGCACAGTGAAGGTGTGGCAGCTGA GCACAGAGTTCACGGAACAAGGGCCCCGGGAAGCTGAGGACCTGGACTGCCTGGCAGCAGAGGTGGCGGCCTGAGGGGTCCCGGGAGGCG >82622_82622_6_SLC25A25-WDR34_SLC25A25_chr9_130864760_ENST00000433501_WDR34_chr9_131403218_ENST00000372715_length(amino acids)=554AA_BP=80 MRLVFKSLDKKNDGRIDAQEIMQSLRDLGVKISEQQAEKILKSMDKNGTMTIDWNEWRDYHLLHPVENIPEIILYWKHSTKSCQTASIAT ASASAQARNHVDAQVQTEAPVPVSVQPPSQYDIPRLAAFLRRVEAMVIRELNKNWQSHAFDGFEVNWTEQQQMVSCLYTLGYPPAQAQGL HVTSISWNSTGSVVACAYGRLDHGDWSTLKSFVCAWNLDRRDLRPQQPSAVVEVPSAVLCLAFHPTQPSHVAGGLYSGEVLVWDLSRLED PLLWRTGLTDDTHTDPVSQVVWLPEPGHSHRFQVLSVATDGKVLLWQGIGVGQLQLTEGFALVMQQLPRSTKLKKHPRGETEVGATAVAF SSFDPRLFILGTEGGFPLKCSLAAGEAALTRMPSSVPLRAPAQFTFSPHGGPIYSVSCSPFHRNLFLSAGTDGHVHLYSMLQAPPLTSLQ LSLKYLFAVRWSPVRPLVFAAASGKGDVQLFDLQKSSQKPTVLIKQTQDESPVYCLEFNSQQTQLLAAGDAQGTVKVWQLSTEFTEQGPR -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SLC25A25-WDR34 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SLC25A25-WDR34 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SLC25A25-WDR34 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
