
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SLC9A8-B4GALT5 (FusionGDB2 ID:83558) |
Fusion Gene Summary for SLC9A8-B4GALT5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SLC9A8-B4GALT5 | Fusion gene ID: 83558 | Hgene | Tgene | Gene symbol | SLC9A8 | B4GALT5 | Gene ID | 23315 | 9334 |
| Gene name | solute carrier family 9 member A8 | beta-1,4-galactosyltransferase 5 | |
| Synonyms | NHE-8|NHE8 | B4Gal-T5|BETA4-GALT-IV|beta4Gal-T5|beta4GalT-V|gt-V | |
| Cytomap | 20q13.13 | 20q13.13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | sodium/hydrogen exchanger 8Na(+)/H(+) exchanger 8solute carrier family 9 (sodium/hydrogen exchanger)solute carrier family 9 (sodium/hydrogen exchanger), member 8solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 | beta-1,4-galactosyltransferase 5UDP-Gal:beta-GlcNAc beta-1,4-galactosyltransferase 5UDP-Gal:betaGlcNAc beta 1,4-galactosyltransferase, polypeptide 5UDP-galactose:beta-N-acetylglucosamine beta-1,4-galactosyltransferase 5beta-1,4-GalT IIbeta-1,4-GalT I | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O43286 | |
| Ensembl transtripts involved in fusion gene | ENST00000539601, ENST00000541138, ENST00000361573, ENST00000417961, ENST00000490250, | ENST00000371711, | |
| Fusion gene scores | * DoF score | 9 X 7 X 6=378 | 12 X 9 X 5=540 |
| # samples | 9 | 13 | |
| ** MAII score | log2(9/378*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/540*10)=-2.05444778402238 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SLC9A8 [Title/Abstract] AND B4GALT5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SLC9A8(48431726)-B4GALT5(48273239), # samples:2 B4GALT5(48273104)-SLC9A8(48430005), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across SLC9A8 (5'-gene) Fusion gene breakpoints across SLC9A8 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
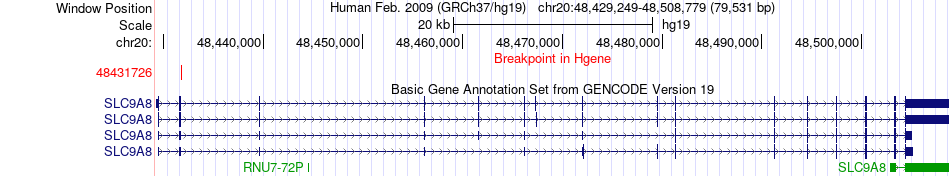 |
 Fusion gene breakpoints across B4GALT5 (3'-gene) Fusion gene breakpoints across B4GALT5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
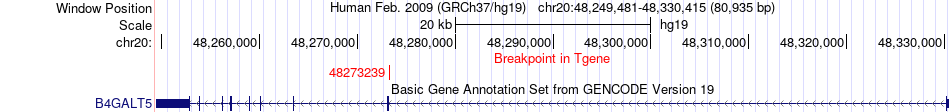 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A1L0-01A | SLC9A8 | chr20 | 48431726 | - | B4GALT5 | chr20 | 48273239 | - |
| ChimerDB4 | SARC | TCGA-DX-A1L0-01A | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - |
Top |
Fusion Gene ORF analysis for SLC9A8-B4GALT5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5UTR-3CDS | ENST00000539601 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - |
| 5UTR-3CDS | ENST00000541138 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - |
| In-frame | ENST00000361573 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - |
| In-frame | ENST00000417961 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - |
| intron-3CDS | ENST00000490250 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000417961 | SLC9A8 | chr20 | 48431726 | + | ENST00000371711 | B4GALT5 | chr20 | 48273239 | - | 4837 | 418 | 63 | 1469 | 468 |
| ENST00000361573 | SLC9A8 | chr20 | 48431726 | + | ENST00000371711 | B4GALT5 | chr20 | 48273239 | - | 4669 | 250 | 42 | 1301 | 419 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000417961 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - | 0.000629505 | 0.9993705 |
| ENST00000361573 | ENST00000371711 | SLC9A8 | chr20 | 48431726 | + | B4GALT5 | chr20 | 48273239 | - | 0.000611568 | 0.99938846 |
Top |
Fusion Genomic Features for SLC9A8-B4GALT5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SLC9A8-B4GALT5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr20:48431726/chr20:48273239) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | B4GALT5 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Catalyzes the synthesis of lactosylceramide (LacCer) via the transfer of galactose from UDP-galactose to glucosylceramide (GlcCer) (PubMed:24498430). LacCer is the starting point in the biosynthesis of all gangliosides (membrane-bound glycosphingolipids) which play pivotal roles in the CNS including neuronal maturation and axonal and myelin formation (By similarity). Plays a role in the glycosylation of BMPR1A and regulation of its protein stability (By similarity). Essential for extraembryonic development during early embryogenesis (By similarity). {ECO:0000250|UniProtKB:Q9JMK0, ECO:0000269|PubMed:24498430}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 169_173 | 38 | 389.0 | Region | UDP-alpha-D-galactose binding | |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 208_210 | 38 | 389.0 | Region | UDP-alpha-D-galactose binding | |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 235_236 | 38 | 389.0 | Region | UDP-alpha-D-galactose binding | |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 298_301 | 38 | 389.0 | Region | N-acetyl-D-glucosamine binding | |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 329_330 | 38 | 389.0 | Region | UDP-alpha-D-galactose binding | |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 36_388 | 38 | 389.0 | Topological domain | Lumenal |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 123_143 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 156_176 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 191_211 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 264_284 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 311_331 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 354_374 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 379_399 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 417_437 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 451_471 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 60_80 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000361573 | + | 2 | 16 | 84_104 | 69 | 582.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 123_143 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 156_176 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 191_211 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 264_284 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 311_331 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 354_374 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 379_399 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 417_437 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 451_471 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 60_80 | 69 | 598.0 | Transmembrane | Helical |
| Hgene | SLC9A8 | chr20:48431726 | chr20:48273239 | ENST00000417961 | + | 2 | 16 | 84_104 | 69 | 598.0 | Transmembrane | Helical |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 1_14 | 38 | 389.0 | Topological domain | Cytoplasmic | |
| Tgene | B4GALT5 | chr20:48431726 | chr20:48273239 | ENST00000371711 | 0 | 9 | 15_35 | 38 | 389.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for SLC9A8-B4GALT5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >83558_83558_1_SLC9A8-B4GALT5_SLC9A8_chr20_48431726_ENST00000361573_B4GALT5_chr20_48273239_ENST00000371711_length(transcript)=4669nt_BP=250nt GCCCCGCGGCTCCGAACTCGGTGGTCCTGGAAGCTCCGCAGGATGGGGGAGAAGATGGCGGAAGAGGAGAGGTTCCCCAATACAACTCAT GAGGGTTTCAATGTCACCCTCCACACCACCCTGGTTGTCACGACGAAACTGGTGCTCCCGACCCCTGGCAAGCCCATCCTCCCCGTGCAG ACAGGGGAGCAGGCCCAGCAAGAGGAGCAGTCCAGCGGCATGACCATTTTCTTCAGCCTCCTTGTCCTAGTGAACACCTACCTCTTCATG ATGCAAGCCCAAGGCATTCTGATCCGGGACAACGTGAGAACAATCGGTGCTCAGGTTTATGAGCAGGTGCTTCGGAGTGCTTATGCCAAG AGGAACAGCAGTGTAAATGACTCAGATTATCCTCTTGACTTGAACCACAGTGAAACCTTCCTGCAAACTACAACATTTCTTCCTGAAGAC TTCACCTACTTTGCAAACCATACCTGCCCTGAAAGACTCCCTTCCATGAAGGGCCCAATAGACATAAACATGAGTGAAATTGGAATGGAT TACATTCATGAACTCTTCTCCAAAGACCCAACCATCAAGCTCGGAGGTCACTGGAAGCCTTCTGATTGCATGCCTCGGTGGAAGGTGGCG ATCCTTATCCCCTTCCGGAACCGCCACGAGCACCTCCCAGTCCTGTTCAGACACCTGCTTCCCATGCTCCAGCGCCAGCGCTTGCAGTTT GCATTTTATGTGGTTGAACAAGTTGGTACCCAACCCTTTAATCGAGCCATGCTTTTCAACGTTGGCTTTCAAGAGGCAATGAAAGACTTG GATTGGGACTGTTTGATTTTTCATGATGTAGATCACATACCGGAAAGTGATCGCAACTATTATGGATGTGGACAGATGCCGAGGCATTTT GCAACCAAATTGGATAAGTATATGTATCTGCTTCCTTATACCGAGTTCTTTGGCGGAGTGAGTGGCTTAACAGTGGAACAATTTCGGAAA ATCAATGGCTTTCCTAATGCTTTCTGGGGTTGGGGTGGAGAAGATGACGACCTCTGGAACAGAGTACAGAATGCAGGCTATTCTGTGAGC CGGCCAGAGGGTGACACAGGAAAGTACAAGTCCATTCCTCATCACCATCGAGGAGAAGTCCAGTTTCTTGGAAGGTATGCTCTGCTGAGG AAGTCAAAAGAACGGCAAGGGCTGGATGGCCTCAACAACCTGAACTACTTTGCAAACATCACATACGACGCCTTGTATAAAAACATAACT GTCAACCTGACACCCGAGCTGGCTCAGGTGAACGAGTACTGAGAGGAGAGAATGTACGTTTGCTTTACCCACCGCCACCAAGAAAGCAGT CCGATGAGATTTTTTTTTGGAGGGGGGAGGGTCTACACAGCAAGAGAACAGAAATACTGTGTCTCATGAAGGATCACAGAGTTCAGGGGG AAAATGTGACAGCACACGCACAAACGCCTTCACTGGATCAGCCGCTGGAACTGAGGGAGTGAGCTTGGGGACTTCCTTCGTCAGCACTGG CTTTCTGTTTTCACAAGACAGACGTCTGTCCCGCTGCTCTCTCCCCATCTCCTACCCCACATCCTGTCTTAGCCGCAGTCTCCAGAACCC ATGATGAACTGTGATCTGCCGTGGTCCTGCCGTGGTCCTGCCGTGGAGCCTGTCCCTACACATGACCTTGGAGCCTCTTGGCCTTCAGAG CAGAGGCAAACCCACCACAGGGCAGCTGCGTTTTAGGAAGAGCAAATGAAACTCCACACCATTCTTCTAGATCTCTGGTGTTCTCTTTGG TTTCATTTTTTTAAAAAATTACCTTCTTTGGGTGGGGATTGAGGGTGGAGGGGAGGGTGTTTGGGAAAGATAAATAGACATAAATATATA ACAATCACTTCTTGAAGAAGTATAATTGTAAATAAGCCATGTAAAATGCCTTTTTAAAATTTAATTTTCTAGCTGGCTCCAATTCAAATT GAGGATTTATGTATTAGGCCACTTACTTGGTTGGCAAGTGCAGGAACTCAGTTAAAATGCAGTTGAAGAATGTCATCTCCCGAATTGCTG TCACTTTGGCGAGGGAGTGGATATAGGGCATGTCACAAAAGAACAAAATAACCCGACCTTTATTGCTGGGAGCTGGCTTCTGTCCCTTTC TTCCCCCCCCCACGAGTCTTGCCCTTGACTTCTGCTCTGGATTCACTCTTCCCTGTCGGCCGCGCATGTGCTCATCCCACTCTCCGCTAA GCGGGAGGCTGCTGTTAGAGCAGGCTGCTTCCTGCCTAAAGCAGGCCCTTCGGGGCTCGCTGCACACACATCTCTGGCTCTCCAGGCTTC GTGTTCTGTCTTTTCATCAGCATGGCGGGGCGGGGGGCGGGGGGCGGGGGTGTGTATGGGAATCCCTCCCCCTCTTACTTTTTCTCTTGT GGAACTTGGCCACAGTTTCTGAACAATGTGCCTACATTACCAGCTGGCTTCAGTGATTCCTCTGTGTCCCTTTTTGGTTTCTGGAAAGAT TCTTTGTCAACATTAGTAACTGATACATAGAACCAAGGAGCACTCAAATAGGGAGCCAGGAGCCAGGGAGCTGGTGACACTTGTGTGCTG TGGGGCAGCTGGGATCCAGGTAAGACCGGATTGAAGCTTTGAAATTAGACTAACAAAGCTCCAGACAGCAAGAGCCCAGGTGCACTGCTC ACACCCCCACCTGCATTTTGAAGTCATATTATTTTTTGTTTTGTTTTTTAAGACGGTCTGGCTCTGTCGCCTAAGCTGGAGTGTGGTGGC ACGATCACAGCTCACTGCAGCCTCCATCTCCTAGGCTCAAGCCATTTTCCCACCTCAGCCTCCCGAGTAGCTGGGACTACAGGTGCACAC CACCACACCTGGCTAATTTTTTGTATTTTTAGTAGAGACAGGGGTTTCTTCCATGTTGCCCAGGCTGGTCTCGAACTCCTGGACTCAAGC AATCCGCCCACCTTGACTTCCCAAAGTGCTGGGATTATGGGCGGGTGTGAGCCATTGCGCCCAGCCTTGAAGTCATGTTCTAAATTGTAT TTGAATTTGTGCCTCTTTGTTTTTCCCCAAACCAAAGCCCTCAAATTGTAGTCTCTGTCGGCTTCTGCAGAATTCTGGAAAATGCCAGTT TTCCTCCCCCGCCCTTGTTTTCCATAAAACATATTTATATATTGTGATGAGGAGTACTTTCTGAAGAGTACTTCGTATTTTTTTTTAATT GCCTTGTTTGCCTTCAACTTCCTTGATTTTCATAGTTTACATGGGTGTGTGTAGGGGTGTGTGTGTGTATGTGTGTGGGTTAGGGCTTTT TTCGTTGCATGTGATGGTTCTGTGGACATATGATCCCCACAAACTGTGGGAGTGATTGGCCAGGCCTTGTTTTGTTTGTTTGTTTGTTTG TGTTTTTGTTCTTTTGAAGAATAGAGTGGTATTTAGAAAATAAATTGCATTGCAAAGCTCTTATCGGCTCATATGAGAGAGCAGGTTCCT GCCCTTGAAAATGCCGGTAAGCTATAGCATATGTTTTTTAAGACTTAAGCATTTCATGCTTTAAAATACCTTCACAAGTGAACATTACAC ACAGAAGTTCATTTGGTTTTCCTTTGTTTTATGGTGCATATAGCAATAAAGACCCCCCTCCACCCTGCAACCCCCATCCCCCACCGGGCC TTTGTCCCTGCCTTGGCTTTTCTCCCCTTCTCATTCTCCTCTCCCCTTTCCTCACTGAAGGCTGTGAGTTGCTTTCAATGTGACAACACT ATGATGTCATTTGGAAGGATTTGCCAGGACAGACTGATTCTGAGTCCTGGGTGCCGTATGTGTATGCGGCAGTGTTGTCAGGCGATCTTG TTTGAAGCTCTATGTTGCCATAATTACCATCAAGTACACACTGTTGGCAAAAGGCTAACACCTGACTTTAGAAAATGCTGATTTGAGAAC AAAAGGAAAGGTCTTTTTTCACTGCTTAAAGTGGGGTCACTTTGATACCTTTGCGGTCATGTCTGTGTCTGATGAGTGTAGAATCTCTGG ATGTGCACTGTCAGTCATGTGTCCACCAGGCCTCGAATATCATATGGGAAATGTCATAGTTAAAAACGTACAGCCAGGCCCGTGTGCTGT TAATAGTGTGAAATTGTCATGTTAAAAAAAAAAACAGGAACCAAATGTGACCTTGTGCATATATTGGTAGCTGAAAATCTTCAAGGCTAC TGATGGGTGGCCCCTTAATCTTGTCTTTGATTGCTGTGTGCAGGGAAAGGTGTCCCCGTTTGTTCATGCTGTTTTGGGGGGTGGGGGGGT ATTTGCAAGAATACTCATTTTGACATAATAGGTCCTCTTGTCAGAGATCCTCTACCACAGACATTAATAGCTGAGCAGGAGCCACATGGA TTGATTGTATCCACTCACCATTGACGATGGCATTGAGCGTAGCTAGCTTATTTCCATCACTACGTGTTTTTGAGCTTGCTCTTACGTTTT AAGAGGTGCCAGGGGTACATTTTTGCACTGAAATCTAAAGATGTTTTAAAAAACACTTTTCACAAAAATAGTCCTTTGTCATTACATTAT >83558_83558_1_SLC9A8-B4GALT5_SLC9A8_chr20_48431726_ENST00000361573_B4GALT5_chr20_48273239_ENST00000371711_length(amino acids)=419AA_BP=65 MGEKMAEEERFPNTTHEGFNVTLHTTLVVTTKLVLPTPGKPILPVQTGEQAQQEEQSSGMTIFFSLLVLVNTYLFMMQAQGILIRDNVRT IGAQVYEQVLRSAYAKRNSSVNDSDYPLDLNHSETFLQTTTFLPEDFTYFANHTCPERLPSMKGPIDINMSEIGMDYIHELFSKDPTIKL GGHWKPSDCMPRWKVAILIPFRNRHEHLPVLFRHLLPMLQRQRLQFAFYVVEQVGTQPFNRAMLFNVGFQEAMKDLDWDCLIFHDVDHIP ESDRNYYGCGQMPRHFATKLDKYMYLLPYTEFFGGVSGLTVEQFRKINGFPNAFWGWGGEDDDLWNRVQNAGYSVSRPEGDTGKYKSIPH -------------------------------------------------------------- >83558_83558_2_SLC9A8-B4GALT5_SLC9A8_chr20_48431726_ENST00000417961_B4GALT5_chr20_48273239_ENST00000371711_length(transcript)=4837nt_BP=418nt CCGAAAGCCCCGGGAGAGACTAAGAAGCAATCCTCCCACGCGCTTTCTCCCACCCTCGGGCCACTGAGACGGAGGGACAGAGGGCCGCCC TCGCGCGGCCGAGGCCCCGCCTCCCGCTCGCCCGCCCGCGCCTCCAGCGGAAGCCGGAAGCAAAAGCGGGTCCTGCTAGCCCCGCGGCTC CGAACTCGGTGGTCCTGGAAGCTCCGCAGGATGGGGGAGAAGATGGCGGAAGAGGAGAGGTTCCCCAATACAACTCATGAGGGTTTCAAT GTCACCCTCCACACCACCCTGGTTGTCACGACGAAACTGGTGCTCCCGACCCCTGGCAAGCCCATCCTCCCCGTGCAGACAGGGGAGCAG GCCCAGCAAGAGGAGCAGTCCAGCGGCATGACCATTTTCTTCAGCCTCCTTGTCCTAGTGAACACCTACCTCTTCATGATGCAAGCCCAA GGCATTCTGATCCGGGACAACGTGAGAACAATCGGTGCTCAGGTTTATGAGCAGGTGCTTCGGAGTGCTTATGCCAAGAGGAACAGCAGT GTAAATGACTCAGATTATCCTCTTGACTTGAACCACAGTGAAACCTTCCTGCAAACTACAACATTTCTTCCTGAAGACTTCACCTACTTT GCAAACCATACCTGCCCTGAAAGACTCCCTTCCATGAAGGGCCCAATAGACATAAACATGAGTGAAATTGGAATGGATTACATTCATGAA CTCTTCTCCAAAGACCCAACCATCAAGCTCGGAGGTCACTGGAAGCCTTCTGATTGCATGCCTCGGTGGAAGGTGGCGATCCTTATCCCC TTCCGGAACCGCCACGAGCACCTCCCAGTCCTGTTCAGACACCTGCTTCCCATGCTCCAGCGCCAGCGCTTGCAGTTTGCATTTTATGTG GTTGAACAAGTTGGTACCCAACCCTTTAATCGAGCCATGCTTTTCAACGTTGGCTTTCAAGAGGCAATGAAAGACTTGGATTGGGACTGT TTGATTTTTCATGATGTAGATCACATACCGGAAAGTGATCGCAACTATTATGGATGTGGACAGATGCCGAGGCATTTTGCAACCAAATTG GATAAGTATATGTATCTGCTTCCTTATACCGAGTTCTTTGGCGGAGTGAGTGGCTTAACAGTGGAACAATTTCGGAAAATCAATGGCTTT CCTAATGCTTTCTGGGGTTGGGGTGGAGAAGATGACGACCTCTGGAACAGAGTACAGAATGCAGGCTATTCTGTGAGCCGGCCAGAGGGT GACACAGGAAAGTACAAGTCCATTCCTCATCACCATCGAGGAGAAGTCCAGTTTCTTGGAAGGTATGCTCTGCTGAGGAAGTCAAAAGAA CGGCAAGGGCTGGATGGCCTCAACAACCTGAACTACTTTGCAAACATCACATACGACGCCTTGTATAAAAACATAACTGTCAACCTGACA CCCGAGCTGGCTCAGGTGAACGAGTACTGAGAGGAGAGAATGTACGTTTGCTTTACCCACCGCCACCAAGAAAGCAGTCCGATGAGATTT TTTTTTGGAGGGGGGAGGGTCTACACAGCAAGAGAACAGAAATACTGTGTCTCATGAAGGATCACAGAGTTCAGGGGGAAAATGTGACAG CACACGCACAAACGCCTTCACTGGATCAGCCGCTGGAACTGAGGGAGTGAGCTTGGGGACTTCCTTCGTCAGCACTGGCTTTCTGTTTTC ACAAGACAGACGTCTGTCCCGCTGCTCTCTCCCCATCTCCTACCCCACATCCTGTCTTAGCCGCAGTCTCCAGAACCCATGATGAACTGT GATCTGCCGTGGTCCTGCCGTGGTCCTGCCGTGGAGCCTGTCCCTACACATGACCTTGGAGCCTCTTGGCCTTCAGAGCAGAGGCAAACC CACCACAGGGCAGCTGCGTTTTAGGAAGAGCAAATGAAACTCCACACCATTCTTCTAGATCTCTGGTGTTCTCTTTGGTTTCATTTTTTT AAAAAATTACCTTCTTTGGGTGGGGATTGAGGGTGGAGGGGAGGGTGTTTGGGAAAGATAAATAGACATAAATATATAACAATCACTTCT TGAAGAAGTATAATTGTAAATAAGCCATGTAAAATGCCTTTTTAAAATTTAATTTTCTAGCTGGCTCCAATTCAAATTGAGGATTTATGT ATTAGGCCACTTACTTGGTTGGCAAGTGCAGGAACTCAGTTAAAATGCAGTTGAAGAATGTCATCTCCCGAATTGCTGTCACTTTGGCGA GGGAGTGGATATAGGGCATGTCACAAAAGAACAAAATAACCCGACCTTTATTGCTGGGAGCTGGCTTCTGTCCCTTTCTTCCCCCCCCCA CGAGTCTTGCCCTTGACTTCTGCTCTGGATTCACTCTTCCCTGTCGGCCGCGCATGTGCTCATCCCACTCTCCGCTAAGCGGGAGGCTGC TGTTAGAGCAGGCTGCTTCCTGCCTAAAGCAGGCCCTTCGGGGCTCGCTGCACACACATCTCTGGCTCTCCAGGCTTCGTGTTCTGTCTT TTCATCAGCATGGCGGGGCGGGGGGCGGGGGGCGGGGGTGTGTATGGGAATCCCTCCCCCTCTTACTTTTTCTCTTGTGGAACTTGGCCA CAGTTTCTGAACAATGTGCCTACATTACCAGCTGGCTTCAGTGATTCCTCTGTGTCCCTTTTTGGTTTCTGGAAAGATTCTTTGTCAACA TTAGTAACTGATACATAGAACCAAGGAGCACTCAAATAGGGAGCCAGGAGCCAGGGAGCTGGTGACACTTGTGTGCTGTGGGGCAGCTGG GATCCAGGTAAGACCGGATTGAAGCTTTGAAATTAGACTAACAAAGCTCCAGACAGCAAGAGCCCAGGTGCACTGCTCACACCCCCACCT GCATTTTGAAGTCATATTATTTTTTGTTTTGTTTTTTAAGACGGTCTGGCTCTGTCGCCTAAGCTGGAGTGTGGTGGCACGATCACAGCT CACTGCAGCCTCCATCTCCTAGGCTCAAGCCATTTTCCCACCTCAGCCTCCCGAGTAGCTGGGACTACAGGTGCACACCACCACACCTGG CTAATTTTTTGTATTTTTAGTAGAGACAGGGGTTTCTTCCATGTTGCCCAGGCTGGTCTCGAACTCCTGGACTCAAGCAATCCGCCCACC TTGACTTCCCAAAGTGCTGGGATTATGGGCGGGTGTGAGCCATTGCGCCCAGCCTTGAAGTCATGTTCTAAATTGTATTTGAATTTGTGC CTCTTTGTTTTTCCCCAAACCAAAGCCCTCAAATTGTAGTCTCTGTCGGCTTCTGCAGAATTCTGGAAAATGCCAGTTTTCCTCCCCCGC CCTTGTTTTCCATAAAACATATTTATATATTGTGATGAGGAGTACTTTCTGAAGAGTACTTCGTATTTTTTTTTAATTGCCTTGTTTGCC TTCAACTTCCTTGATTTTCATAGTTTACATGGGTGTGTGTAGGGGTGTGTGTGTGTATGTGTGTGGGTTAGGGCTTTTTTCGTTGCATGT GATGGTTCTGTGGACATATGATCCCCACAAACTGTGGGAGTGATTGGCCAGGCCTTGTTTTGTTTGTTTGTTTGTTTGTGTTTTTGTTCT TTTGAAGAATAGAGTGGTATTTAGAAAATAAATTGCATTGCAAAGCTCTTATCGGCTCATATGAGAGAGCAGGTTCCTGCCCTTGAAAAT GCCGGTAAGCTATAGCATATGTTTTTTAAGACTTAAGCATTTCATGCTTTAAAATACCTTCACAAGTGAACATTACACACAGAAGTTCAT TTGGTTTTCCTTTGTTTTATGGTGCATATAGCAATAAAGACCCCCCTCCACCCTGCAACCCCCATCCCCCACCGGGCCTTTGTCCCTGCC TTGGCTTTTCTCCCCTTCTCATTCTCCTCTCCCCTTTCCTCACTGAAGGCTGTGAGTTGCTTTCAATGTGACAACACTATGATGTCATTT GGAAGGATTTGCCAGGACAGACTGATTCTGAGTCCTGGGTGCCGTATGTGTATGCGGCAGTGTTGTCAGGCGATCTTGTTTGAAGCTCTA TGTTGCCATAATTACCATCAAGTACACACTGTTGGCAAAAGGCTAACACCTGACTTTAGAAAATGCTGATTTGAGAACAAAAGGAAAGGT CTTTTTTCACTGCTTAAAGTGGGGTCACTTTGATACCTTTGCGGTCATGTCTGTGTCTGATGAGTGTAGAATCTCTGGATGTGCACTGTC AGTCATGTGTCCACCAGGCCTCGAATATCATATGGGAAATGTCATAGTTAAAAACGTACAGCCAGGCCCGTGTGCTGTTAATAGTGTGAA ATTGTCATGTTAAAAAAAAAAACAGGAACCAAATGTGACCTTGTGCATATATTGGTAGCTGAAAATCTTCAAGGCTACTGATGGGTGGCC CCTTAATCTTGTCTTTGATTGCTGTGTGCAGGGAAAGGTGTCCCCGTTTGTTCATGCTGTTTTGGGGGGTGGGGGGGTATTTGCAAGAAT ACTCATTTTGACATAATAGGTCCTCTTGTCAGAGATCCTCTACCACAGACATTAATAGCTGAGCAGGAGCCACATGGATTGATTGTATCC ACTCACCATTGACGATGGCATTGAGCGTAGCTAGCTTATTTCCATCACTACGTGTTTTTGAGCTTGCTCTTACGTTTTAAGAGGTGCCAG GGGTACATTTTTGCACTGAAATCTAAAGATGTTTTAAAAAACACTTTTCACAAAAATAGTCCTTTGTCATTACATTATTTACTCATGTGT >83558_83558_2_SLC9A8-B4GALT5_SLC9A8_chr20_48431726_ENST00000417961_B4GALT5_chr20_48273239_ENST00000371711_length(amino acids)=468AA_BP=114 MRRRDRGPPSRGRGPASRSPARASSGSRKQKRVLLAPRLRTRWSWKLRRMGEKMAEEERFPNTTHEGFNVTLHTTLVVTTKLVLPTPGKP ILPVQTGEQAQQEEQSSGMTIFFSLLVLVNTYLFMMQAQGILIRDNVRTIGAQVYEQVLRSAYAKRNSSVNDSDYPLDLNHSETFLQTTT FLPEDFTYFANHTCPERLPSMKGPIDINMSEIGMDYIHELFSKDPTIKLGGHWKPSDCMPRWKVAILIPFRNRHEHLPVLFRHLLPMLQR QRLQFAFYVVEQVGTQPFNRAMLFNVGFQEAMKDLDWDCLIFHDVDHIPESDRNYYGCGQMPRHFATKLDKYMYLLPYTEFFGGVSGLTV EQFRKINGFPNAFWGWGGEDDDLWNRVQNAGYSVSRPEGDTGKYKSIPHHHRGEVQFLGRYALLRKSKERQGLDGLNNLNYFANITYDAL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SLC9A8-B4GALT5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SLC9A8-B4GALT5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SLC9A8-B4GALT5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
