
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SMDT1-NFAM1 (FusionGDB2 ID:84043) |
Fusion Gene Summary for SMDT1-NFAM1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SMDT1-NFAM1 | Fusion gene ID: 84043 | Hgene | Tgene | Gene symbol | SMDT1 | NFAM1 | Gene ID | 91689 | 150372 |
| Gene name | single-pass membrane protein with aspartate rich tail 1 | NFAT activating protein with ITAM motif 1 | |
| Synonyms | C22orf32|DDDD|EMRE | CNAIP | |
| Cytomap | 22q13.2 | 22q13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | essential MCU regulator, mitochondrialUPF0466 protein C22orf32, mitochondrialsingle-pass membrane protein with aspartate-rich tail 1, mitochondrial | NFAT activation molecule 1calcineurin/NFAT-activating ITAM-containing protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q9H4I9 | Q8NET5 | |
| Ensembl transtripts involved in fusion gene | ENST00000331479, | ENST00000329021, | |
| Fusion gene scores | * DoF score | 2 X 2 X 2=8 | 4 X 3 X 4=48 |
| # samples | 2 | 4 | |
| ** MAII score | log2(2/8*10)=1.32192809488736 | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SMDT1 [Title/Abstract] AND NFAM1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SMDT1(42475958)-NFAM1(42783084), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | NFAM1 | GO:0007165 | signal transduction | 12615919 |
| Tgene | NFAM1 | GO:0035556 | intracellular signal transduction | 12615919 |
| Tgene | NFAM1 | GO:0051091 | positive regulation of DNA-binding transcription factor activity | 12615919 |
 Fusion gene breakpoints across SMDT1 (5'-gene) Fusion gene breakpoints across SMDT1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
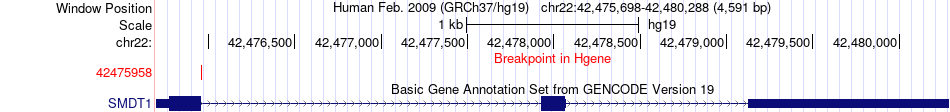 |
 Fusion gene breakpoints across NFAM1 (3'-gene) Fusion gene breakpoints across NFAM1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
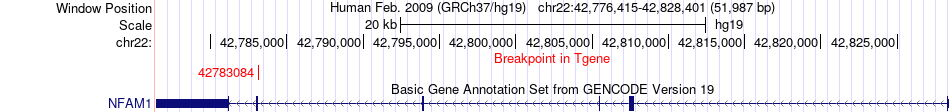 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-44-6147-01A | SMDT1 | chr22 | 42475958 | + | NFAM1 | chr22 | 42783084 | - |
Top |
Fusion Gene ORF analysis for SMDT1-NFAM1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000331479 | ENST00000329021 | SMDT1 | chr22 | 42475958 | + | NFAM1 | chr22 | 42783084 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000331479 | SMDT1 | chr22 | 42475958 | + | ENST00000329021 | NFAM1 | chr22 | 42783084 | - | 5161 | 260 | 1039 | 2100 | 353 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000331479 | ENST00000329021 | SMDT1 | chr22 | 42475958 | + | NFAM1 | chr22 | 42783084 | - | 0.43706408 | 0.56293595 |
Top |
Fusion Genomic Features for SMDT1-NFAM1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SMDT1-NFAM1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:42475958/chr22:42783084) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| SMDT1 | NFAM1 |
| FUNCTION: Essential regulatory subunit of the mitochondrial calcium uniporter complex (uniplex), a complex that mediates calcium uptake into mitochondria (PubMed:24231807, PubMed:26774479, PubMed:27099988). Required to bridge the calcium-sensing proteins MICU1 and MICU2 with the calcium-conducting subunit MCU (PubMed:24231807). Plays a central role in regulating the uniplex complex response to intracellular calcium signaling (PubMed:27099988). Acts by mediating activation of MCU and retention of MICU1 to the MCU pore, in order to ensure tight regulation of the uniplex complex and appropriate responses to intracellular calcium signaling (PubMed:27099988). {ECO:0000269|PubMed:24231807, ECO:0000269|PubMed:26774479, ECO:0000269|PubMed:27099988}. | FUNCTION: May function in immune system as a receptor which activates via the calcineurin/NFAT-signaling pathway the downstream cytokine gene promoters. Activates the transcription of IL-13 and TNF-alpha promoters. May be involved in the regulation of B-cell, but not T-cell, development. Overexpression activates downstream effectors without ligand binding or antibody cross-linking. {ECO:0000269|PubMed:12615919, ECO:0000269|PubMed:15143214}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SMDT1 | chr22:42475958 | chr22:42783084 | ENST00000331479 | + | 1 | 3 | 53_64 | 62 | 410.0 | Topological domain | Mitochondrial matrix |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SMDT1 | chr22:42475958 | chr22:42783084 | ENST00000331479 | + | 1 | 3 | 102_107 | 62 | 410.0 | Compositional bias | Poly-Asp |
| Hgene | SMDT1 | chr22:42475958 | chr22:42783084 | ENST00000331479 | + | 1 | 3 | 81_85 | 62 | 410.0 | Motif | GXXXX[G/A/S] |
| Hgene | SMDT1 | chr22:42475958 | chr22:42783084 | ENST00000331479 | + | 1 | 3 | 85_107 | 62 | 410.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SMDT1 | chr22:42475958 | chr22:42783084 | ENST00000331479 | + | 1 | 3 | 65_84 | 62 | 410.0 | Transmembrane | Helical |
| Tgene | NFAM1 | chr22:42475958 | chr22:42783084 | ENST00000329021 | 3 | 6 | 209_237 | 221 | 271.0 | Domain | ITAM | |
| Tgene | NFAM1 | chr22:42475958 | chr22:42783084 | ENST00000329021 | 3 | 6 | 50_150 | 221 | 271.0 | Domain | Note=Ig-like V-type | |
| Tgene | NFAM1 | chr22:42475958 | chr22:42783084 | ENST00000329021 | 3 | 6 | 185_270 | 221 | 271.0 | Topological domain | Cytoplasmic | |
| Tgene | NFAM1 | chr22:42475958 | chr22:42783084 | ENST00000329021 | 3 | 6 | 43_163 | 221 | 271.0 | Topological domain | Extracellular | |
| Tgene | NFAM1 | chr22:42475958 | chr22:42783084 | ENST00000329021 | 3 | 6 | 164_184 | 221 | 271.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for SMDT1-NFAM1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >84043_84043_1_SMDT1-NFAM1_SMDT1_chr22_42475958_ENST00000331479_NFAM1_chr22_42783084_ENST00000329021_length(transcript)=5161nt_BP=260nt GCTTTCTTCCCGAGGGCGGCACGAGGGCTGGGCGGTGGGGTGCGGGTGCCCGGGTGAGGGGCGGAGCTGGGGGCATGGCGTCCGGAGCGG CTCGCTGGCTAGTATTGGCACCCGTCAGGTCCGGGGCTCTCCGGAGCGGGCCTAGCTTGAGGAAAGATGGCGATGTCTCCGCCGCATGGA GCGGCTCAGGCCGGAGCCTGGTACCGTCGAGGTCAGTCATCGTTACCCGCAGCGGCGCCATTTTGCCCAAACCGGTGAAAGCTCTGCAGC GCCGCGAGACCGAGGTCTATGCCTGCATCGAGAATGAGGATGGCAGCTCACCCACCGCCAAGCAGAGCCCCCTCTCCCAGGAGAGACCGC ATAGATTCGAAGATGATGGCGAACTTAACCTGGTCTATGAAAATCTCTAGGATGGGCTCCACCGCTCATAGAGCTTGCCCTGGGTCAGAG GACCGGGGCAGCCCCTGCCACCAAAGGACTTGATCTGAGTTGGGAGTAAGGCCCCCAGGGACACCCCATCATTTCACCCTCACATTCAAG GCCCTTCCTGTCTTGGCCGGCCCCACTGCCCCCCCATTCTATGCCCCAACCACCAAGGCTTTCCCATCTTGGGGCCTTTGCCCAGGCTGT TCCTTCTGCCAGGCAGGCCCTTCCTCCCTTGGCCCAACCAGCAAAGCGCCCCTCAGTCTTCAAGGGCTTTCCTGGATGCCACCCTTGCCC AGAGCCCTGCTCACTGCCTCATGCTGACAGAGCAGCATGCTCTCTTGGCACCAGCCTGCACCTCAGCCAGTAGGGCAGCCTCTGAGCTGA GGGAGGACCCTGCAGGGGCGCCTAGCAGTGCTGTGTGGGGAGCCGAGGTATGGAGGAGCTGGGGAGCAGCTATGCAAATCCCTACCTACC CAGCTGGGCCAGAGGCACGAGGCAGCCAGGCAGGGTGGGAGCAGGGGCTGGCGTGGGAGGTCTGGGGGACAGACAAGGATGATGGACCCC TCAGAGGCAAGACTAGGATCAGGGACAGATCTGGAGGCCCAGGACATGGCTGAGAGGTGGTCAGCAGGCTGGGGGCATGGAGCAGAGAGG GCTTGGGTACAAGCTGGAGAGGAAGGAGATTCTGGCATGTTGGGGTGAGGGCAGAGGGACAGATGAGGGGAGGAGGGTGCTGGCTAAACA GGAACTGAATGAATGGCCTGAAGCTCCCTCTCCCAAGGGTTCCCACTCCCATTGGCCTGGCCCTGTACGTGCACTCTGTCATCTCCTTGG TCACCCTCTGCTCCAGGCCTTCTCCACCACAGGACCTCAGCTGTCCCTCCTTCTACCCAAGGTGGAGTCCCCCAAAGTCCCTTCTGTCTG GCCTATCCTGTGCCCACCACCTGTGGCTTCATCCAGCCCTCCTCCTCTATCTTTATCTGCCATAGGCCGCAGCCCTGGCCCCCTGTGGAT TGAGGTCCCAAGTGGTCCAGCTGTGGGGTTTGGTGCTGGGACCCTCAATGGCAGCCGGAAGGTCCCTCTGAGGCAGAAGCTCAAGCCCCA TGGGGACAGTGGCTCCCCAGTGCCTGGCCCTGAGCCATTTCTGCTGTTTCAGGTGCCCACATCACCCTCATTTCCTCCTCGCAGTAGCCC TGCACCGTGGCCAGAAACCCATCCACAGGCAGGCAGTGGCTGGGGTCCGGGTGCCCCGGGCTGTTGGGCTCTGCTTCCTCCCTCTGCTCC CCCATGTCTCCCTGAGTTCTCCCACCTTTGCTCATTTGTAAAATGGGCAGGTCAATCTTCCCACCCCACCGTGTGGGCTGCCCCAACCCC AAATGTGCCCAGGGAGGGGGGGGAACCCAAGGCACAGGCCCCGCCCCTGCCCCCAAGCAGCACTGGGCTACTGGCTCACAGAAGCACCTT TGGTTCTGCAGGTCATGCCCCTCCCTGCAGGGAAGGGAAAGCTGGTGGTAGAGGCTGGGCACAGACTGACGTCCAGAAGAAGTCCCTTGG CCCTGCTAGGGTGGCACCAGACTTCTCTGGTCCCACAGGACAGCCAGTGGCCTCCCAACTTGGTTCTGGGCCCCCGCCTGCAAGCACACA TGGCAAGGAGGGCTTTAGCAGGCTCACATAACCGAGTGGCAAGGCCCCTGCTCCCAGAGGCTGGATTTGAATGAACTCCGGGCCTGTTTG GGGAGGTGAAGCTGCCAGGTGTGCACAGGGGCCTCCCAGGCTCCTGGCTCCACCAGCCAAGGAGGTGGGGGCAAGGGTCAGGGCAATGGG GGACAATGCCGCTCAGGGTGCAGTGAGCTCCCCTGGGAAAACCGAAGCCAGCAAGACCAGGAGAGGGCCACCTCACTGGGCCTGCCTCCC CCACCTCCTGGGCCTCTCTGACCTCAGCTCCTAGACCCTGGACCCTTGGCTGGGACTCAGCATTTTCCAGTCTTTTTCAGGGGTAGACAG GGGAGCCTGGGTTAAACTTTACCACTGTGCCCTGGCCCAGGCATCACCAACACGGCCTCCCTGCAGCTGCCCTGCCACCGACAGACCCCG ATTAAACCCACACCTGGCCTTGGCCTCCCCTGCTCTATACCTTGCATGGTTTTTGCCTCACTTGGAATAAAATTCCAACTCCTCCCTGTG GCGTAGGAGGTTCTTGGGGGCTGACCCATCACCCCTCTGGTGGACAGAGACATTGCCAGCGTGGTGGTCGTCTGTGCTTGGTTCAGGGCA AGGCTGGGAGGCAGTGGGTGCTGAGGTCTTCAGTGAGCAAGGAGAGAGGCAGGGAGGAGGAGGAGGAGGAGGAGAGCACAGCATGAAAGG ACGGGACCCTAAAGGGCTTTGGCCCAAGGGAGGGTGGGGCAAGAATGTTCTGAGACTTCTGTGGGCAGCAAAGAGGACTGAGCACCTGTT CTCCAGCCCCCAGGACTGAGAGCCTGCAGAGACAAGCTGGCACAGGGCGAGGGGTGGCAGGGAACTTCAGGGTGGGAGGTGCATGGGGCA GGCGGGAGGAGGAAGAACAGAGAGGGGACAGAGAGGGTATGAATGGGAGCTCCTGGGGTGCAGGCTTGCTTTCCACTATGAATGGGTGGA GGAGGGAAGAGAAGAAAATTGCTTTTGAGACCCACCCAGGCTGGGTGGCCTCAGGGCCTCCCTTGAATCCCTGCTCCACCTCCCACAGCC TTGTGGACTTGGGCAAGTCACTGGCCACCTGAGCCTCAGTTTGCAGTGACAGTCAAGACAGCTCAGGCCAGGCGTAGTGGCTTATGCCTG TAATCCCAGCATGTTGGGAGGCCGAGGCGGGTGGATCACTTGAGGCCAGGAGTTTGAGACCAGCCTGGCCAACGTGGTGAAACCCCGTCT CTACTAAAAATACAAAAATTAGCCTGGTGTGGTGGCACGCACCTGTAGTCCCAGCTACTCGAGAGGCTAAGGCAGGAGAATCTCTTGAAC CTGGGAGACAGAGGTTGCAGTGAGCCGAAGTGGCACCACTGCACACCAGCCTGGGCTACAGAGCACGCTCTGTCCAAATAAACAAACAAA CAAACACTACTCAGTGTCTCAGAAAGGACAGAAGGGGACTCCCTGGCTCAGGCAGAAGCTCAGTGGGTGTGCATTTCCTTCAAGACTCGT CCTTCAGAAGGAATAGGCTTCTGGGGCAGTACAGAGGAGTCTAAGTGAGAGGCAGGCCTCCCTGGGAGCTGGGCCCAGGCTCTCTGCAGC CATGCCTTTAAGGAGTGCCTACGGGGTAGCACGATCAGATGGACAGGCCAGGGCTGCCCAGAGCTGGGGGTGGGAGCAAGGTAAGCAGAG AGGTGAGGGAAGGAGGGTGCTGGCTTAGCAGGAGCTGAATGAATGGGCCTGGAGGGGAAAAGCTAAGGGGGATGGGGTGGCTTGAGTGGA GTAGGTGAGGCCTAGGTGAAGGGGAGCTGTGGACCTTCCAGGACGGACGGATGTGAGCAGTCCTGAGTGGGGACTGGAGTTAGGGTGGGG AAGGGAAGCCCCAGGCAGCCCCTGCAGGCCCCTGCCCCTGGGCCCTACCGCAGCAGGGCAGCCCTGTCTCTGGGTCTCAGCTGATGCTTC CCTCTGCCCACTGGCCCCTGACCTGGTGGGATTTCCCACCATCTCTCCCGGCCCAGCCTGCCCTGCCCTCCTAGGTCTTCCCAACCTCCC CTCTGCAGAAACATCTCAGCCACGTCTGGCACCCCCAGGCATCCCTGCCTGCCTCTAGGAAGTTGACCCCTTCAATGGGTTTCAGCCTCC AACCCCACCCCCGCTTGACTCCTGCTCTGTCAGCACTTGCTTGAGAAAATCATCCTGGGAAACCAGCTTCTGGGAGCAGGGGCCTTGCCA CTCGGTTCTGCGAGCCTGCTGAAGCCCTCCTTGCCACGTGTGCTTGCAGGCGGGGGCCCAGAACTAAGCTAGGAGGCCACTGGCTGTCCT GTGGGACCAGGGAAGTCTGGTGCCACCCTAGCAGAGGGCCCATGGACTTCTTCTGGAGGTCAGTCTGTGCCCAGCCTCTACCACCAGTCC AGAACTTGCAGTGGGTGCGGTAAACGGTGGCCCCTCCTCAGCAAACATTTCCAGGGGATTCTGGCAGCAGAGTTTCCATGGGAACATGTG CTGGTTCTGCGCTGAACCCGGTGTGGCTGGGGTTCCCTTCACAGCCAATATTGCAGGAACTGAGCCTGGTGGCCTCGGCCAGGGTAGGCA GTGCTCTTTGGCCACCCAGTGTCACAAACCTGAACTCAAGTTACAGGCAAAACCTGCTTTTCCTTTTTCTTTCTCTCTCTCTCTCTTTTT TTTTTTGTTAAAGCACATTCTAATGTAAAGTCCATGCTCCATTCTAAAGATGCCTCTGGGATTTCAGGGATGGATTTTTATGTAGTTCAA TAGTTTTCTTGGCAGACAGCTTATGCAGGAGAGGTGGGCCGTTGGCTCAGTCCAGCCAGGCTGAGTGGCGTCACTCCATTCCTTGCAATG CCAGGGGATGTCATTTGTCCACCTGAGGGGGCTTCCCCAGTCTCCCTTCCCTGACCTGTGGTCAGAATGATGGCCTCAGCCTTGACTGAG CGCGGGTCAGGCCAGACTCTCTGAACTTGGTGTTGGCGTGTGTTTTGTGTGTGTCTGGAGGTGACATATGTGGGTGACAGAAGCATATGT >84043_84043_1_SMDT1-NFAM1_SMDT1_chr22_42475958_ENST00000331479_NFAM1_chr22_42783084_ENST00000329021_length(amino acids)=353AA_BP= MRGGQQAGGMEQRGLGYKLERKEILACWGEGRGTDEGRRVLAKQELNEWPEAPSPKGSHSHWPGPVRALCHLLGHPLLQAFSTTGPQLSL LLPKVESPKVPSVWPILCPPPVASSSPPPLSLSAIGRSPGPLWIEVPSGPAVGFGAGTLNGSRKVPLRQKLKPHGDSGSPVPGPEPFLLF QVPTSPSFPPRSSPAPWPETHPQAGSGWGPGAPGCWALLPPSAPPCLPEFSHLCSFVKWAGQSSHPTVWAAPTPNVPREGGEPKAQAPPL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SMDT1-NFAM1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | SMDT1 | chr22:42475958 | chr22:42783084 | ENST00000331479 | + | 1 | 3 | 1_52 | 62.0 | 410.0 | MAIP1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SMDT1-NFAM1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SMDT1-NFAM1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
