
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SNX6-ADAMTS12 (FusionGDB2 ID:85017) |
Fusion Gene Summary for SNX6-ADAMTS12 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SNX6-ADAMTS12 | Fusion gene ID: 85017 | Hgene | Tgene | Gene symbol | SNX6 | ADAMTS12 | Gene ID | 58533 | 81792 |
| Gene name | sorting nexin 6 | ADAM metallopeptidase with thrombospondin type 1 motif 12 | |
| Synonyms | MSTP010|TFAF2 | PRO4389 | |
| Cytomap | 14q13.1 | 5p13.3-p13.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | sorting nexin-6TRAF4-associated factor 2tumor necrosis factor receptor-associated factor 4(TRAF4)-associated factor 2 | A disintegrin and metalloproteinase with thrombospondin motifs 12ADAM-TS 12ADAM-TS12ADAMTS-12a disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motif, 12 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | P58397 | |
| Ensembl transtripts involved in fusion gene | ENST00000362031, ENST00000355110, ENST00000396526, ENST00000396534, | ENST00000504582, ENST00000515401, ENST00000352040, ENST00000504830, | |
| Fusion gene scores | * DoF score | 13 X 10 X 6=780 | 9 X 7 X 7=441 |
| # samples | 15 | 9 | |
| ** MAII score | log2(15/780*10)=-2.37851162325373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/441*10)=-2.29278174922785 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SNX6 [Title/Abstract] AND ADAMTS12 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SNX6(35099113)-ADAMTS12(33751653), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | SNX6 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway | 11279102 |
| Hgene | SNX6 | GO:0045892 | negative regulation of transcription, DNA-templated | 11279102|20830743 |
| Tgene | ADAMTS12 | GO:0030167 | proteoglycan catabolic process | 17895370 |
| Tgene | ADAMTS12 | GO:0032331 | negative regulation of chondrocyte differentiation | 22247065 |
| Tgene | ADAMTS12 | GO:0050727 | regulation of inflammatory response | 23019333 |
| Tgene | ADAMTS12 | GO:0051603 | proteolysis involved in cellular protein catabolic process | 16611630 |
| Tgene | ADAMTS12 | GO:0071773 | cellular response to BMP stimulus | 22247065 |
| Tgene | ADAMTS12 | GO:1901509 | regulation of endothelial tube morphogenesis | 17895370 |
| Tgene | ADAMTS12 | GO:1902203 | negative regulation of hepatocyte growth factor receptor signaling pathway | 17895370 |
| Tgene | ADAMTS12 | GO:1902548 | negative regulation of cellular response to vascular endothelial growth factor stimulus | 17895370 |
| Tgene | ADAMTS12 | GO:2001113 | negative regulation of cellular response to hepatocyte growth factor stimulus | 17895370 |
 Fusion gene breakpoints across SNX6 (5'-gene) Fusion gene breakpoints across SNX6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
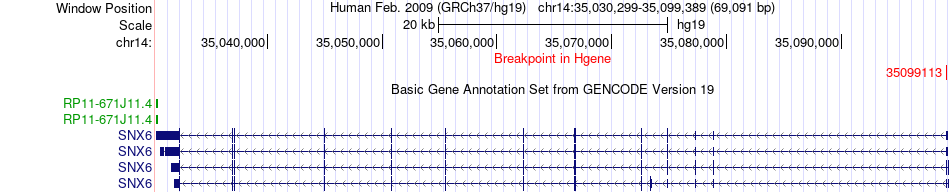 |
 Fusion gene breakpoints across ADAMTS12 (3'-gene) Fusion gene breakpoints across ADAMTS12 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
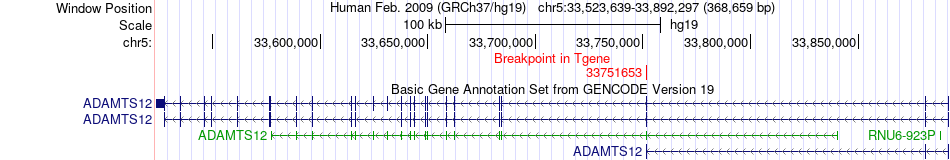 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | COAD | TCGA-CM-6172-01A | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
Top |
Fusion Gene ORF analysis for SNX6-ADAMTS12 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000362031 | ENST00000504582 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5CDS-5UTR | ENST00000362031 | ENST00000515401 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-3CDS | ENST00000355110 | ENST00000352040 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-3CDS | ENST00000355110 | ENST00000504830 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-3CDS | ENST00000396526 | ENST00000352040 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-3CDS | ENST00000396526 | ENST00000504830 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-3CDS | ENST00000396534 | ENST00000352040 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-3CDS | ENST00000396534 | ENST00000504830 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-5UTR | ENST00000355110 | ENST00000504582 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-5UTR | ENST00000355110 | ENST00000515401 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-5UTR | ENST00000396526 | ENST00000504582 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-5UTR | ENST00000396526 | ENST00000515401 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-5UTR | ENST00000396534 | ENST00000504582 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| 5UTR-5UTR | ENST00000396534 | ENST00000515401 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| In-frame | ENST00000362031 | ENST00000352040 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
| In-frame | ENST00000362031 | ENST00000504830 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000362031 | SNX6 | chr14 | 35099113 | - | ENST00000504830 | ADAMTS12 | chr5 | 33751653 | - | 8070 | 121 | 31 | 4416 | 1461 |
| ENST00000362031 | SNX6 | chr14 | 35099113 | - | ENST00000352040 | ADAMTS12 | chr5 | 33751653 | - | 4169 | 121 | 31 | 4161 | 1376 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000362031 | ENST00000504830 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - | 0.000298647 | 0.9997013 |
| ENST00000362031 | ENST00000352040 | SNX6 | chr14 | 35099113 | - | ADAMTS12 | chr5 | 33751653 | - | 0.001221543 | 0.9987784 |
Top |
Fusion Genomic Features for SNX6-ADAMTS12 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
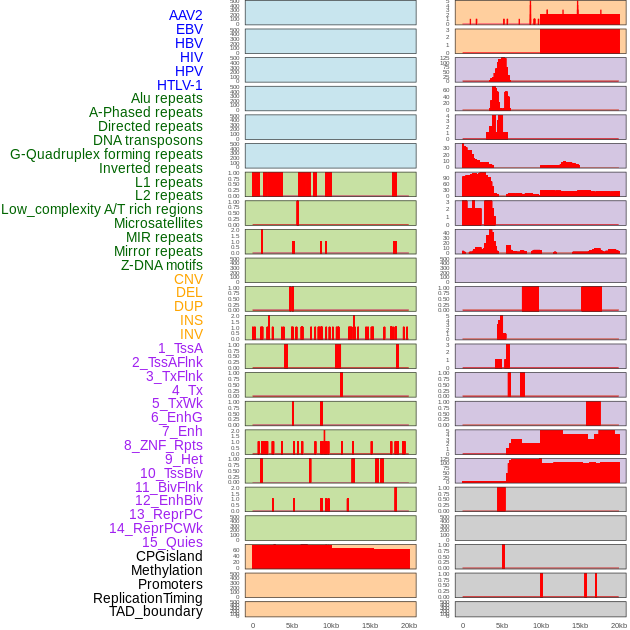 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SNX6-ADAMTS12 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr14:35099113/chr5:33751653) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ADAMTS12 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Metalloprotease that may play a role in the degradation of COMP. Cleaves also alpha-2 macroglobulin and aggregan. Has anti-tumorigenic properties. {ECO:0000269|PubMed:16611630, ECO:0000269|PubMed:17895370, ECO:0000269|PubMed:18485748}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 302_305 | 163 | 1510.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 597_700 | 163 | 1510.0 | Compositional bias | Note=Cys-rich | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 302_305 | 163 | 1595.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 597_700 | 163 | 1595.0 | Compositional bias | Note=Cys-rich | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 1313_1366 | 163 | 1510.0 | Domain | TSP type-1 5 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 1368_1422 | 163 | 1510.0 | Domain | TSP type-1 6 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 1423_1471 | 163 | 1510.0 | Domain | TSP type-1 7 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 1472_1532 | 163 | 1510.0 | Domain | TSP type-1 8 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 1535_1575 | 163 | 1510.0 | Domain | PLAC | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 246_456 | 163 | 1510.0 | Domain | Peptidase M12B | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 465_544 | 163 | 1510.0 | Domain | Note=Disintegrin | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 542_597 | 163 | 1510.0 | Domain | TSP type-1 1 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 823_883 | 163 | 1510.0 | Domain | TSP type-1 2 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 887_943 | 163 | 1510.0 | Domain | TSP type-1 3 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 944_997 | 163 | 1510.0 | Domain | TSP type-1 4 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 1313_1366 | 163 | 1595.0 | Domain | TSP type-1 5 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 1368_1422 | 163 | 1595.0 | Domain | TSP type-1 6 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 1423_1471 | 163 | 1595.0 | Domain | TSP type-1 7 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 1472_1532 | 163 | 1595.0 | Domain | TSP type-1 8 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 1535_1575 | 163 | 1595.0 | Domain | PLAC | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 246_456 | 163 | 1595.0 | Domain | Peptidase M12B | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 465_544 | 163 | 1595.0 | Domain | Note=Disintegrin | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 542_597 | 163 | 1595.0 | Domain | TSP type-1 1 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 823_883 | 163 | 1595.0 | Domain | TSP type-1 2 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 887_943 | 163 | 1595.0 | Domain | TSP type-1 3 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 944_997 | 163 | 1595.0 | Domain | TSP type-1 4 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 208_213 | 163 | 1510.0 | Motif | Cysteine switch | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 208_213 | 163 | 1595.0 | Motif | Cysteine switch | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 701_827 | 163 | 1510.0 | Region | Note=Spacer 1 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000352040 | 1 | 22 | 997_1316 | 163 | 1510.0 | Region | Note=Spacer 2 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 701_827 | 163 | 1595.0 | Region | Note=Spacer 1 | |
| Tgene | ADAMTS12 | chr14:35099113 | chr5:33751653 | ENST00000504830 | 1 | 24 | 997_1316 | 163 | 1595.0 | Region | Note=Spacer 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 203_406 | 0 | 291.0 | Domain | BAR |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 26_173 | 0 | 291.0 | Domain | PX |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 203_406 | 0 | 755.3333333333334 | Domain | BAR |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 26_173 | 0 | 755.3333333333334 | Domain | PX |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 100_106 | 0 | 291.0 | Region | Phosphatidylinositol bisphosphate binding |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 114_117 | 0 | 291.0 | Region | Phosphatidylinositol bisphosphate binding |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 182_199 | 0 | 291.0 | Region | Membrane-binding amphipathic helix |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 41_47 | 0 | 291.0 | Region | Phosphatidylinositol bisphosphate binding |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 100_106 | 0 | 755.3333333333334 | Region | Phosphatidylinositol bisphosphate binding |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 114_117 | 0 | 755.3333333333334 | Region | Phosphatidylinositol bisphosphate binding |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 182_199 | 0 | 755.3333333333334 | Region | Membrane-binding amphipathic helix |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 41_47 | 0 | 755.3333333333334 | Region | Phosphatidylinositol bisphosphate binding |
Top |
Fusion Gene Sequence for SNX6-ADAMTS12 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >85017_85017_1_SNX6-ADAMTS12_SNX6_chr14_35099113_ENST00000362031_ADAMTS12_chr5_33751653_ENST00000352040_length(transcript)=4169nt_BP=121nt CTGGCCGCCGGCTCCCTCCGGCAGCAGGGAGATGCGCGCCTGCGCCGGCCCTCGCCTCGGAGCAGCCATGATGGAAGGCCTGGACGACGG CCCGGACTTCCTCTCAGAAGAGGACCGCGGAACTGGATTTTTCCAACTACCACATGGAGACTTTTTCATTGAACCCGTGAAGAAGCATCC ACTGGTTGAGGGAGGGTACCACCCGCACATCGTTTACAGGAGGCAGAAAGTTCCAGAAACCAAGGAGCCAACCTGTGGATTAAAGGACAG TGTTAACATCTCCCAGAAGCAAGAGCTATGGCGGGAGAAGTGGGAGAGGCACAACTTGCCAAGCAGAAGCCTCTCTCGGCGTTCCATCAG CAAGGAGAGATGGGTGGAGACACTGGTGGTGGCCGACACAAAGATGATTGAATACCATGGGAGTGAGAATGTGGAGTCCTACATCCTCAC CATCATGAACATGGTCACTGGGTTGTTCCATAACCCAAGCATTGGCAATGCAATTCACATTGTTGTGGTTCGGCTCATTCTACTCGAAGA AGAAGAGCAAGGACTGAAAATAGTTCACCATGCAGAAAAGACACTGTCTAGCTTCTGCAAGTGGCAGAAGAGTATCAATCCCAAGAGTGA CCTCAATCCTGTTCATCACGACGTGGCTGTCCTTCTCACCAGAAAGGACATCTGTGCTGGTTTCAATCGCCCCTGCGAGACCCTGGGCCT GTCTCACCTTTCAGGAATGTGTCAGCCTCACCGCAGTTGTAACATCAATGAAGATTCGGGACTCCCTCTGGCTTTCACAATTGCCCATGA GCTAGGACACAGCTTCGGCATCCAGCATGATGGGAAAGAAAATGACTGTGAGCCTGTGGGCAGACATCCGTACATCATGTCCCGCCAGCT CCAGTACGATCCCACTCCGCTGACATGGTCCAAGTGCAGCGAGGAGTACATCACCCGCTTCTTGGACCGAGGCTGGGGGTTCTGTCTTGA TGACATACCTAAAAAGAAAGGCTTGAAGTCCAAGGTCATTGCCCCCGGAGTGATCTATGATGTTCACCACCAGTGCCAGCTACAATATGG ACCCAATGCTACCTTCTGCCAGGAAGTAGAAAACGTCTGCCAGACACTGTGGTGCTCCGTGAAGGGCTTTTGTCGCTCTAAGCTGGACGC TGCTGCAGATGGAACTCAATGTGGTGAGAAGAAGTGGTGTATGGCAGGCAAGTGCATCACAGTGGGGAAGAAACCAGAGAGCATTCCTGG AGGCTGGGGCCGCTGGTCACCCTGGTCCCACTGTTCCAGGACCTGTGGGGCTGGAGTCCAGAGCGCAGAGAGGCTCTGCAACAACCCCGA GCCAAAGTTTGGAGGGAAATATTGCACTGGAGAAAGAAAACGCTATCGCTTGTGCAACGTCCACCCCTGTCGCTCAGAGGCACCAACATT TCGGCAGATGCAGTGCAGTGAATTTGACACTGTTCCCTACAAGAATGAACTCTACCACTGGTTTCCCATTTTTAACCCAGGTTATGTTGA CATTGGGCTCATTCCAAAAGGAGCAAGGGACATAAGAGTGATGGAAATTGAGGGAGCTGGAAACTTCCTGGCCATCAGGAGTGAAGATCC TGAAAAATATTACCTGAATGGAGGGTTTATTATCCAGTGGAACGGGAACTATAAGCTGGCAGGGACTGTCTTTCAGTATGACAGGAAAGG AGACCTGGAAAAGCTGATGGCCACAGGTCCCACCAATGAGTCTGTGTGGATCCAGCTTCTATTCCAGGTGACTAACCCTGGCATCAAGTA TGAGTACACAATCCAGAAAGATGGCCTTGACAATGATGTTGAGCAGCAGATGTACTTCTGGCAGTACGGCCACTGGACAGAGTGCAGTGT GACCTGCGGGACAGGTATCCGCCGCCAAACTGCCCATTGCATAAAGAAGGGCCGCGGGATGGTGAAAGCTACATTCTGTGACCCAGAAAC ACAGCCCAATGGGAGACAGAAGAAGTGCCATGAAAAGGCTTGTCCACCCAGGTGGTGGGCAGGGGAGTGGGAAGCATGCTCGGCGACATG CGGGCCCCACGGGGAGAAGAAGCGAACCGTGCTGTGCATCCAGACCATGGTCTCTGACGAGCAGGCTCTCCCGCCCACAGACTGCCAGCA CCTGCTGAAGCCCAAGACCCTCCTTTCCTGCAACAGAGACATCCTGTGCCCCTCGGACTGGACAGTGGGCAACTGGAGTGAGTGTTCTGT TTCCTGTGGTGGTGGAGTGCGGATTCGCAGTGTCACATGTGCCAAGAACCATGATGAACCTTGCGATGTGACAAGGAAACCCAACAGCCG AGCTCTGTGTGGCCTCCAGCAATGCCCTTCTAGCCGGAGAGTTCTGAAACCAAACAAAGGCACTATTTCCAATGGAAAAAACCCACCAAC ACTAAAGCCCGTCCCTCCACCTACATCCAGGCCCAGAATGCTGACCACACCCACAGGGCCTGAGTCTATGAGCACAAGCACTCCAGCAAT CAGCAGCCCTAGTCCTACCACAGCCTCCAAAGAAGGAGACCTGGGTGGGAAACAGTGGCAAGATAGCTCAACCCAACCTGAGCTGAGCTC TCGCTATCTCATTTCCACTGGAAGCACTTCCCAGCCCATCCTCACTTCCCAATCCTTGAGCATTCAGCCAAGTGAGGAAAATGTTTCCAG TTCAGATACTGGTCCTACCTCGGAGGGAGGCCTTGTAGCTACAACAACAAGTGGTTCTGGCTTGTCATCTTCCCGCAACCCTATCACTTG GCCTGTGACTCCATTTTACAATACCTTGACCAAAGGTCCAGAAATGGAGATTCACAGTGGCTCAGGGGAAGAAAGAGAACAGCCTGAGGA CAAAGATGAAAGCAATCCTGTAATATGGACCAAGATCAGAGTACCTGGAAATGACGCTCCAGTGGAAAGTACAGAAATGCCACTTGCACC TCCACTAACACCAGATCTCAGCAGGGAGTCCTGGTGGCCACCCTTCAGCACAGTAATGGAAGGACTGCTCCCCAGCCAAAGGCCCACTAC TTCCGAAACTGGGACACCCAGAGTTGAGGGGATGGTTACTGAAAAGCCAGCCAACACTCTGCTCCCTCTGGGAGGAGACCACCAGCCAGA ACCCTCAGGAAAGACGGCAAACCGTAACCACCTGAAACTTCCAAACAACATGAACCAAACAAAAAGTTCTGAACCAGTCCTGACTGAGGA GGATGCAACAAGTCTGATTACTGAGGGCTTTTTGCTAAATGCCTCCAATTACAAGCAGCTCACAAACGGCCACGGCTCTGCACACTGGAT CGTCGGAAACTGGAGCGAGTGCTCCACCACATGTGGCCTGGGGGCCTACTGGAGAAGGGTGGAGTGCAGCACCCAGATGGATTCTGACTG TGCGGCCATCCAGAGACCTGACCCTGCAAAAAGATGCCACCTCCGTCCCTGTGCTGGCTGGAAAGTGGGAAACTGGAGCAAGTGCTCCAG AAACTGCAGTGGGGGCTTCAAGATACGCGAGATTCAGTGCGTGGACAGCCGGGACCACCGGAACCTGAGGCCATTTCACTGCCAGTTCCT GGCCGGCATTCCTCCCCCATTGAGCATGAGCTGTAACCCGGAGCCCTGTGAGGCGTGGCAGGTGGAGCCTTGGAGCCAGTGCTCCAGGTC CTGTGGAGGTGGAGTTCAGGAGAGAGGAGTGTTCTGTCCAGGAGGCCTCTGTGATTGGACAAAAAGACCCACATCCACCATGTCTTGCAA TGAGCACCTGTGCTGTCACTGGGCCACTGGGAACTGGGACCTGTGTTCCACTTCCTGTGGAGGTGGCTTTCAGAAGAGGACTGTCCAATG TGTGCCCTCAGAGGGCAATAAAACTGAAGACCAAGACCAATGTCTATGTGATCACAAACCCAGACCTCCAGAATTCAAAAAATGCAACCA GCAGGCCTGCAAGAAAAGTGCCGATTTACTTTGCACTAAGGACAAACTGTCAGCCAGTTTCTGCCAGACACTGAAAGCCATGAAGAAATG TTCTGTGCCCACCGTGAGGGCTGAGTGCTGCTTCTCGTGTCCCCAGACACACATCACACACACCCAAAGGCAAAGAAGGCAACGGTTGCT >85017_85017_1_SNX6-ADAMTS12_SNX6_chr14_35099113_ENST00000362031_ADAMTS12_chr5_33751653_ENST00000352040_length(amino acids)=1376AA_BP=30 MRACAGPRLGAAMMEGLDDGPDFLSEEDRGTGFFQLPHGDFFIEPVKKHPLVEGGYHPHIVYRRQKVPETKEPTCGLKDSVNISQKQELW REKWERHNLPSRSLSRRSISKERWVETLVVADTKMIEYHGSENVESYILTIMNMVTGLFHNPSIGNAIHIVVVRLILLEEEEQGLKIVHH AEKTLSSFCKWQKSINPKSDLNPVHHDVAVLLTRKDICAGFNRPCETLGLSHLSGMCQPHRSCNINEDSGLPLAFTIAHELGHSFGIQHD GKENDCEPVGRHPYIMSRQLQYDPTPLTWSKCSEEYITRFLDRGWGFCLDDIPKKKGLKSKVIAPGVIYDVHHQCQLQYGPNATFCQEVE NVCQTLWCSVKGFCRSKLDAAADGTQCGEKKWCMAGKCITVGKKPESIPGGWGRWSPWSHCSRTCGAGVQSAERLCNNPEPKFGGKYCTG ERKRYRLCNVHPCRSEAPTFRQMQCSEFDTVPYKNELYHWFPIFNPGYVDIGLIPKGARDIRVMEIEGAGNFLAIRSEDPEKYYLNGGFI IQWNGNYKLAGTVFQYDRKGDLEKLMATGPTNESVWIQLLFQVTNPGIKYEYTIQKDGLDNDVEQQMYFWQYGHWTECSVTCGTGIRRQT AHCIKKGRGMVKATFCDPETQPNGRQKKCHEKACPPRWWAGEWEACSATCGPHGEKKRTVLCIQTMVSDEQALPPTDCQHLLKPKTLLSC NRDILCPSDWTVGNWSECSVSCGGGVRIRSVTCAKNHDEPCDVTRKPNSRALCGLQQCPSSRRVLKPNKGTISNGKNPPTLKPVPPPTSR PRMLTTPTGPESMSTSTPAISSPSPTTASKEGDLGGKQWQDSSTQPELSSRYLISTGSTSQPILTSQSLSIQPSEENVSSSDTGPTSEGG LVATTTSGSGLSSSRNPITWPVTPFYNTLTKGPEMEIHSGSGEEREQPEDKDESNPVIWTKIRVPGNDAPVESTEMPLAPPLTPDLSRES WWPPFSTVMEGLLPSQRPTTSETGTPRVEGMVTEKPANTLLPLGGDHQPEPSGKTANRNHLKLPNNMNQTKSSEPVLTEEDATSLITEGF LLNASNYKQLTNGHGSAHWIVGNWSECSTTCGLGAYWRRVECSTQMDSDCAAIQRPDPAKRCHLRPCAGWKVGNWSKCSRNCSGGFKIRE IQCVDSRDHRNLRPFHCQFLAGIPPPLSMSCNPEPCEAWQVEPWSQCSRSCGGGVQERGVFCPGGLCDWTKRPTSTMSCNEHLCCHWATG NWDLCSTSCGGGFQKRTVQCVPSEGNKTEDQDQCLCDHKPRPPEFKKCNQQACKKSADLLCTKDKLSASFCQTLKAMKKCSVPTVRAECC -------------------------------------------------------------- >85017_85017_2_SNX6-ADAMTS12_SNX6_chr14_35099113_ENST00000362031_ADAMTS12_chr5_33751653_ENST00000504830_length(transcript)=8070nt_BP=121nt CTGGCCGCCGGCTCCCTCCGGCAGCAGGGAGATGCGCGCCTGCGCCGGCCCTCGCCTCGGAGCAGCCATGATGGAAGGCCTGGACGACGG CCCGGACTTCCTCTCAGAAGAGGACCGCGGAACTGGATTTTTCCAACTACCACATGGAGACTTTTTCATTGAACCCGTGAAGAAGCATCC ACTGGTTGAGGGAGGGTACCACCCGCACATCGTTTACAGGAGGCAGAAAGTTCCAGAAACCAAGGAGCCAACCTGTGGATTAAAGGACAG TGTTAACATCTCCCAGAAGCAAGAGCTATGGCGGGAGAAGTGGGAGAGGCACAACTTGCCAAGCAGAAGCCTCTCTCGGCGTTCCATCAG CAAGGAGAGATGGGTGGAGACACTGGTGGTGGCCGACACAAAGATGATTGAATACCATGGGAGTGAGAATGTGGAGTCCTACATCCTCAC CATCATGAACATGGTCACTGGGTTGTTCCATAACCCAAGCATTGGCAATGCAATTCACATTGTTGTGGTTCGGCTCATTCTACTCGAAGA AGAAGAGCAAGGACTGAAAATAGTTCACCATGCAGAAAAGACACTGTCTAGCTTCTGCAAGTGGCAGAAGAGTATCAATCCCAAGAGTGA CCTCAATCCTGTTCATCACGACGTGGCTGTCCTTCTCACCAGAAAGGACATCTGTGCTGGTTTCAATCGCCCCTGCGAGACCCTGGGCCT GTCTCACCTTTCAGGAATGTGTCAGCCTCACCGCAGTTGTAACATCAATGAAGATTCGGGACTCCCTCTGGCTTTCACAATTGCCCATGA GCTAGGACACAGCTTCGGCATCCAGCATGATGGGAAAGAAAATGACTGTGAGCCTGTGGGCAGACATCCGTACATCATGTCCCGCCAGCT CCAGTACGATCCCACTCCGCTGACATGGTCCAAGTGCAGCGAGGAGTACATCACCCGCTTCTTGGACCGAGGCTGGGGGTTCTGTCTTGA TGACATACCTAAAAAGAAAGGCTTGAAGTCCAAGGTCATTGCCCCCGGAGTGATCTATGATGTTCACCACCAGTGCCAGCTACAATATGG ACCCAATGCTACCTTCTGCCAGGAAGTAGAAAACGTCTGCCAGACACTGTGGTGCTCCGTGAAGGGCTTTTGTCGCTCTAAGCTGGACGC TGCTGCAGATGGAACTCAATGTGGTGAGAAGAAGTGGTGTATGGCAGGCAAGTGCATCACAGTGGGGAAGAAACCAGAGAGCATTCCTGG AGGCTGGGGCCGCTGGTCACCCTGGTCCCACTGTTCCAGGACCTGTGGGGCTGGAGTCCAGAGCGCAGAGAGGCTCTGCAACAACCCCGA GCCAAAGTTTGGAGGGAAATATTGCACTGGAGAAAGAAAACGCTATCGCTTGTGCAACGTCCACCCCTGTCGCTCAGAGGCACCAACATT TCGGCAGATGCAGTGCAGTGAATTTGACACTGTTCCCTACAAGAATGAACTCTACCACTGGTTTCCCATTTTTAACCCAGCACATCCTTG TGAGCTCTACTGCCGACCCATAGATGGCCAGTTTTCTGAGAAAATGCTGGATGCTGTCATTGATGGTACCCCTTGCTTTGAAGGCGGCAA CAGCAGAAATGTCTGTATTAATGGCATATGTAAGATGGTTGGCTGTGACTATGAGATCGATTCCAATGCCACCGAGGATCGCTGCGGTGT GTGCCTGGGAGATGGCTCTTCCTGCCAGACTGTGAGAAAGATGTTTAAGCAGAAGGAAGGATCTGGTTATGTTGACATTGGGCTCATTCC AAAAGGAGCAAGGGACATAAGAGTGATGGAAATTGAGGGAGCTGGAAACTTCCTGGCCATCAGGAGTGAAGATCCTGAAAAATATTACCT GAATGGAGGGTTTATTATCCAGTGGAACGGGAACTATAAGCTGGCAGGGACTGTCTTTCAGTATGACAGGAAAGGAGACCTGGAAAAGCT GATGGCCACAGGTCCCACCAATGAGTCTGTGTGGATCCAGCTTCTATTCCAGGTGACTAACCCTGGCATCAAGTATGAGTACACAATCCA GAAAGATGGCCTTGACAATGATGTTGAGCAGCAGATGTACTTCTGGCAGTACGGCCACTGGACAGAGTGCAGTGTGACCTGCGGGACAGG TATCCGCCGCCAAACTGCCCATTGCATAAAGAAGGGCCGCGGGATGGTGAAAGCTACATTCTGTGACCCAGAAACACAGCCCAATGGGAG ACAGAAGAAGTGCCATGAAAAGGCTTGTCCACCCAGGTGGTGGGCAGGGGAGTGGGAAGCATGCTCGGCGACATGCGGGCCCCACGGGGA GAAGAAGCGAACCGTGCTGTGCATCCAGACCATGGTCTCTGACGAGCAGGCTCTCCCGCCCACAGACTGCCAGCACCTGCTGAAGCCCAA GACCCTCCTTTCCTGCAACAGAGACATCCTGTGCCCCTCGGACTGGACAGTGGGCAACTGGAGTGAGTGTTCTGTTTCCTGTGGTGGTGG AGTGCGGATTCGCAGTGTCACATGTGCCAAGAACCATGATGAACCTTGCGATGTGACAAGGAAACCCAACAGCCGAGCTCTGTGTGGCCT CCAGCAATGCCCTTCTAGCCGGAGAGTTCTGAAACCAAACAAAGGCACTATTTCCAATGGAAAAAACCCACCAACACTAAAGCCCGTCCC TCCACCTACATCCAGGCCCAGAATGCTGACCACACCCACAGGGCCTGAGTCTATGAGCACAAGCACTCCAGCAATCAGCAGCCCTAGTCC TACCACAGCCTCCAAAGAAGGAGACCTGGGTGGGAAACAGTGGCAAGATAGCTCAACCCAACCTGAGCTGAGCTCTCGCTATCTCATTTC CACTGGAAGCACTTCCCAGCCCATCCTCACTTCCCAATCCTTGAGCATTCAGCCAAGTGAGGAAAATGTTTCCAGTTCAGATACTGGTCC TACCTCGGAGGGAGGCCTTGTAGCTACAACAACAAGTGGTTCTGGCTTGTCATCTTCCCGCAACCCTATCACTTGGCCTGTGACTCCATT TTACAATACCTTGACCAAAGGTCCAGAAATGGAGATTCACAGTGGCTCAGGGGAAGAAAGAGAACAGCCTGAGGACAAAGATGAAAGCAA TCCTGTAATATGGACCAAGATCAGAGTACCTGGAAATGACGCTCCAGTGGAAAGTACAGAAATGCCACTTGCACCTCCACTAACACCAGA TCTCAGCAGGGAGTCCTGGTGGCCACCCTTCAGCACAGTAATGGAAGGACTGCTCCCCAGCCAAAGGCCCACTACTTCCGAAACTGGGAC ACCCAGAGTTGAGGGGATGGTTACTGAAAAGCCAGCCAACACTCTGCTCCCTCTGGGAGGAGACCACCAGCCAGAACCCTCAGGAAAGAC GGCAAACCGTAACCACCTGAAACTTCCAAACAACATGAACCAAACAAAAAGTTCTGAACCAGTCCTGACTGAGGAGGATGCAACAAGTCT GATTACTGAGGGCTTTTTGCTAAATGCCTCCAATTACAAGCAGCTCACAAACGGCCACGGCTCTGCACACTGGATCGTCGGAAACTGGAG CGAGTGCTCCACCACATGTGGCCTGGGGGCCTACTGGAGAAGGGTGGAGTGCAGCACCCAGATGGATTCTGACTGTGCGGCCATCCAGAG ACCTGACCCTGCAAAAAGATGCCACCTCCGTCCCTGTGCTGGCTGGAAAGTGGGAAACTGGAGCAAGTGCTCCAGAAACTGCAGTGGGGG CTTCAAGATACGCGAGATTCAGTGCGTGGACAGCCGGGACCACCGGAACCTGAGGCCATTTCACTGCCAGTTCCTGGCCGGCATTCCTCC CCCATTGAGCATGAGCTGTAACCCGGAGCCCTGTGAGGCGTGGCAGGTGGAGCCTTGGAGCCAGTGCTCCAGGTCCTGTGGAGGTGGAGT TCAGGAGAGAGGAGTGTTCTGTCCAGGAGGCCTCTGTGATTGGACAAAAAGACCCACATCCACCATGTCTTGCAATGAGCACCTGTGCTG TCACTGGGCCACTGGGAACTGGGACCTGTGTTCCACTTCCTGTGGAGGTGGCTTTCAGAAGAGGACTGTCCAATGTGTGCCCTCAGAGGG CAATAAAACTGAAGACCAAGACCAATGTCTATGTGATCACAAACCCAGACCTCCAGAATTCAAAAAATGCAACCAGCAGGCCTGCAAGAA AAGTGCCGATTTACTTTGCACTAAGGACAAACTGTCAGCCAGTTTCTGCCAGACACTGAAAGCCATGAAGAAATGTTCTGTGCCCACCGT GAGGGCTGAGTGCTGCTTCTCGTGTCCCCAGACACACATCACACACACCCAAAGGCAAAGAAGGCAACGGTTGCTCCAAAAGTCAAAAGA ACTCTAAGCCCAAAAGGAAGCCAGCTTCCACCAGACTGCAGCAGCCTGCTGACCATGACATGCTTTAGCCCTGGGAGCTGCTTCGTTGAG GTTTTGCTTCATATATTTCAGATTCAGCTTTGGGTCATGATGAAACAAAATCCATTGTGTTCCTCTAGCCACCGAATGTCCCTCACAGGA GGCCTAGGCTGCCGTACTTGCTGCTCCCTGATAGGAAGGTGAAATAGCTAGGAGAATCAGGCAGCACCTGGTATCGGCTCCTGGGAAAGG TGCCAGGTTAGACAGATTTGTGAAATACTGTATTTTTGTTCTCCCTTAAAGATAAAAACAAAGAGACCAGCTATTTCCCTGACTACCTCA AGAGTATCAGGCAACACTTACCCAGGGGACAAATAATCTTCTTAATTTATCTTGATGAATACCTTTCTTTTTTTTTTCTTGCCTCCCTGC CTGGGGCAGCTGCTATTTAAAATGTCACAGCGGAGCTTTAGGAACATTCCTTTAATTCATCCAGGCAGAGGACTGCTTTCTTCACCCCAT GTTCAACTAGGGCAGGAAGTGCTGGTTACATCCCTCAGTCTCTTCCTCCATCCCTGTAGGGCTCCATATCTGGGCTGCTTAGAGTCTCCT GCCAACCTTGGGACACTAGACTAAGAGGAAACAGGCATAAGAAACACAGCCTAAGGGGTTTAGGTCACTTACACAGAAAACACTTCTGGT TTGAAGAGCATTTCTATGAAAACTCGTAAAATGCAAATGAGCAGAAATCAATTTCTTAATATTACAGAAAAGGAACTGTGGGCCTTTATT CTTCCCACTTTCCTGGCAAATACACACTCCTTTGCAGAAGGGAAAATGTGTCTTCCTGGCTGACTTCCTCCCATCCTCCAGTCCCCACAG GAGGCCTGAGCTCAGGTCCCACCATCGTCTTGCTGCAGGGGCTGTATATGCAAAGACCACTATGACCAGCTCTAGGGCTGCCACCTGTCC TCACCTCTTCTCAGGTTGGAAAATGGAGGTTCCATATTAAAATATTTCAAAGAACCAGGTAAATTGTACTAAGTACAAGAACATAGTGCA AACTTCTAATCTAGAGAAAAAACATTTCTAAAAAGTAAGGTAAAGGGGATCTACTCCTCAGTGTACCCAATATGTAGTTAGTCTTAAAGA TCTTTTACTTTGGGGAGAACAGGTGAATCAGGGAGTCCAAAGAAACTGAAATGAAAGTTAGAATGCTTGTTCTGGATGTTCCTACCTCCC ACCAGAAGTCAAAGTAGGCTTGGTCTCAAATTCAGAGCAAATTTCTAGACAAAGTGATTTCTAAGGGAAGGTACTTACATGAACTGAACC CAGAGAAAGAGGGTAAGTGGGTGGGTGGAGGGGCATAAGCTTCAAATGAAAGAAGATATCAGAACCAATTCTTGACAGGGAGAAAGGCAA GGATGTGGCCTTCATGTGATGGGCAGCATATTTTCTGACTCATGCACAGATACACTTAATACTCCTCATGGAACACAATATCCCACGTCT CCCTTTACCTATCCGTTCAGCAGAATGGTGCTATTAAAAAAATGGTGCTAATATCTAGATGGTGGCAGAATTAATGTAAACATAGATAGG CATACAAATGGGTAAGTGTATGTGAAACTCTCCATATACATCTTCTGTTAAACATGTAACGGTGTTGCTGACCAATGCCAGTTTTATTGC TTGGGTACCTTGAATTTAAGATTTAATATTATGTGCTTTTTATTTTTTTCCAAGCCATTAACGTGTATTACTCTCATGAATTCCTGTGAA AAATAAAACAGTGGGATTGGGATGAAATTCAAGATAGACCAGAAACAGGAGGAAAGTACAAATGTTTACCTAAAGCCAGAGAATTTATTT CTAGATGATAATTACATTATTGTATTCTCTGCCTTTAAACATAAACATTGCTGGCCGATAGCAATCTCTGGTGAATAATGGTTTTCGAGG AGAGGAGACACTTAAATTGTTAGTGACATGAGTGGAGGAGAATCTGATATTTACTAATGAGGCAGTTTGCAAAGACTGTCCCTGAAGTGT AGTGTAGTCTTTCAGGGGGATTCATTTAGTAAGCTAGATTGATTTAACCTGGTACTGTACTAGCATAGGGTCAAATACGTGTCATCAGAG ACCTGGGTATGACCAGGCTTACAAAACTCAGGAACAAACTCAGATTCCTACTGACCTCAACCAACTAAACTAGGCCAAATTTCTCCGTGT ACAAAATGGACCACGTTATTTACAATCCACGTGGTGGAGAAGATGGTAGACATGTGGAGAGAGTTTGGCCAGGTGCTCCATTCTAGGTCT TTTTCCAGTTTCTCAAAGGCAGAACATTGGCTCCTAATTATTTGCCATGGATATTTGCTTCCTGTTCTGGAGCTATGTTGTAAGACAGCT GTGTGATGTCCATCATTCTTGACTCCAGAATGAAGACAGGGCTTGCTGTTTTCTGTCCTGTTGGTATCATCTGTTGCCTTGGCGATCACT CAGTGATGAGTCGTCAGTATTTTGACATGTCCCAGTGCTTGCTGTACAAGAGGGGACTCAGATCAACAGGAAGACTCTGAAGACAGGAAC CTGCATGGTATCTTACATCTTTGATACTTGGGTGCTGATATGAAGCAGAGTTGTTGATTTACTTTATCTAGGCCCTTCTTTCTTCTCACC TGGATCAAGCAACTGAGAAGTGTATCAGGAGACACTGGATACATATCTCTATGAAATAGAGAGGGCCATCGCAGGGCCTGGTACTTAACT ACATTAACGTTCTAAAACCCAGTTTGGTTTACGTTGTCTTTCACAGTAGTATATTTAGCTCTTCTCTGGAAAGTTGTGGGTTAATATAAT TCTTAAACATGAAAATGTAATTAAACACACCACGAGAGAACAATATTCCAGGAGACTTAATAGTGATTACTTTCTTCAATCAGGAAATCG TTTCAGTGCCTCCTTTGTAGGAATGCTTTGTTTTGTGATGGGTTTTCTTAAAGAAGAGCACACCTCCGTCCAATCTCCTGAGACAGCCAC GTCTCCGCTGACATCCCACTGTGATGCTTTCAGATAGTCAGTGAATGTTTCTGATAACCTTCATCCAGTATCTGAAACACAATGTGAGAG ATTATATTGTTTTAGATAATAACATCCCATTTAGTTGACTAAAATCTTCCAAACTCTGAAAGCTGCACACTGCTACTCCAGAGAGTGCAG GTCTTAGCTCTTCTCCTTTCTGACTTCAAGATGAATCTTTGGGACGATGTTTCTGGTGCTTGGTCCACAGTGATTCACTTTTGAAGGAGA GGCCACATGACATGAACTGCCTGGTGTTACAACCTAGCTAACATATTTGATGCTACTCCTGTTGTCTGTACTGCTTATTCAAGTAGTATT CTAAGTTATGTTACTAAAAAACATGGTGGGTAAAGCACAATCCTACCCATCATTGTCCTCCAAAATAATTGTATGACATACACGGCCCAG CCCATTGCCCTCCCTGCATCTCTGTGCTGCTTTGCCATTTCCCCTTCTACCCAGCCTCCTCAAGGGGTACCTTGGTGGATATTTCAGTAC >85017_85017_2_SNX6-ADAMTS12_SNX6_chr14_35099113_ENST00000362031_ADAMTS12_chr5_33751653_ENST00000504830_length(amino acids)=1461AA_BP=30 MRACAGPRLGAAMMEGLDDGPDFLSEEDRGTGFFQLPHGDFFIEPVKKHPLVEGGYHPHIVYRRQKVPETKEPTCGLKDSVNISQKQELW REKWERHNLPSRSLSRRSISKERWVETLVVADTKMIEYHGSENVESYILTIMNMVTGLFHNPSIGNAIHIVVVRLILLEEEEQGLKIVHH AEKTLSSFCKWQKSINPKSDLNPVHHDVAVLLTRKDICAGFNRPCETLGLSHLSGMCQPHRSCNINEDSGLPLAFTIAHELGHSFGIQHD GKENDCEPVGRHPYIMSRQLQYDPTPLTWSKCSEEYITRFLDRGWGFCLDDIPKKKGLKSKVIAPGVIYDVHHQCQLQYGPNATFCQEVE NVCQTLWCSVKGFCRSKLDAAADGTQCGEKKWCMAGKCITVGKKPESIPGGWGRWSPWSHCSRTCGAGVQSAERLCNNPEPKFGGKYCTG ERKRYRLCNVHPCRSEAPTFRQMQCSEFDTVPYKNELYHWFPIFNPAHPCELYCRPIDGQFSEKMLDAVIDGTPCFEGGNSRNVCINGIC KMVGCDYEIDSNATEDRCGVCLGDGSSCQTVRKMFKQKEGSGYVDIGLIPKGARDIRVMEIEGAGNFLAIRSEDPEKYYLNGGFIIQWNG NYKLAGTVFQYDRKGDLEKLMATGPTNESVWIQLLFQVTNPGIKYEYTIQKDGLDNDVEQQMYFWQYGHWTECSVTCGTGIRRQTAHCIK KGRGMVKATFCDPETQPNGRQKKCHEKACPPRWWAGEWEACSATCGPHGEKKRTVLCIQTMVSDEQALPPTDCQHLLKPKTLLSCNRDIL CPSDWTVGNWSECSVSCGGGVRIRSVTCAKNHDEPCDVTRKPNSRALCGLQQCPSSRRVLKPNKGTISNGKNPPTLKPVPPPTSRPRMLT TPTGPESMSTSTPAISSPSPTTASKEGDLGGKQWQDSSTQPELSSRYLISTGSTSQPILTSQSLSIQPSEENVSSSDTGPTSEGGLVATT TSGSGLSSSRNPITWPVTPFYNTLTKGPEMEIHSGSGEEREQPEDKDESNPVIWTKIRVPGNDAPVESTEMPLAPPLTPDLSRESWWPPF STVMEGLLPSQRPTTSETGTPRVEGMVTEKPANTLLPLGGDHQPEPSGKTANRNHLKLPNNMNQTKSSEPVLTEEDATSLITEGFLLNAS NYKQLTNGHGSAHWIVGNWSECSTTCGLGAYWRRVECSTQMDSDCAAIQRPDPAKRCHLRPCAGWKVGNWSKCSRNCSGGFKIREIQCVD SRDHRNLRPFHCQFLAGIPPPLSMSCNPEPCEAWQVEPWSQCSRSCGGGVQERGVFCPGGLCDWTKRPTSTMSCNEHLCCHWATGNWDLC STSCGGGFQKRTVQCVPSEGNKTEDQDQCLCDHKPRPPEFKKCNQQACKKSADLLCTKDKLSASFCQTLKAMKKCSVPTVRAECCFSCPQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SNX6-ADAMTS12 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396526 | - | 1 | 13 | 2_179 | 0 | 291.0 | PIM1 |
| Hgene | SNX6 | chr14:35099113 | chr5:33751653 | ENST00000396534 | - | 1 | 14 | 2_179 | 0 | 755.3333333333334 | PIM1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SNX6-ADAMTS12 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SNX6-ADAMTS12 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
