
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SPG7-MOB1A (FusionGDB2 ID:85761) |
Fusion Gene Summary for SPG7-MOB1A |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SPG7-MOB1A | Fusion gene ID: 85761 | Hgene | Tgene | Gene symbol | SPG7 | MOB1A | Gene ID | 6687 | 55233 |
| Gene name | SPG7 matrix AAA peptidase subunit, paraplegin | MOB kinase activator 1A | |
| Synonyms | CAR|CMAR|PGN|SPG5C | C2orf6|MATS1|MOB1|MOBK1B|MOBKL1B|Mob4B | |
| Cytomap | 16q24.3 | 2p13.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | parapleginSPG7, paraplegin matrix AAA peptidase subunitcell matrix adhesion regulatorspastic paraplegia 7 (pure and complicated autosomal recessive)spastic paraplegia 7 protein | MOB kinase activator 1AMOB1 Mps One Binder homolog AMOB1, Mps One Binder kinase activator-like 1Bmob1 alphamob1 homolog 1Bmps one binder kinase activator-like 1B | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | Q9H8S9 | |
| Ensembl transtripts involved in fusion gene | ENST00000268704, ENST00000341316, ENST00000565891, | ENST00000409969, ENST00000497054, ENST00000396049, | |
| Fusion gene scores | * DoF score | 10 X 7 X 6=420 | 6 X 6 X 4=144 |
| # samples | 10 | 6 | |
| ** MAII score | log2(10/420*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/144*10)=-1.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: SPG7 [Title/Abstract] AND MOB1A [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SPG7(89579445)-MOB1A(74394234), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MOB1A | GO:0035329 | hippo signaling | 19739119 |
 Fusion gene breakpoints across SPG7 (5'-gene) Fusion gene breakpoints across SPG7 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
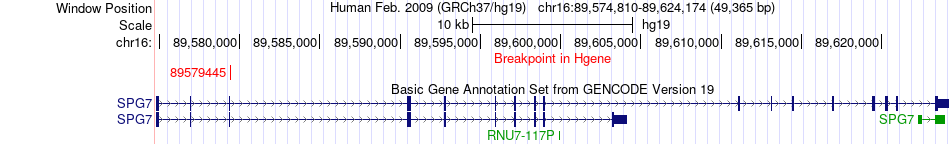 |
 Fusion gene breakpoints across MOB1A (3'-gene) Fusion gene breakpoints across MOB1A (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
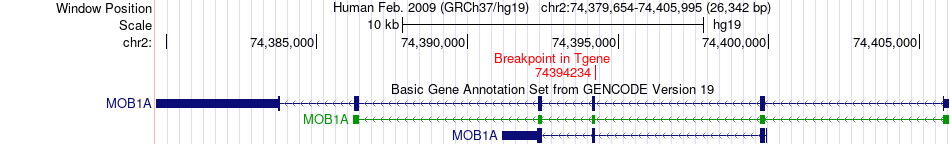 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GN-01A | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
Top |
Fusion Gene ORF analysis for SPG7-MOB1A |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000268704 | ENST00000409969 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| 5CDS-5UTR | ENST00000268704 | ENST00000497054 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| 5CDS-5UTR | ENST00000341316 | ENST00000409969 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| 5CDS-5UTR | ENST00000341316 | ENST00000497054 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| In-frame | ENST00000268704 | ENST00000396049 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| In-frame | ENST00000341316 | ENST00000396049 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| intron-3CDS | ENST00000565891 | ENST00000396049 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| intron-5UTR | ENST00000565891 | ENST00000409969 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
| intron-5UTR | ENST00000565891 | ENST00000497054 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000268704 | SPG7 | chr16 | 89579445 | + | ENST00000396049 | MOB1A | chr2 | 74394234 | - | 4901 | 391 | 15 | 860 | 281 |
| ENST00000341316 | SPG7 | chr16 | 89579445 | + | ENST00000396049 | MOB1A | chr2 | 74394234 | - | 4893 | 383 | 7 | 852 | 281 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000268704 | ENST00000396049 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - | 0.000354864 | 0.9996451 |
| ENST00000341316 | ENST00000396049 | SPG7 | chr16 | 89579445 | + | MOB1A | chr2 | 74394234 | - | 0.000352178 | 0.99964786 |
Top |
Fusion Genomic Features for SPG7-MOB1A |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
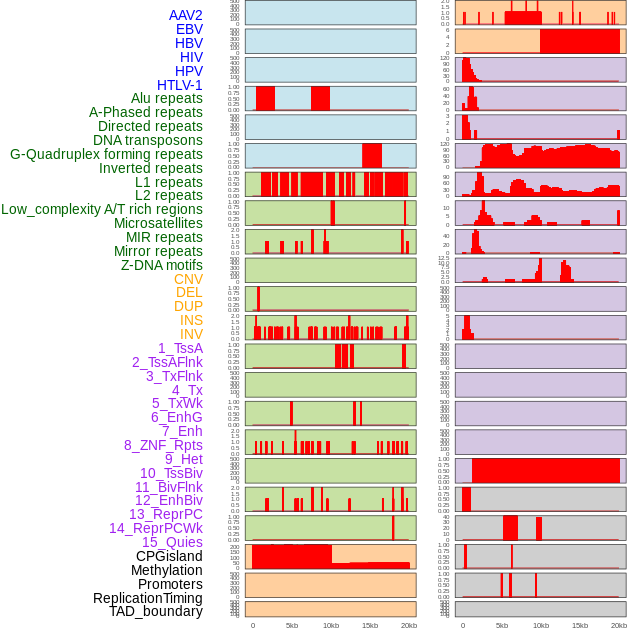 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for SPG7-MOB1A |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:89579445/chr2:74394234) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MOB1A |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Activator of LATS1/2 in the Hippo signaling pathway which plays a pivotal role in organ size control and tumor suppression by restricting proliferation and promoting apoptosis. The core of this pathway is composed of a kinase cascade wherein STK3/MST2 and STK4/MST1, in complex with its regulatory protein SAV1, phosphorylates and activates LATS1/2 in complex with its regulatory protein MOB1, which in turn phosphorylates and inactivates YAP1 oncoprotein and WWTR1/TAZ. Phosphorylation of YAP1 by LATS1/2 inhibits its translocation into the nucleus to regulate cellular genes important for cell proliferation, cell death, and cell migration. Stimulates the kinase activity of STK38 and STK38L. Acts cooperatively with STK3/MST2 to activate STK38. {ECO:0000269|PubMed:15197186, ECO:0000269|PubMed:18362890, ECO:0000269|PubMed:19739119}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 349_357 | 125 | 796.0 | Nucleotide binding | ATP |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 349_357 | 125 | 490.0 | Nucleotide binding | ATP |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 106_144 | 125 | 796.0 | Topological domain | Mitochondrial matrix |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 166_248 | 125 | 796.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 270_795 | 125 | 796.0 | Topological domain | Mitochondrial matrix |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 106_144 | 125 | 490.0 | Topological domain | Mitochondrial matrix |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 166_248 | 125 | 490.0 | Topological domain | Mitochondrial intermembrane |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 270_795 | 125 | 490.0 | Topological domain | Mitochondrial matrix |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 145_165 | 125 | 796.0 | Transmembrane | Helical |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 249_269 | 125 | 796.0 | Transmembrane | Helical |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 145_165 | 125 | 490.0 | Transmembrane | Helical |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 249_269 | 125 | 490.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for SPG7-MOB1A |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >85761_85761_1_SPG7-MOB1A_SPG7_chr16_89579445_ENST00000268704_MOB1A_chr2_74394234_ENST00000396049_length(transcript)=4901nt_BP=391nt GGCTTTCAGGCCAACATGGCCGTGCTGCTGCTGCTGCTCCGTGCCCTCCGCCGGGGTCCAGGCCCGGGTCCTCGGCCGCTGTGGGGCCCA GGCCCGGCCTGGAGTCCAGGGTTCCCCGCCAGGCCCGGGAGGGGGCGGCCGTACATGGCCAGCAGGCCTCCGGGGGACCTCGCCGAGGCT GGAGGCCGAGCTCTGCAGAGCTTACAATTGAGACTGCTAACCCCTACCTTTGAAGGGATCAACGGATTGTTGTTGAAACAACATTTAGTT CAGAATCCAGTCAGACTCTGGCAACTTTTAGGTGGTACTTTCTATTTTAACACCTCAAGGTTGAAGCAGAAGAATAAGGAGAAGGATAAG TCGAAGGGGAAGGCGCCTGAAGAGGACGAAGCTGTGGATTTCTTTAACCAGATCAACATGTTATATGGAACTATTACAGAATTCTGCACT GAAGCAAGCTGTCCAGTCATGTCTGCAGGTCCGAGATATGAATATCACTGGGCAGATGGTACTAATATTAAAAAGCCAATCAAATGTTCT GCACCAAAATACATTGACTATTTGATGACTTGGGTTCAAGATCAGCTTGATGATGAAACTCTTTTTCCTTCTAAGATTGGTGTCCCATTT CCCAAAAACTTTATGTCTGTGGCAAAGACTATTCTAAAGCGTCTGTTCAGGGTTTATGCCCATATTTATCACCAGCACTTTGATTCTGTG ATGCAGCTGCAAGAGGAGGCCCACCTCAACACCTCCTTTAAGCACTTTATTTTCTTTGTTCAGGAGTTTAATCTGATTGATAGGCGTGAG CTGGCACCTCTTCAAGAATTAATAGAGAAACTTGGATCAAAAGACAGATAAATGTTTCTTCTAGAACACAGTTACCCCCTTGCTTCATCT ATTGCTAGAACTATCTCATTGCTATCTGTTATAGACTAGTGATACAAACTTTAAGAAAACAGGATAAAAAGATACCCATTGCCTGTGTCT ACTGATAAAATTATCCCAAAGGTAGGTTGGTGTGATAGTTTCCGAGTAAGACCTTAAGGACACAGCCAAATCTTAAGTACTGTGTGACCA CTCTTGTTGTTATCACATAGTCATACTTGGTTGTAATATGTGATGGTTAACCTGTAGCTTATAAATTTACTTATTATTCTTTTACTCATT TACTCAGTCATTTCTTTACAAGAAAATGATTGAATCTGTTTTAGGTGACAGCACAATGGACATTAAGAATTTCCATCAATAATTTATGAA TAAGTTTCCAGAACAAATTTCCTAATAACACAATCAGATTGGTTTTATTCTTTTATTTTACGAATAAAAAATGTATTTTTCAGTATCCTT GAGATTTAGAACATCTGTGTCACTTCAGATAACATTTTAGTTTCAAGTTTGTATGGTAGTGTTTTTATAGATAAGATACGTCTATTTTTT CAAAATTCATGATTGCAGTTTAAATCATCATATGACGTGTGGGTGGGAGCAACCAAAGTTATTTTTACAGGGACTTTATTTTTTGATCTT TATTTGAGATTGTTTTCATATCTATCTAAATTATTAGGAGTGTGTGTATCAGAAGTAATTTTTTAATGTCTTCTAAGGATGGTCTTCCAG GCTTTTAAACTGAAAAGCTTAATTCAGATAGTAGCTTTTGGCTGAGAAAAGGAATCCAAAATATTAATAAATTTAGATCTCAAAACCACT ATTTTTATTATTTCATTATTTTTCAGAGGCCTTAAAATTCTGGATAAGAGAATGGAGGAAAATACTCAGAGTACTTGATTATTTTATTTC CTTTTATTAAAAAATTACTTCTATGTTTTTATTGTCTCTTGAGCCTTAGTTAAGAGTAGTGTAGAAATGCATGAACTTCATCCTAATAAG GATAAAACTTAAGGAAAACCACAATAAACCATGAAGGTGTACACATCTTATAACACAGATAAAGTTTTGGTGTGCTACCTATTCTTGAGA GAGTGAGTGAGTGTATGTGTTTAAAGGAAACAAAATGGGAGAAATAAGTTTTAAAAAAATCCTCATTTTGTTAATATTCAAAAGATGGAC TGAGCTTCCACTTGGGTTTTATCTTGTTTTAATTGTTTTTGTATCAAAACTTGAAATTCCTCTATTTCTATTGGGATATAAAAGCCTTCC CCTTCAGTGAAGAAAACATTTATTTTTTATTTGATTCCTAGGATTTAGTAAACTCTAGCTGTCTATTTAAAATGTACTGAGGCACAACAA GTATTATACTGGAAGACTTGCCAAACTGGCAAAGCTTTAAGTTCATCAGCATTCTATGTGGTTCAGAGCTGTGATTTTTGCAAAGTATTT TACCAACCTCCTCGATGGCTTTGATAAAGGTTAGATTTGATGTTTTTTTTTAGATTTATTTTTCTTACTCCACTAAACTATAAAGAAAAT AATTACTTAGAAACTCCATTTTAAATAATCATTTCCTAGAAATTCTTAAATATATACAGAATTTTAAAGAAAACATTTCATCTGATTTAG TTAGCATCCACATATCATTGAGGAATTAAAGTGTGGGACAGTCATTATTAAAAAAAAGAGAGAAAAGCCCTCTATTAGACATTCCACAAT CCATGTTTTAAGCTTATCCAAAGGTCCAAATGTCAGCCATTCTGTATGTTCATGTTGATCATTTGCGAACAAGAAAGCAGGTTTCTAGGT ATCACTTAGGATGTGAACTGCCTCTCAACTTTAAACCCTGTTAGCTTTACTTTTTTAAGTCCACAAGTGATGAAACTAGTTTCTCAGCTA GGCTTGTACTTTCCTCATTATTTCTAGTATTTCAAATATTCTCAAACAAAAGAGTTACCACTTTTCTCCATTTATTTTCAGTTATGGAAA TGTTCCCTCTCTTCACCACTAAGCTCCAAAGCAAATGAAAGACGATCACATGTCAGGACAGTAGTAAAGGCAGCTTATAAATGGGACATA AATCAGAGATGTGTTGGTATTTTGAGACTCAAGACTGTCCTTTTTTAAAATAAAAATAAAAACATTACCAGGTTCCCAAGCCAATCTGGC TTAACCAACAGTGCACTGAAATATTAGTGTTTACCTCCAAGGCTAGGGAGCCAAGGGGAGGAGGAGAATTGGAGGAAGGGGAGATAATGG GAAGAGGATGGCGCCTTCCTGAGTTGGCTAGAGGGCCAACCTTTGATAACAGTTTGACGAAATCAATCTTTTTTTTTTTTTTTTTTGAGA CAGAGTCTCACTCTGTCACCCAGGCTGGAGTACAGTGGTGGGATCTCGGCTCACTGCAAGCTCTGCCTCCCAGGTTCATGCCATTCTCCT GCCCCAGCCTCTCCTGAGTAGCTGGAACTACAGGTGCCCGCCACCACGCCCAGCTAATTTTTTCTATTTTTAGTCATGCCTGGCTAATTT TTTGTATTTTTAGTAGAGATGGGGTTTCACCGTGTTAGCCAGGATGGTCTCCATCTCCTTACCTCGCAATCCGCCCGCCTCAGCCTCCCA AAGTGTTGGGATTACAGGCATGAGCCACCGTGCCCGGCCTATAGAAATCAATCTTTTTGACTCTTCTCACTTTTATCTCCCCATGCCCAA GGTTTGCCTGTTCCATAACACTCACTCCCTTCCCCCTTGCTAATCAGAAGCCATCTCCTCTCAGTGTCTGATCTCTGCTCTTCATACATG ATTACAGTCATGGGGTAGAGAGTGCTTGCTAAATTATGCAGTTAATCCTATGGTGCTTTAATTTTCAGGCCTTCAAAAAACACTTGTACA GTGATGTGCAGATTTTTAAACAGTTGAACTTCCTTGTACTACAGTTTTTGTATTGACAGCCAAATTTGTCTTTCATTCTTCAGATTGTGA ATAAAGTGATTTTTACAGGGCTTCCAGCAAAGTTTTTCCTTTCATCTAAGGCTTGTAGAACCCTAGCTTATATAGCTGCTTACATGAGAA ATGCAAAATCTGTATTCACCATGACTTTAGTAACAAAGGTAAAGTTTTTTAGTAGTGCCAAGGCAAGAGGAACAATCTTGGTGGTAGTAC TAAGTTTTGTCAATATTGTGGTTTCCTGATTGTATTGTTGGCTTTCTCTCTGAGCATTGAGGTATACTAGAAGTAGAGCTTCTCAAACAT AATATCATTACCTCATAAGCATTAACAAATCAGGCCCAAAGAGCGTAAGTCCTAGAAATTTGTTTTAAAGCAGCCCTAGTCATGGTGCTG GTGCTACCGCCTTGTTTTAGGAGCCTGCCTCCTGTCAGTATGAAACCCTCACCTGAAAAATGCCAGCCTGGACACCAAACACTGAGCCCC TTCAACAGGCACATTATTTCCCCCTGAGATCCATAAGGGAATTTAGTTTCTACTATTGTAGAGTTCTGAAAAGAGGTAAAATAGTAGTCC TTTGGTCATCCTATTTTTGCTTTCAATTTTGATATTTCAGACTGTAAAAGGCCTTGGGGGATGATAGTACATGTGGTAGCAGTAATTTTT TTGAAGCAACTGCACTGACATTCATTTGAGTTTTCTCTCATTATCAGATTCTGTTCCAAACAAGTATTCTGTAGATCCAAATGGATTACC AGTGTGCTACAGACTTCTTATTATAGAACAGCATTCTATTCTACATCAAAAATAGTTTGTGTAAGTTAGTTTTGGTTACCATCTAAAATA TTTTTAAATGTTCTTTACATAAAAATTTATGTTGTGTTTTAAAATCCTTAGGGGCTTTATCTATTTTTCTAAGTCAGTTAACTGTACTTC TAAAAAAAGTATTTTGTATCTACTTTTGTAACTTCGTCAGAATAAAATATATTGAAAGCAAGAAATGACTGATTAAAGTATGTATATCAA >85761_85761_1_SPG7-MOB1A_SPG7_chr16_89579445_ENST00000268704_MOB1A_chr2_74394234_ENST00000396049_length(amino acids)=281AA_BP=125 MAVLLLLLRALRRGPGPGPRPLWGPGPAWSPGFPARPGRGRPYMASRPPGDLAEAGGRALQSLQLRLLTPTFEGINGLLLKQHLVQNPVR LWQLLGGTFYFNTSRLKQKNKEKDKSKGKAPEEDEAVDFFNQINMLYGTITEFCTEASCPVMSAGPRYEYHWADGTNIKKPIKCSAPKYI DYLMTWVQDQLDDETLFPSKIGVPFPKNFMSVAKTILKRLFRVYAHIYHQHFDSVMQLQEEAHLNTSFKHFIFFVQEFNLIDRRELAPLQ -------------------------------------------------------------- >85761_85761_2_SPG7-MOB1A_SPG7_chr16_89579445_ENST00000341316_MOB1A_chr2_74394234_ENST00000396049_length(transcript)=4893nt_BP=383nt GGCCAACATGGCCGTGCTGCTGCTGCTGCTCCGTGCCCTCCGCCGGGGTCCAGGCCCGGGTCCTCGGCCGCTGTGGGGCCCAGGCCCGGC CTGGAGTCCAGGGTTCCCCGCCAGGCCCGGGAGGGGGCGGCCGTACATGGCCAGCAGGCCTCCGGGGGACCTCGCCGAGGCTGGAGGCCG AGCTCTGCAGAGCTTACAATTGAGACTGCTAACCCCTACCTTTGAAGGGATCAACGGATTGTTGTTGAAACAACATTTAGTTCAGAATCC AGTCAGACTCTGGCAACTTTTAGGTGGTACTTTCTATTTTAACACCTCAAGGTTGAAGCAGAAGAATAAGGAGAAGGATAAGTCGAAGGG GAAGGCGCCTGAAGAGGACGAAGCTGTGGATTTCTTTAACCAGATCAACATGTTATATGGAACTATTACAGAATTCTGCACTGAAGCAAG CTGTCCAGTCATGTCTGCAGGTCCGAGATATGAATATCACTGGGCAGATGGTACTAATATTAAAAAGCCAATCAAATGTTCTGCACCAAA ATACATTGACTATTTGATGACTTGGGTTCAAGATCAGCTTGATGATGAAACTCTTTTTCCTTCTAAGATTGGTGTCCCATTTCCCAAAAA CTTTATGTCTGTGGCAAAGACTATTCTAAAGCGTCTGTTCAGGGTTTATGCCCATATTTATCACCAGCACTTTGATTCTGTGATGCAGCT GCAAGAGGAGGCCCACCTCAACACCTCCTTTAAGCACTTTATTTTCTTTGTTCAGGAGTTTAATCTGATTGATAGGCGTGAGCTGGCACC TCTTCAAGAATTAATAGAGAAACTTGGATCAAAAGACAGATAAATGTTTCTTCTAGAACACAGTTACCCCCTTGCTTCATCTATTGCTAG AACTATCTCATTGCTATCTGTTATAGACTAGTGATACAAACTTTAAGAAAACAGGATAAAAAGATACCCATTGCCTGTGTCTACTGATAA AATTATCCCAAAGGTAGGTTGGTGTGATAGTTTCCGAGTAAGACCTTAAGGACACAGCCAAATCTTAAGTACTGTGTGACCACTCTTGTT GTTATCACATAGTCATACTTGGTTGTAATATGTGATGGTTAACCTGTAGCTTATAAATTTACTTATTATTCTTTTACTCATTTACTCAGT CATTTCTTTACAAGAAAATGATTGAATCTGTTTTAGGTGACAGCACAATGGACATTAAGAATTTCCATCAATAATTTATGAATAAGTTTC CAGAACAAATTTCCTAATAACACAATCAGATTGGTTTTATTCTTTTATTTTACGAATAAAAAATGTATTTTTCAGTATCCTTGAGATTTA GAACATCTGTGTCACTTCAGATAACATTTTAGTTTCAAGTTTGTATGGTAGTGTTTTTATAGATAAGATACGTCTATTTTTTCAAAATTC ATGATTGCAGTTTAAATCATCATATGACGTGTGGGTGGGAGCAACCAAAGTTATTTTTACAGGGACTTTATTTTTTGATCTTTATTTGAG ATTGTTTTCATATCTATCTAAATTATTAGGAGTGTGTGTATCAGAAGTAATTTTTTAATGTCTTCTAAGGATGGTCTTCCAGGCTTTTAA ACTGAAAAGCTTAATTCAGATAGTAGCTTTTGGCTGAGAAAAGGAATCCAAAATATTAATAAATTTAGATCTCAAAACCACTATTTTTAT TATTTCATTATTTTTCAGAGGCCTTAAAATTCTGGATAAGAGAATGGAGGAAAATACTCAGAGTACTTGATTATTTTATTTCCTTTTATT AAAAAATTACTTCTATGTTTTTATTGTCTCTTGAGCCTTAGTTAAGAGTAGTGTAGAAATGCATGAACTTCATCCTAATAAGGATAAAAC TTAAGGAAAACCACAATAAACCATGAAGGTGTACACATCTTATAACACAGATAAAGTTTTGGTGTGCTACCTATTCTTGAGAGAGTGAGT GAGTGTATGTGTTTAAAGGAAACAAAATGGGAGAAATAAGTTTTAAAAAAATCCTCATTTTGTTAATATTCAAAAGATGGACTGAGCTTC CACTTGGGTTTTATCTTGTTTTAATTGTTTTTGTATCAAAACTTGAAATTCCTCTATTTCTATTGGGATATAAAAGCCTTCCCCTTCAGT GAAGAAAACATTTATTTTTTATTTGATTCCTAGGATTTAGTAAACTCTAGCTGTCTATTTAAAATGTACTGAGGCACAACAAGTATTATA CTGGAAGACTTGCCAAACTGGCAAAGCTTTAAGTTCATCAGCATTCTATGTGGTTCAGAGCTGTGATTTTTGCAAAGTATTTTACCAACC TCCTCGATGGCTTTGATAAAGGTTAGATTTGATGTTTTTTTTTAGATTTATTTTTCTTACTCCACTAAACTATAAAGAAAATAATTACTT AGAAACTCCATTTTAAATAATCATTTCCTAGAAATTCTTAAATATATACAGAATTTTAAAGAAAACATTTCATCTGATTTAGTTAGCATC CACATATCATTGAGGAATTAAAGTGTGGGACAGTCATTATTAAAAAAAAGAGAGAAAAGCCCTCTATTAGACATTCCACAATCCATGTTT TAAGCTTATCCAAAGGTCCAAATGTCAGCCATTCTGTATGTTCATGTTGATCATTTGCGAACAAGAAAGCAGGTTTCTAGGTATCACTTA GGATGTGAACTGCCTCTCAACTTTAAACCCTGTTAGCTTTACTTTTTTAAGTCCACAAGTGATGAAACTAGTTTCTCAGCTAGGCTTGTA CTTTCCTCATTATTTCTAGTATTTCAAATATTCTCAAACAAAAGAGTTACCACTTTTCTCCATTTATTTTCAGTTATGGAAATGTTCCCT CTCTTCACCACTAAGCTCCAAAGCAAATGAAAGACGATCACATGTCAGGACAGTAGTAAAGGCAGCTTATAAATGGGACATAAATCAGAG ATGTGTTGGTATTTTGAGACTCAAGACTGTCCTTTTTTAAAATAAAAATAAAAACATTACCAGGTTCCCAAGCCAATCTGGCTTAACCAA CAGTGCACTGAAATATTAGTGTTTACCTCCAAGGCTAGGGAGCCAAGGGGAGGAGGAGAATTGGAGGAAGGGGAGATAATGGGAAGAGGA TGGCGCCTTCCTGAGTTGGCTAGAGGGCCAACCTTTGATAACAGTTTGACGAAATCAATCTTTTTTTTTTTTTTTTTTGAGACAGAGTCT CACTCTGTCACCCAGGCTGGAGTACAGTGGTGGGATCTCGGCTCACTGCAAGCTCTGCCTCCCAGGTTCATGCCATTCTCCTGCCCCAGC CTCTCCTGAGTAGCTGGAACTACAGGTGCCCGCCACCACGCCCAGCTAATTTTTTCTATTTTTAGTCATGCCTGGCTAATTTTTTGTATT TTTAGTAGAGATGGGGTTTCACCGTGTTAGCCAGGATGGTCTCCATCTCCTTACCTCGCAATCCGCCCGCCTCAGCCTCCCAAAGTGTTG GGATTACAGGCATGAGCCACCGTGCCCGGCCTATAGAAATCAATCTTTTTGACTCTTCTCACTTTTATCTCCCCATGCCCAAGGTTTGCC TGTTCCATAACACTCACTCCCTTCCCCCTTGCTAATCAGAAGCCATCTCCTCTCAGTGTCTGATCTCTGCTCTTCATACATGATTACAGT CATGGGGTAGAGAGTGCTTGCTAAATTATGCAGTTAATCCTATGGTGCTTTAATTTTCAGGCCTTCAAAAAACACTTGTACAGTGATGTG CAGATTTTTAAACAGTTGAACTTCCTTGTACTACAGTTTTTGTATTGACAGCCAAATTTGTCTTTCATTCTTCAGATTGTGAATAAAGTG ATTTTTACAGGGCTTCCAGCAAAGTTTTTCCTTTCATCTAAGGCTTGTAGAACCCTAGCTTATATAGCTGCTTACATGAGAAATGCAAAA TCTGTATTCACCATGACTTTAGTAACAAAGGTAAAGTTTTTTAGTAGTGCCAAGGCAAGAGGAACAATCTTGGTGGTAGTACTAAGTTTT GTCAATATTGTGGTTTCCTGATTGTATTGTTGGCTTTCTCTCTGAGCATTGAGGTATACTAGAAGTAGAGCTTCTCAAACATAATATCAT TACCTCATAAGCATTAACAAATCAGGCCCAAAGAGCGTAAGTCCTAGAAATTTGTTTTAAAGCAGCCCTAGTCATGGTGCTGGTGCTACC GCCTTGTTTTAGGAGCCTGCCTCCTGTCAGTATGAAACCCTCACCTGAAAAATGCCAGCCTGGACACCAAACACTGAGCCCCTTCAACAG GCACATTATTTCCCCCTGAGATCCATAAGGGAATTTAGTTTCTACTATTGTAGAGTTCTGAAAAGAGGTAAAATAGTAGTCCTTTGGTCA TCCTATTTTTGCTTTCAATTTTGATATTTCAGACTGTAAAAGGCCTTGGGGGATGATAGTACATGTGGTAGCAGTAATTTTTTTGAAGCA ACTGCACTGACATTCATTTGAGTTTTCTCTCATTATCAGATTCTGTTCCAAACAAGTATTCTGTAGATCCAAATGGATTACCAGTGTGCT ACAGACTTCTTATTATAGAACAGCATTCTATTCTACATCAAAAATAGTTTGTGTAAGTTAGTTTTGGTTACCATCTAAAATATTTTTAAA TGTTCTTTACATAAAAATTTATGTTGTGTTTTAAAATCCTTAGGGGCTTTATCTATTTTTCTAAGTCAGTTAACTGTACTTCTAAAAAAA GTATTTTGTATCTACTTTTGTAACTTCGTCAGAATAAAATATATTGAAAGCAAGAAATGACTGATTAAAGTATGTATATCAACACACATT >85761_85761_2_SPG7-MOB1A_SPG7_chr16_89579445_ENST00000341316_MOB1A_chr2_74394234_ENST00000396049_length(amino acids)=281AA_BP=125 MAVLLLLLRALRRGPGPGPRPLWGPGPAWSPGFPARPGRGRPYMASRPPGDLAEAGGRALQSLQLRLLTPTFEGINGLLLKQHLVQNPVR LWQLLGGTFYFNTSRLKQKNKEKDKSKGKAPEEDEAVDFFNQINMLYGTITEFCTEASCPVMSAGPRYEYHWADGTNIKKPIKCSAPKYI DYLMTWVQDQLDDETLFPSKIGVPFPKNFMSVAKTILKRLFRVYAHIYHQHFDSVMQLQEEAHLNTSFKHFIFFVQEFNLIDRRELAPLQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SPG7-MOB1A |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000268704 | + | 3 | 17 | 701_795 | 125.33333333333333 | 796.0 | PPIF |
| Hgene | SPG7 | chr16:89579445 | chr2:74394234 | ENST00000341316 | + | 3 | 10 | 701_795 | 125.33333333333333 | 490.0 | PPIF |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SPG7-MOB1A |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SPG7-MOB1A |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
