
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:SPTBN1-C2orf73 (FusionGDB2 ID:86121) |
Fusion Gene Summary for SPTBN1-C2orf73 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: SPTBN1-C2orf73 | Fusion gene ID: 86121 | Hgene | Tgene | Gene symbol | SPTBN1 | C2orf73 | Gene ID | 6711 | 129852 |
| Gene name | spectrin beta, non-erythrocytic 1 | chromosome 2 open reading frame 73 | |
| Synonyms | ELF|HEL102|SPTB2|betaSpII | - | |
| Cytomap | 2p16.2 | 2p16.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | spectrin beta chain, non-erythrocytic 1beta-G spectrinbeta-II spectrinbeta-fodrinbeta-spectrin 2beta-spectrin IIbeta-spectrin non-erythrocytic 1embryonic liver beta-fodrinepididymis luminal protein 102fodrin beta chainspectrin beta chain, brain | uncharacterized protein C2orf73 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000333896, ENST00000356805, | ENST00000491538, ENST00000398634, ENST00000405749, | |
| Fusion gene scores | * DoF score | 24 X 20 X 9=4320 | 1 X 1 X 1=1 |
| # samples | 26 | 1 | |
| ** MAII score | log2(26/4320*10)=-4.05444778402238 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: SPTBN1 [Title/Abstract] AND C2orf73 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | SPTBN1(54864940)-C2orf73(54608983), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across SPTBN1 (5'-gene) Fusion gene breakpoints across SPTBN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
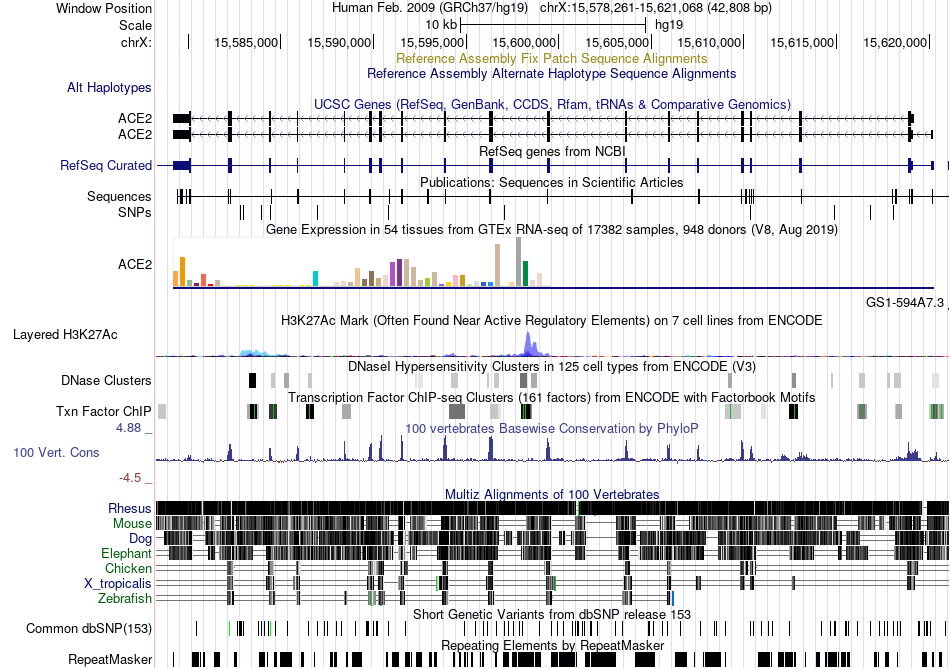 |
 Fusion gene breakpoints across C2orf73 (3'-gene) Fusion gene breakpoints across C2orf73 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
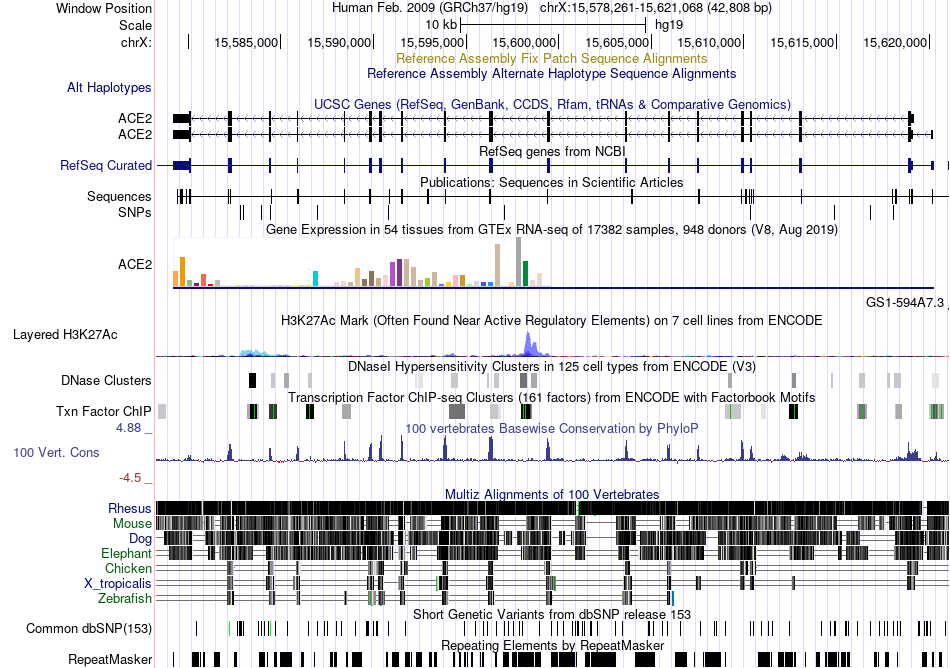 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GH-01A | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
Top |
Fusion Gene ORF analysis for SPTBN1-C2orf73 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000333896 | ENST00000491538 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
| 5CDS-3UTR | ENST00000356805 | ENST00000491538 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
| 5CDS-intron | ENST00000333896 | ENST00000398634 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
| 5CDS-intron | ENST00000356805 | ENST00000398634 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
| In-frame | ENST00000333896 | ENST00000405749 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
| In-frame | ENST00000356805 | ENST00000405749 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000356805 | SPTBN1 | chr2 | 54864940 | + | ENST00000405749 | C2orf73 | chr2 | 54608983 | + | 4427 | 4139 | 266 | 4150 | 1294 |
| ENST00000333896 | SPTBN1 | chr2 | 54864940 | + | ENST00000405749 | C2orf73 | chr2 | 54608983 | + | 4492 | 4204 | 313 | 4215 | 1300 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000356805 | ENST00000405749 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + | 0.011304446 | 0.9886955 |
| ENST00000333896 | ENST00000405749 | SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608983 | + | 0.011084297 | 0.9889157 |
Top |
Fusion Genomic Features for SPTBN1-C2orf73 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608982 | + | 0.053265978 | 0.9467341 |
| SPTBN1 | chr2 | 54864940 | + | C2orf73 | chr2 | 54608982 | + | 0.053265978 | 0.9467341 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
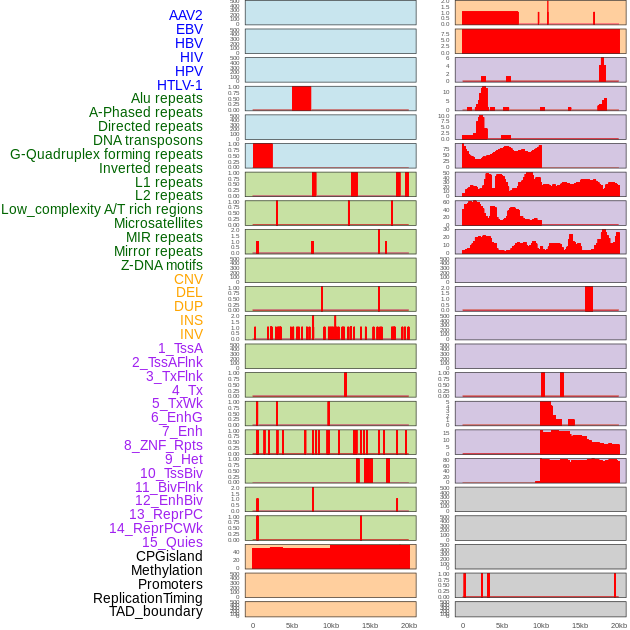 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
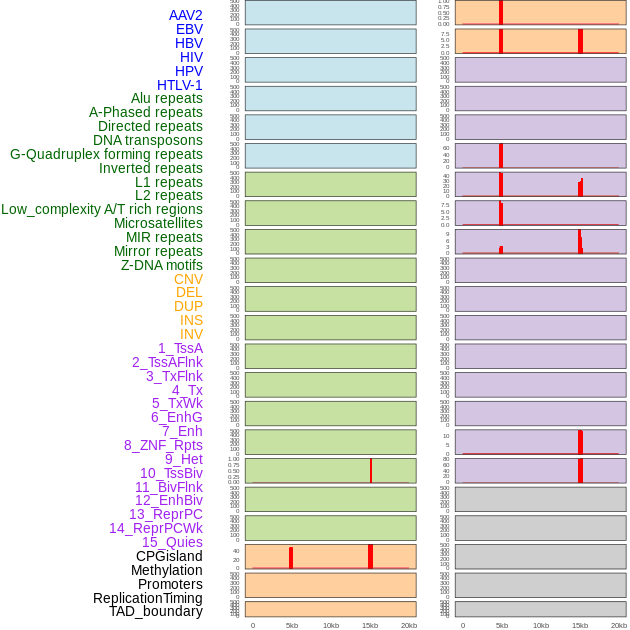 |
Top |
Fusion Protein Features for SPTBN1-C2orf73 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr2:54864940/chr2:54608983) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 173_278 | 1273 | 2156.0 | Domain | Calponin-homology (CH) 2 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 54_158 | 1273 | 2156.0 | Domain | Calponin-homology (CH) 1 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 173_278 | 1286 | 2365.0 | Domain | Calponin-homology (CH) 2 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 54_158 | 1286 | 2365.0 | Domain | Calponin-homology (CH) 1 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 2_275 | 1273 | 2156.0 | Region | Note=Actin-binding |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 2_275 | 1286 | 2365.0 | Region | Note=Actin-binding |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1063_1166 | 1273 | 2156.0 | Repeat | Spectrin 8 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1170_1258 | 1273 | 2156.0 | Repeat | Spectrin 9 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 303_411 | 1273 | 2156.0 | Repeat | Spectrin 1 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 423_525 | 1273 | 2156.0 | Repeat | Spectrin 2 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 530_636 | 1273 | 2156.0 | Repeat | Spectrin 3 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 639_742 | 1273 | 2156.0 | Repeat | Spectrin 4 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 745_847 | 1273 | 2156.0 | Repeat | Spectrin 5 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 850_952 | 1273 | 2156.0 | Repeat | Spectrin 6 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 957_1060 | 1273 | 2156.0 | Repeat | Spectrin 7 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1063_1166 | 1286 | 2365.0 | Repeat | Spectrin 8 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1170_1258 | 1286 | 2365.0 | Repeat | Spectrin 9 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 303_411 | 1286 | 2365.0 | Repeat | Spectrin 1 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 423_525 | 1286 | 2365.0 | Repeat | Spectrin 2 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 530_636 | 1286 | 2365.0 | Repeat | Spectrin 3 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 639_742 | 1286 | 2365.0 | Repeat | Spectrin 4 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 745_847 | 1286 | 2365.0 | Repeat | Spectrin 5 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 850_952 | 1286 | 2365.0 | Repeat | Spectrin 6 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 957_1060 | 1286 | 2365.0 | Repeat | Spectrin 7 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 2197_2307 | 1273 | 2156.0 | Domain | PH |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 2197_2307 | 1286 | 2365.0 | Domain | PH |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1276_1376 | 1273 | 2156.0 | Repeat | Spectrin 10 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1381_1482 | 1273 | 2156.0 | Repeat | Spectrin 11 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1486_1590 | 1273 | 2156.0 | Repeat | Spectrin 12 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1592_1696 | 1273 | 2156.0 | Repeat | Spectrin 13 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1698_1801 | 1273 | 2156.0 | Repeat | Spectrin 14 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1805_1907 | 1273 | 2156.0 | Repeat | Spectrin 15 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1914_2014 | 1273 | 2156.0 | Repeat | Spectrin 16 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 2018_2097 | 1273 | 2156.0 | Repeat | Spectrin 17 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1276_1376 | 1286 | 2365.0 | Repeat | Spectrin 10 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1381_1482 | 1286 | 2365.0 | Repeat | Spectrin 11 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1486_1590 | 1286 | 2365.0 | Repeat | Spectrin 12 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1592_1696 | 1286 | 2365.0 | Repeat | Spectrin 13 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1698_1801 | 1286 | 2365.0 | Repeat | Spectrin 14 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1805_1907 | 1286 | 2365.0 | Repeat | Spectrin 15 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1914_2014 | 1286 | 2365.0 | Repeat | Spectrin 16 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 2018_2097 | 1286 | 2365.0 | Repeat | Spectrin 17 |
Top |
Fusion Gene Sequence for SPTBN1-C2orf73 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >86121_86121_1_SPTBN1-C2orf73_SPTBN1_chr2_54864940_ENST00000333896_C2orf73_chr2_54608983_ENST00000405749_length(transcript)=4492nt_BP=4204nt GGTGTGCGCGCTGCGCCCGCGAGCTCCCGGGCTCGGCAACCGTGGCATGCTTAGGATTGGCCATATTTAAAAGTTCTGAAGTAGAACCTG CGCCTCCTCTCTGCTTCTCCCTCCTCCTCAGTAATTTATTTCGAGCTTCCAGGCAAGGGCCACGGAAGAAGGGAAAGCAAGAAATTAGAT GCCTGTGTGGTAACTCCTCGCGGAGCTAAGGTGGACTCTCTTGCAGCCAACTTCCCATCAGATCACCCTGCTGGACTTGCAGACCGGAAT GGGGCTCGCCTAAGGAGCCGAGCGCTGCGGAGGCTGCTGCGTGTTGGATGGGAGTGGGAGGGGGCTGGAGCGAGATTTCCAGGGCGCAGT CCTCCGGGGCGTTACGCCGGGCATAATGGAATTGCAGAGGACGTCTAGTATCTCCGGGCCGCTGTCGCCGGCGTACACGGGGCAGGTGCC TTACAACTACAACCAGCTGGAAGGCAGATTCAAGCAGCTGCAAGATGAGCGTGAAGCCGTGCAGAAGAAGACCTTCACCAAGTGGGTCAA TTCCCACCTTGCCCGTGTGTCCTGCCGGATCACAGACCTGTACACTGACCTTCGAGATGGACGGATGCTCATCAAGCTGCTGGAGGTCCT CTCTGGAGAGAGGCTGCCTAAACCCACCAAGGGACGAATGCGCATCCACTGCTTAGAGAATGTGGACAAGGCCCTTCAGTTCCTGAAGGA GCAGAGAGTCCATCTTGAGAACATGGGGTCCCATGACATCGTGGATGGAAACCACCGGCTGACCCTTGGCCTCATCTGGACCATCATCCT GCGCTTCCAGATCCAGGATATCAGTGTGGAAACTGAAGACAACAAAGAGAAGAAATCTGCCAAGGATGCATTGCTGTTGTGGTGCCAGAT GAAGACAGCTGGGTACCCCAATGTCAACATTCACAATTTCACCACTAGCTGGAGGGACGGCATGGCCTTCAATGCACTGATACACAAACA CCGGCCTGACCTGATAGATTTTGACAAACTAAAGAAATCTAACGCACACTACAACCTGCAGAATGCATTTAATCTGGCAGAACAGCACCT CGGCCTCACTAAACTGTTGGACCCCGAAGACATCAGCGTGGACCATCCTGATGAGAAGTCCATAATCACTTATGTGGTGACTTATTACCA CTACTTCTCTAAGATGAAGGCCTTAGCTGTTGAAGGAAAACGAATTGGAAAGGTGCTTGACAATGCTATTGAAACAGAAAAAATGATTGA AAAGTATGAATCACTTGCCTCTGACCTTCTGGAATGGATTGAACAAACCATCATCATTCTGAACAATCGCAAATTTGCCAATTCACTGGT CGGGGTTCAACAGCAGCTTCAGGCATTCAACACTTACCGCACTGTGGAGAAACCACCCAAATTTACTGAGAAGGGGAACTTGGAAGTGCT GCTCTTCACCATTCAGAGCAAGATGAGGGCCAACAACCAGAAGGTCTACATGCCCCGGGAGGGGAAGCTCATCTCTGACATCAACAAGGC CTGGGAAAGACTGGAAAAAGCGGAACACGAAAGAGAACTGGCTTTGCGGAATGAGCTCATAAGACAGGAGAAACTGGAACAGCTCGCCCG CAGATTTGATCGCAAGGCAGCTATGAGGGAGACTTGGCTGAGCGAAAACCAGCGTCTGGTGTCTCAGGACAACTTTGGGTTTGACCTTCC TGCAGTTGAGGCCGCCACAAAAAAGCACGAGGCCATTGAGACAGACATTGCCGCATACGAGGAGCGTGTGCAGGCTGTGGTAGCCGTGGC CAGGGAGCTCGAGGCCGAGAATTACCACGACATCAAGCGCATCACAGCGAGGAAGGACAATGTCATCCGGCTCTGGGAATACCTACTGGA ACTGCTCAGGGCCCGGAGACAGCGGCTCGAGATGAACCTGGGGCTGCAGAAGATATTCCAGGAAATGCTCTACATTATGGACTGGATGGA TGAAATGAAGGTGCTAGTATTGTCTCAAGACTATGGCAAACACTTACTTGGTGTGGAAGACCTGTTACAGAAGCACACCCTGGTTGAAGC AGACATTGGCATCCAGGCAGAGCGGGTGAGAGGTGTCAATGCCTCCGCCCAGAAGTTCGCAACAGACGGGGAAGGTTACAAGCCCTGTGA CCCCCAGGTGATCCGAGACCGCGTGGCCCACATGGAGTTCTGTTATCAAGAGCTTTGCCAGCTGGCGGCTGAGCGCAGGGCCCGTCTGGA AGAGTCCCGCCGCCTCTGGAAGTTCTTCTGGGAGATGGCAGAAGAGGAAGGCTGGATACGGGAGAAGGAGAAGATCCTGTCCTCGGACGA TTACGGGAAAGACCTGACCAGCGTCATGCGCCTGCTCAGCAAGCACCGGGCGTTCGAGGACGAGATGAGCGGCCGCAGTGGCCACTTTGA GCAGGCCATCAAGGAAGGCGAAGACATGATCGCGGAGGAGCACTTCGGGTCGGAGAAGATCCGTGAGAGGATCATTTACATCCGGGAGCA GTGGGCCAACCTAGAGCAGCTCTCGGCCATTCGGAAGAAGCGCCTGGAGGAGGCCTCCCTGCTGCACCAGTTCCAGGCAGATGCTGATGA CATTGATGCCTGGATGCTGGACATCCTCAAGATTGTCTCCAGCAGCGACGTGGGCCACGATGAGTATTCCACACAGTCTCTGGTCAAGAA ACACAAGGACGTGGCGGAAGAGATCGCCAATTACAGGCCCACCCTTGACACGCTGCACGAACAAGCCAGCGCCCTCCCCCAGGAGCATGC CGAGTCTCCAGACGTGAGGGGCAGGCTGTCGGGCATCGAGGAGCGGTATAAGGAGGTGGCAGAGCTGACGCGGCTGCGGAAGCAGGCACT CCAGGACACTCTGGCCCTGTACAAGATGTTCAGCGAGGCTGATGCCTGTGAGCTCTGGATCGACGAGAAGGAGCAGTGGCTCAACAACAT GCAGATCCCAGAGAAGCTGGAGGATCTGGAGGTCATCCAGCACAGATTTGAGAGCCTAGAACCAGAAATGAACAACCAGGCTTCCCGGGT TGCAGTGGTGAACCAGATTGCACGCCAGCTGATGCACAGCGGCCACCCAAGTGAGAAGGAAATCAAAGCCCAGCAGGACAAACTCAACAC AAGGTGGAGCCAGTTCAGAGAACTGGTTGACAGGAAGAAGGATGCCCTCCTGTCTGCCCTGAGCATCCAGAACTACCACCTCGAGTGCAA TGAAACCAAATCCTGGATTCGGGAAAAGACCAAGGTCATCGAGTCCACCCAGGACCTGGGCAATGACCTGGCTGGCGTCATGGCCCTGCA GCGCAAGCTGACCGGCATGGAGCGGGACTTGGTGGCCATTGAGGCAAAGCTGAGTGACCTGCAGAAGGAGGCGGAGAAGCTGGAGTCCGA GCACCCCGACCAGGCCCAGGCCATCCTGTCTCGGCTGGCCGAGATCAGCGACGTGTGGGAGGAGATGAAGACCACCCTGAAAAACCGAGA GGCCTCCCTGGGAGAGGCCAGCAAGCTGCAGCAGTTCCTACGGGACTTGGACGACTTCCAGTCCTGGCTCTCTAGGACCCAGACAGCGAT CGCCTCGGAGGACATGCCAAACACCCTGACCGAGGCTGAGAAGCTGCTCACGCAGCACGAGAACATCAAGAACGAGATCGACAACTACGA GGAGGACTACCAGAAGATGAGGGACATGGGCGAGATGGTCACCCAGGGGCAGACCGATGCCCAGTACATGTTTCTGCGGCAGCGGCTGCA GGCCCTGGACACTGGATGGAACGAGCTCCACAAGATGTGGGAGAACAGACAAAATCTCCTATCCCAGTCACATGCCTACCAGCAGTTCCT CAGAGACACGAAGCAAGCCGAAGCCTTTCTTAACAACCAGGAGTATGTTCTGGCTCACACTGAAATGCCTACCACCTTGGAAGGAGCTGA AGCAGCAATTAAAAAGCAAGAGGACTTCATGACCACCATGGACGCCAATGAGGAGAAGATCAATGCTGTGGTGGAGACTGGCCGGAGGCT GGTGAGCGATGGGAACATCAACTCAGATCGCATCCAGGAGAAGGTGGACTCTATTGATGACAGACATAGGAAGAATCGTGAGACAGCCAG TGAACTTTTGATGAGGTTGAAGGACAACAGGGATCTACAGAAATTCCTGCAAGATTGTCAAGAGATGAAACCTTGAGAAAAGGAGCCAAG ACTGTAACAGTGGTCCTGTGCCACAGTGCAGGAAGAAAAGGAACAGAGAGATCACGGGCATGTTTTTTTAGGTGGTTTCATGCCATCTTA TTTTCTCCTTGAAATACATTCACAATGAGGTGACTGGCTCTGTGGGAAGCCTATGGCCATGTTGTAATGACACTCAATCAGCCCAGTGGA >86121_86121_1_SPTBN1-C2orf73_SPTBN1_chr2_54864940_ENST00000333896_C2orf73_chr2_54608983_ENST00000405749_length(amino acids)=1300AA_BP= MDGSGRGLERDFQGAVLRGVTPGIMELQRTSSISGPLSPAYTGQVPYNYNQLEGRFKQLQDEREAVQKKTFTKWVNSHLARVSCRITDLY TDLRDGRMLIKLLEVLSGERLPKPTKGRMRIHCLENVDKALQFLKEQRVHLENMGSHDIVDGNHRLTLGLIWTIILRFQIQDISVETEDN KEKKSAKDALLLWCQMKTAGYPNVNIHNFTTSWRDGMAFNALIHKHRPDLIDFDKLKKSNAHYNLQNAFNLAEQHLGLTKLLDPEDISVD HPDEKSIITYVVTYYHYFSKMKALAVEGKRIGKVLDNAIETEKMIEKYESLASDLLEWIEQTIIILNNRKFANSLVGVQQQLQAFNTYRT VEKPPKFTEKGNLEVLLFTIQSKMRANNQKVYMPREGKLISDINKAWERLEKAEHERELALRNELIRQEKLEQLARRFDRKAAMRETWLS ENQRLVSQDNFGFDLPAVEAATKKHEAIETDIAAYEERVQAVVAVARELEAENYHDIKRITARKDNVIRLWEYLLELLRARRQRLEMNLG LQKIFQEMLYIMDWMDEMKVLVLSQDYGKHLLGVEDLLQKHTLVEADIGIQAERVRGVNASAQKFATDGEGYKPCDPQVIRDRVAHMEFC YQELCQLAAERRARLEESRRLWKFFWEMAEEEGWIREKEKILSSDDYGKDLTSVMRLLSKHRAFEDEMSGRSGHFEQAIKEGEDMIAEEH FGSEKIRERIIYIREQWANLEQLSAIRKKRLEEASLLHQFQADADDIDAWMLDILKIVSSSDVGHDEYSTQSLVKKHKDVAEEIANYRPT LDTLHEQASALPQEHAESPDVRGRLSGIEERYKEVAELTRLRKQALQDTLALYKMFSEADACELWIDEKEQWLNNMQIPEKLEDLEVIQH RFESLEPEMNNQASRVAVVNQIARQLMHSGHPSEKEIKAQQDKLNTRWSQFRELVDRKKDALLSALSIQNYHLECNETKSWIREKTKVIE STQDLGNDLAGVMALQRKLTGMERDLVAIEAKLSDLQKEAEKLESEHPDQAQAILSRLAEISDVWEEMKTTLKNREASLGEASKLQQFLR DLDDFQSWLSRTQTAIASEDMPNTLTEAEKLLTQHENIKNEIDNYEEDYQKMRDMGEMVTQGQTDAQYMFLRQRLQALDTGWNELHKMWE NRQNLLSQSHAYQQFLRDTKQAEAFLNNQEYVLAHTEMPTTLEGAEAAIKKQEDFMTTMDANEEKINAVVETGRRLVSDGNINSDRIQEK -------------------------------------------------------------- >86121_86121_2_SPTBN1-C2orf73_SPTBN1_chr2_54864940_ENST00000356805_C2orf73_chr2_54608983_ENST00000405749_length(transcript)=4427nt_BP=4139nt GGAATCAGTGCCGCTGTTGCCGTGCAGGCTGCGGATTCCTCCAGTCCCTCCCTCGGCCGCCTCTCCTCCCGGAGCGAGCGCGCAGCCCTG CGCAGCAGCGCCCACTGGTCCCGTCCTGTGAGCCCCGGCCCCAGCCGCGGACAGACCCGCGGAGTCGCCTCCCGGCCCACCCGCCCGGCC GCCGAGGAGCGGGAGGAGGACGGGACCCCGGCGCCCCCACCCCATCCCCGGGAGAACTCTAAGAAGGAGCTGATGTGGAGGAGCAGCTGA GACAGTTCAAGATGACGACCACAGTAGCCACAGACTATGACAACATTGAGATCCAGCAGCAGTACAGTGATGTCAACAACCGCTGGGATG TCGACGACTGGGACAATGAGAACAGCTCTGCGCGGCTTTTTGAGCGGTCCCGCATCAAGGCTCTGGCAGATGAGCGTGAAGCCGTGCAGA AGAAGACCTTCACCAAGTGGGTCAATTCCCACCTTGCCCGTGTGTCCTGCCGGATCACAGACCTGTACACTGACCTTCGAGATGGACGGA TGCTCATCAAGCTGCTGGAGGTCCTCTCTGGAGAGAGGCTGCCTAAACCCACCAAGGGACGAATGCGCATCCACTGCTTAGAGAATGTGG ACAAGGCCCTTCAGTTCCTGAAGGAGCAGAGAGTCCATCTTGAGAACATGGGGTCCCATGACATCGTGGATGGAAACCACCGGCTGACCC TTGGCCTCATCTGGACCATCATCCTGCGCTTCCAGATCCAGGATATCAGTGTGGAAACTGAAGACAACAAAGAGAAGAAATCTGCCAAGG ATGCATTGCTGTTGTGGTGCCAGATGAAGACAGCTGGGTACCCCAATGTCAACATTCACAATTTCACCACTAGCTGGAGGGACGGCATGG CCTTCAATGCACTGATACACAAACACCGGCCTGACCTGATAGATTTTGACAAACTAAAGAAATCTAACGCACACTACAACCTGCAGAATG CATTTAATCTGGCAGAACAGCACCTCGGCCTCACTAAACTGTTGGACCCCGAAGACATCAGCGTGGACCATCCTGATGAGAAGTCCATAA TCACTTATGTGGTGACTTATTACCACTACTTCTCTAAGATGAAGGCCTTAGCTGTTGAAGGAAAACGAATTGGAAAGGTGCTTGACAATG CTATTGAAACAGAAAAAATGATTGAAAAGTATGAATCACTTGCCTCTGACCTTCTGGAATGGATTGAACAAACCATCATCATTCTGAACA ATCGCAAATTTGCCAATTCACTGGTCGGGGTTCAACAGCAGCTTCAGGCATTCAACACTTACCGCACTGTGGAGAAACCACCCAAATTTA CTGAGAAGGGGAACTTGGAAGTGCTGCTCTTCACCATTCAGAGCAAGATGAGGGCCAACAACCAGAAGGTCTACATGCCCCGGGAGGGGA AGCTCATCTCTGACATCAACAAGGCCTGGGAAAGACTGGAAAAAGCGGAACACGAAAGAGAACTGGCTTTGCGGAATGAGCTCATAAGAC AGGAGAAACTGGAACAGCTCGCCCGCAGATTTGATCGCAAGGCAGCTATGAGGGAGACTTGGCTGAGCGAAAACCAGCGTCTGGTGTCTC AGGACAACTTTGGGTTTGACCTTCCTGCAGTTGAGGCCGCCACAAAAAAGCACGAGGCCATTGAGACAGACATTGCCGCATACGAGGAGC GTGTGCAGGCTGTGGTAGCCGTGGCCAGGGAGCTCGAGGCCGAGAATTACCACGACATCAAGCGCATCACAGCGAGGAAGGACAATGTCA TCCGGCTCTGGGAATACCTACTGGAACTGCTCAGGGCCCGGAGACAGCGGCTCGAGATGAACCTGGGGCTGCAGAAGATATTCCAGGAAA TGCTCTACATTATGGACTGGATGGATGAAATGAAGGTGCTAGTATTGTCTCAAGACTATGGCAAACACTTACTTGGTGTGGAAGACCTGT TACAGAAGCACACCCTGGTTGAAGCAGACATTGGCATCCAGGCAGAGCGGGTGAGAGGTGTCAATGCCTCCGCCCAGAAGTTCGCAACAG ACGGGGAAGGTTACAAGCCCTGTGACCCCCAGGTGATCCGAGACCGCGTGGCCCACATGGAGTTCTGTTATCAAGAGCTTTGCCAGCTGG CGGCTGAGCGCAGGGCCCGTCTGGAAGAGTCCCGCCGCCTCTGGAAGTTCTTCTGGGAGATGGCAGAAGAGGAAGGCTGGATACGGGAGA AGGAGAAGATCCTGTCCTCGGACGATTACGGGAAAGACCTGACCAGCGTCATGCGCCTGCTCAGCAAGCACCGGGCGTTCGAGGACGAGA TGAGCGGCCGCAGTGGCCACTTTGAGCAGGCCATCAAGGAAGGCGAAGACATGATCGCGGAGGAGCACTTCGGGTCGGAGAAGATCCGTG AGAGGATCATTTACATCCGGGAGCAGTGGGCCAACCTAGAGCAGCTCTCGGCCATTCGGAAGAAGCGCCTGGAGGAGGCCTCCCTGCTGC ACCAGTTCCAGGCAGATGCTGATGACATTGATGCCTGGATGCTGGACATCCTCAAGATTGTCTCCAGCAGCGACGTGGGCCACGATGAGT ATTCCACACAGTCTCTGGTCAAGAAACACAAGGACGTGGCGGAAGAGATCGCCAATTACAGGCCCACCCTTGACACGCTGCACGAACAAG CCAGCGCCCTCCCCCAGGAGCATGCCGAGTCTCCAGACGTGAGGGGCAGGCTGTCGGGCATCGAGGAGCGGTATAAGGAGGTGGCAGAGC TGACGCGGCTGCGGAAGCAGGCACTCCAGGACACTCTGGCCCTGTACAAGATGTTCAGCGAGGCTGATGCCTGTGAGCTCTGGATCGACG AGAAGGAGCAGTGGCTCAACAACATGCAGATCCCAGAGAAGCTGGAGGATCTGGAGGTCATCCAGCACAGATTTGAGAGCCTAGAACCAG AAATGAACAACCAGGCTTCCCGGGTTGCAGTGGTGAACCAGATTGCACGCCAGCTGATGCACAGCGGCCACCCAAGTGAGAAGGAAATCA AAGCCCAGCAGGACAAACTCAACACAAGGTGGAGCCAGTTCAGAGAACTGGTTGACAGGAAGAAGGATGCCCTCCTGTCTGCCCTGAGCA TCCAGAACTACCACCTCGAGTGCAATGAAACCAAATCCTGGATTCGGGAAAAGACCAAGGTCATCGAGTCCACCCAGGACCTGGGCAATG ACCTGGCTGGCGTCATGGCCCTGCAGCGCAAGCTGACCGGCATGGAGCGGGACTTGGTGGCCATTGAGGCAAAGCTGAGTGACCTGCAGA AGGAGGCGGAGAAGCTGGAGTCCGAGCACCCCGACCAGGCCCAGGCCATCCTGTCTCGGCTGGCCGAGATCAGCGACGTGTGGGAGGAGA TGAAGACCACCCTGAAAAACCGAGAGGCCTCCCTGGGAGAGGCCAGCAAGCTGCAGCAGTTCCTACGGGACTTGGACGACTTCCAGTCCT GGCTCTCTAGGACCCAGACAGCGATCGCCTCGGAGGACATGCCAAACACCCTGACCGAGGCTGAGAAGCTGCTCACGCAGCACGAGAACA TCAAGAACGAGATCGACAACTACGAGGAGGACTACCAGAAGATGAGGGACATGGGCGAGATGGTCACCCAGGGGCAGACCGATGCCCAGT ACATGTTTCTGCGGCAGCGGCTGCAGGCCCTGGACACTGGATGGAACGAGCTCCACAAGATGTGGGAGAACAGACAAAATCTCCTATCCC AGTCACATGCCTACCAGCAGTTCCTCAGAGACACGAAGCAAGCCGAAGCCTTTCTTAACAACCAGGAGTATGTTCTGGCTCACACTGAAA TGCCTACCACCTTGGAAGGAGCTGAAGCAGCAATTAAAAAGCAAGAGGACTTCATGACCACCATGGACGCCAATGAGGAGAAGATCAATG CTGTGGTGGAGACTGGCCGGAGGCTGGTGAGCGATGGGAACATCAACTCAGATCGCATCCAGGAGAAGGTGGACTCTATTGATGACAGAC ATAGGAAGAATCGTGAGACAGCCAGTGAACTTTTGATGAGGTTGAAGGACAACAGGGATCTACAGAAATTCCTGCAAGATTGTCAAGAGA TGAAACCTTGAGAAAAGGAGCCAAGACTGTAACAGTGGTCCTGTGCCACAGTGCAGGAAGAAAAGGAACAGAGAGATCACGGGCATGTTT TTTTAGGTGGTTTCATGCCATCTTATTTTCTCCTTGAAATACATTCACAATGAGGTGACTGGCTCTGTGGGAAGCCTATGGCCATGTTGT AATGACACTCAATCAGCCCAGTGGAGAGGGCCACATGGCAAGGAACTAAGGCCTCCCACCAACAGCCAGCAAAGAACTGAGACCTTCTGC >86121_86121_2_SPTBN1-C2orf73_SPTBN1_chr2_54864940_ENST00000356805_C2orf73_chr2_54608983_ENST00000405749_length(amino acids)=1294AA_BP= MRQFKMTTTVATDYDNIEIQQQYSDVNNRWDVDDWDNENSSARLFERSRIKALADEREAVQKKTFTKWVNSHLARVSCRITDLYTDLRDG RMLIKLLEVLSGERLPKPTKGRMRIHCLENVDKALQFLKEQRVHLENMGSHDIVDGNHRLTLGLIWTIILRFQIQDISVETEDNKEKKSA KDALLLWCQMKTAGYPNVNIHNFTTSWRDGMAFNALIHKHRPDLIDFDKLKKSNAHYNLQNAFNLAEQHLGLTKLLDPEDISVDHPDEKS IITYVVTYYHYFSKMKALAVEGKRIGKVLDNAIETEKMIEKYESLASDLLEWIEQTIIILNNRKFANSLVGVQQQLQAFNTYRTVEKPPK FTEKGNLEVLLFTIQSKMRANNQKVYMPREGKLISDINKAWERLEKAEHERELALRNELIRQEKLEQLARRFDRKAAMRETWLSENQRLV SQDNFGFDLPAVEAATKKHEAIETDIAAYEERVQAVVAVARELEAENYHDIKRITARKDNVIRLWEYLLELLRARRQRLEMNLGLQKIFQ EMLYIMDWMDEMKVLVLSQDYGKHLLGVEDLLQKHTLVEADIGIQAERVRGVNASAQKFATDGEGYKPCDPQVIRDRVAHMEFCYQELCQ LAAERRARLEESRRLWKFFWEMAEEEGWIREKEKILSSDDYGKDLTSVMRLLSKHRAFEDEMSGRSGHFEQAIKEGEDMIAEEHFGSEKI RERIIYIREQWANLEQLSAIRKKRLEEASLLHQFQADADDIDAWMLDILKIVSSSDVGHDEYSTQSLVKKHKDVAEEIANYRPTLDTLHE QASALPQEHAESPDVRGRLSGIEERYKEVAELTRLRKQALQDTLALYKMFSEADACELWIDEKEQWLNNMQIPEKLEDLEVIQHRFESLE PEMNNQASRVAVVNQIARQLMHSGHPSEKEIKAQQDKLNTRWSQFRELVDRKKDALLSALSIQNYHLECNETKSWIREKTKVIESTQDLG NDLAGVMALQRKLTGMERDLVAIEAKLSDLQKEAEKLESEHPDQAQAILSRLAEISDVWEEMKTTLKNREASLGEASKLQQFLRDLDDFQ SWLSRTQTAIASEDMPNTLTEAEKLLTQHENIKNEIDNYEEDYQKMRDMGEMVTQGQTDAQYMFLRQRLQALDTGWNELHKMWENRQNLL SQSHAYQQFLRDTKQAEAFLNNQEYVLAHTEMPTTLEGAEAAIKKQEDFMTTMDANEEKINAVVETGRRLVSDGNINSDRIQEKVDSIDD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for SPTBN1-C2orf73 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000333896 | + | 17 | 31 | 1563_2093 | 1273.0 | 2156.0 | ANK2 |
| Hgene | SPTBN1 | chr2:54864940 | chr2:54608983 | ENST00000356805 | + | 18 | 36 | 1563_2093 | 1286.0 | 2365.0 | ANK2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for SPTBN1-C2orf73 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for SPTBN1-C2orf73 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
