
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:STT3A-CKB (FusionGDB2 ID:87746) |
Fusion Gene Summary for STT3A-CKB |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: STT3A-CKB | Fusion gene ID: 87746 | Hgene | Tgene | Gene symbol | STT3A | CKB | Gene ID | 3703 | 1152 |
| Gene name | STT3 oligosaccharyltransferase complex catalytic subunit A | creatine kinase B | |
| Synonyms | ITM1|STT3-A|TMC | B-CK|BCK|CKBB|CPK-B|HEL-211|HEL-S-29 | |
| Cytomap | 11q24.2 | 14q32.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3AB5STT3, subunit of the oligosaccharyltransferase complex, homolog ASTT3A, catalytic subunit of the oligosaccharyltransferase complexSTT3A, cataylic subunit of the oligosacchar | creatine kinase B-typebrain creatine kinasecreatine kinase B chaincreatine kinase braincreatine kinase brain-typecreatine phosphokinase B-typeepididymis luminal protein 211epididymis secretory protein Li 29 | |
| Modification date | 20200327 | 20200313 | |
| UniProtAcc | . | P12277 | |
| Ensembl transtripts involved in fusion gene | ENST00000392708, ENST00000529196, ENST00000531491, ENST00000526364, | ENST00000348956, | |
| Fusion gene scores | * DoF score | 12 X 11 X 4=528 | 19 X 21 X 5=1995 |
| # samples | 11 | 22 | |
| ** MAII score | log2(11/528*10)=-2.26303440583379 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(22/1995*10)=-3.18081331758505 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: STT3A [Title/Abstract] AND CKB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | STT3A(125488374)-CKB(103986376), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across STT3A (5'-gene) Fusion gene breakpoints across STT3A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
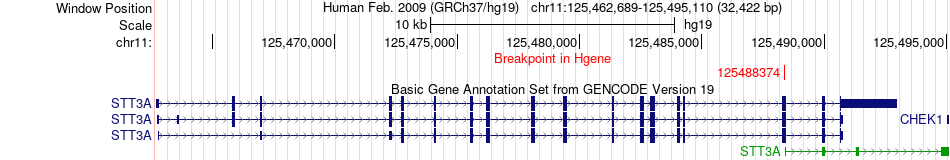 |
 Fusion gene breakpoints across CKB (3'-gene) Fusion gene breakpoints across CKB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
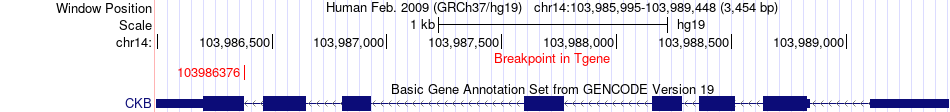 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | EC545946 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - |
Top |
Fusion Gene ORF analysis for STT3A-CKB |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000392708 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - |
| In-frame | ENST00000529196 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - |
| In-frame | ENST00000531491 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - |
| intron-3CDS | ENST00000526364 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000392708 | STT3A | chr11 | 125488374 | + | ENST00000348956 | CKB | chr14 | 103986376 | - | 2399 | 2015 | 138 | 1937 | 599 |
| ENST00000529196 | STT3A | chr11 | 125488374 | + | ENST00000348956 | CKB | chr14 | 103986376 | - | 2446 | 2062 | 164 | 1984 | 606 |
| ENST00000531491 | STT3A | chr11 | 125488374 | + | ENST00000348956 | CKB | chr14 | 103986376 | - | 2204 | 1820 | 66 | 1742 | 558 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000392708 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - | 0.001446831 | 0.9985532 |
| ENST00000529196 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - | 0.001334208 | 0.9986658 |
| ENST00000531491 | ENST00000348956 | STT3A | chr11 | 125488374 | + | CKB | chr14 | 103986376 | - | 0.002101428 | 0.9978986 |
Top |
Fusion Genomic Features for STT3A-CKB |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
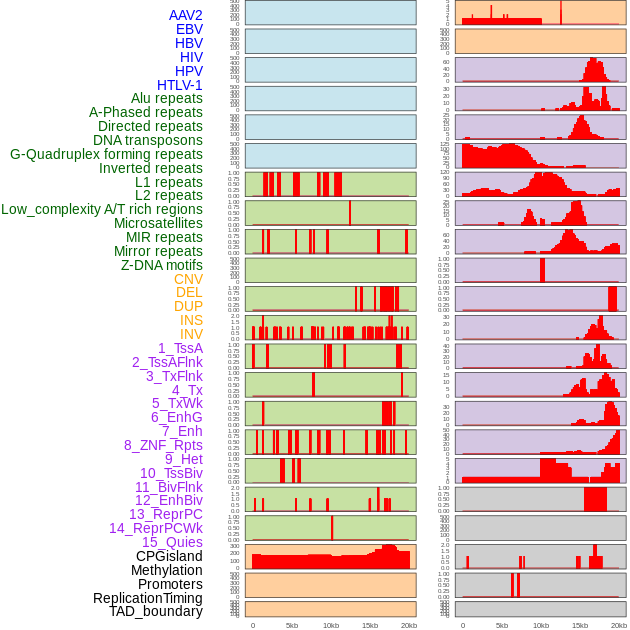 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for STT3A-CKB |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr11:125488374/chr14:103986376) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CKB |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Reversibly catalyzes the transfer of phosphate between ATP and various phosphogens (e.g. creatine phosphate). Creatine kinase isoenzymes play a central role in energy transduction in tissues with large, fluctuating energy demands, such as skeletal muscle, heart, brain and spermatozoa. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CKB | chr11:125488374 | chr14:103986376 | ENST00000348956 | 6 | 8 | 320_325 | 322 | 382.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 167_169 | 0 | 706.0 | Motif | DXD motif 2 |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 348_351 | 0 | 706.0 | Motif | SVSE motif |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 525_529 | 0 | 706.0 | Motif | WWDYG motif |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 592_599 | 0 | 706.0 | Motif | DK motif |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 167_169 | 0 | 706.0 | Motif | DXD motif 2 |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 348_351 | 0 | 706.0 | Motif | SVSE motif |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 525_529 | 0 | 706.0 | Motif | WWDYG motif |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 592_599 | 0 | 706.0 | Motif | DK motif |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 525_527 | 0 | 706.0 | Region | Interacts with target acceptor peptide in protein substrate |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 525_527 | 0 | 706.0 | Region | Interacts with target acceptor peptide in protein substrate |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 139_140 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 159_169 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 190_191 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 1_17 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 207_211 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 229_233 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 260_267 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 288_300 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 322_356 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 380_385 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 39_119 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 403_406 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 429_453 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 474_705 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 139_140 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 159_169 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 190_191 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 1_17 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 207_211 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 229_233 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 260_267 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 288_300 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 322_356 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 380_385 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 39_119 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 403_406 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 429_453 | 0 | 706.0 | Topological domain | Cytoplasmic |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 474_705 | 0 | 706.0 | Topological domain | Lumenal |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 120_138 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 141_158 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 170_189 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 18_38 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 192_206 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 212_228 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 234_259 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 268_287 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 301_321 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 357_379 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 407_428 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000392708 | + | 1 | 18 | 454_473 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 120_138 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 141_158 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 170_189 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 18_38 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 192_206 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 212_228 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 234_259 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 268_287 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 301_321 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 357_379 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 407_428 | 0 | 706.0 | Transmembrane | Helical |
| Hgene | STT3A | chr11:125488374 | chr14:103986376 | ENST00000529196 | + | 1 | 19 | 454_473 | 0 | 706.0 | Transmembrane | Helical |
| Tgene | CKB | chr11:125488374 | chr14:103986376 | ENST00000348956 | 6 | 8 | 11_98 | 322 | 382.0 | Domain | Phosphagen kinase N-terminal | |
| Tgene | CKB | chr11:125488374 | chr14:103986376 | ENST00000348956 | 6 | 8 | 125_367 | 322 | 382.0 | Domain | Phosphagen kinase C-terminal | |
| Tgene | CKB | chr11:125488374 | chr14:103986376 | ENST00000348956 | 6 | 8 | 128_132 | 322 | 382.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for STT3A-CKB |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >87746_87746_1_STT3A-CKB_STT3A_chr11_125488374_ENST00000392708_CKB_chr14_103986376_ENST00000348956_length(transcript)=2399nt_BP=2015nt ATTGGCCTGGAGCCAGCCAACGGGGGAGCGGTCCGGGAGGCGGACCCGGCTCCTGCCAGGGTTGGGTGCGCCGCTGAACGGATGGCTGAG GGAGCCCCGCGGATCGTTTAGGAAAGCCGGCCAGCTGATCGTCGTGTGTTGCCACCCATTCATGTCAAGATGACTAAGTTTGGATTTTTG CGATTGTCCTATGAGAAGCAGGACACACTTTTGAAGCTTCTCATTCTGTCAATGGCTGCTGTATTATCCTTCTCCACTCGTCTGTTTGCT GTCCTGAGATTTGAAAGTGTTATCCATGAGTTTGATCCGTACTTTAATTATCGGACTACCAGGTTCCTGGCTGAGGAGGGGTTTTATAAA TTCCATAACTGGTTTGATGACCGAGCCTGGTACCCTTTGGGACGAATCATTGGAGGAACAATTTACCCAGGTTTAATGATCACCTCTGCT GCAATCTACCATGTACTCCATTTTTTCCACATCACCATCGACATTCGGAATGTCTGTGTGTTCCTGGCCCCTCTCTTCTCCTCCTTCACC ACCATCGTCACGTACCACCTTACCAAAGAGCTCAAGGATGCAGGGGCTGGGCTTCTTGCTGCTGCCATGATTGCTGTAGTTCCTGGATAT ATCTCCCGATCTGTGGCTGGCTCCTATGATAATGAAGGGATTGCCATCTTTTGCATGCTACTCACCTACTACATGTGGATCAAGGCAGTA AAGACTGGTTCCATCTGTTGGGCAGCTAAGTGTGCCCTTGCTTATTTCTACATGGTCTCGTCATGGGGAGGTTATGTGTTCCTGATCAAC TTAATTCCTCTCCACGTCCTCGTGCTGATGCTCACAGGCCGTTTCTCTCACCGGATCTATGTGGCCTACTGTACTGTTTACTGCCTGGGC ACTATACTTTCTATGCAGATCTCCTTTGTGGGTTTCCAGCCTGTCCTTTCATCAGAGCACATGGCAGCCTTTGGGGTCTTTGGTCTCTGC CAGATCCATGCCTTTGTGGATTACCTGCGCAGCAAGTTGAATCCACAACAATTTGAAGTTCTTTTCCGGAGCGTCATCTCTCTGGTAGGC TTTGTCCTTCTCACCGTGGGAGCTCTCCTCATGCTGACAGGAAAAATATCTCCCTGGACGGGGCGTTTCTACTCGCTGCTGGATCCCTCT TATGCTAAGAACAACATCCCCATCATTGCTTCTGTGTCTGAGCATCAGCCCACAACCTGGTCCTCATACTATTTTGACCTGCAGCTCCTC GTCTTCATGTTTCCAGTTGGCCTCTATTACTGCTTTAGCAACCTGTCTGATGCCCGGATTTTTATCATCATGTATGGTGTGACCAGCATG TACTTTTCAGCTGTAATGGTGCGTCTAATGCTAGTGTTGGCACCTGTTATGTGCATTCTCTCTGGCATTGGAGTCTCCCAGGTGCTGTCC ACATACATGAAGAATCTGGACATAAGTCGTCCAGACAAGAAGAGCAAGAAGCAACAGGATTCCACCTACCCTATTAAGAATGAAGTGGCA AGTGGGATGATACTGGTCATGGCTTTCTTTCTCATCACCTACACCTTTCATTCAACCTGGGTGACCAGTGAGGCCTACTCTTCTCCGTCC ATTGTACTATCTGCCCGTGGTGGGGATGGCAGTAGGATCATATTTGATGACTTCCGAGAAGCATATTATTGGCTTCGTCATAATACTCCA GAGGATGCGAAGGTCATGTCCTGGTGGGATTATGGCTATCAGATTACAGCTATGGCAAACCGAACAATTTTAGTGGACAATAACACATGG AATAATACCCATATTTCTCGAGTAGGGCAGGCAATGGCGTCCACAGAGGAAAAAGCCTATGAGATCATGAGGGAGCTCGATGTCAGCTAT GTGCTGGTCATTTTTGGAGGCCTCACTGGGTATTCCTCTGATGCGTGAAGGTTCTCCAGTGCTGCTCAACTGCCTCATGTACAAGATGTG TTACTATCGCTTTGGACAGGTTTACACAGAAGCCAGCGGTGTGGACACGGCTGCGGTGGGCGGGGTCTTCGACGTCTCCAACGCTGACCG CCTGGGCTTCTCAGAGGTGGAGCTGGTGCAGATGGTGGTGGACGGAGTGAAGCTGCTCATCGAGATGGAGCAGCGGCTGGAGCAGGGCCA GGCCATCGACGACCTCATGCCTGCCCAGAAATGAAGCCCGGCCCACACCCGACACCAGCCCTGCTGCTTCCTAACTTATTGCCTGGGCAG TGCCCACCATGCACCCCTGATGTTCGCCGTCTGGCGAGCCCTTAGCCTTGCTGTAGAGACTTCCGTCACCCTTGGTAGAGTTTATTTTTT >87746_87746_1_STT3A-CKB_STT3A_chr11_125488374_ENST00000392708_CKB_chr14_103986376_ENST00000348956_length(amino acids)=599AA_BP= MPPIHVKMTKFGFLRLSYEKQDTLLKLLILSMAAVLSFSTRLFAVLRFESVIHEFDPYFNYRTTRFLAEEGFYKFHNWFDDRAWYPLGRI IGGTIYPGLMITSAAIYHVLHFFHITIDIRNVCVFLAPLFSSFTTIVTYHLTKELKDAGAGLLAAAMIAVVPGYISRSVAGSYDNEGIAI FCMLLTYYMWIKAVKTGSICWAAKCALAYFYMVSSWGGYVFLINLIPLHVLVLMLTGRFSHRIYVAYCTVYCLGTILSMQISFVGFQPVL SSEHMAAFGVFGLCQIHAFVDYLRSKLNPQQFEVLFRSVISLVGFVLLTVGALLMLTGKISPWTGRFYSLLDPSYAKNNIPIIASVSEHQ PTTWSSYYFDLQLLVFMFPVGLYYCFSNLSDARIFIIMYGVTSMYFSAVMVRLMLVLAPVMCILSGIGVSQVLSTYMKNLDISRPDKKSK KQQDSTYPIKNEVASGMILVMAFFLITYTFHSTWVTSEAYSSPSIVLSARGGDGSRIIFDDFREAYYWLRHNTPEDAKVMSWWDYGYQIT -------------------------------------------------------------- >87746_87746_2_STT3A-CKB_STT3A_chr11_125488374_ENST00000529196_CKB_chr14_103986376_ENST00000348956_length(transcript)=2446nt_BP=2062nt GGACCCGGCTCCTGCCAGGGTTGGGTGCGCCGCTGAACGGATGGCTGAGGGAGCCCCGCGGATCGTTTAGGAAAGCCGGCCAGGCATTCC AGAAAACAGCTGCAAACCCTGAGAGATATGGCTGGATTGTTGACCTCCTGGAGTAGTTAGGCCTTTATTTTTAGTTGCAAACTGATCGTC GTGTGTTGCCACCCATTCATGTCAAGATGACTAAGTTTGGATTTTTGCGATTGTCCTATGAGAAGCAGGACACACTTTTGAAGCTTCTCA TTCTGTCAATGGCTGCTGTATTATCCTTCTCCACTCGTCTGTTTGCTGTCCTGAGATTTGAAAGTGTTATCCATGAGTTTGATCCGTACT TTAATTATCGGACTACCAGGTTCCTGGCTGAGGAGGGGTTTTATAAATTCCATAACTGGTTTGATGACCGAGCCTGGTACCCTTTGGGAC GAATCATTGGAGGAACAATTTACCCAGGTTTAATGATCACCTCTGCTGCAATCTACCATGTACTCCATTTTTTCCACATCACCATCGACA TTCGGAATGTCTGTGTGTTCCTGGCCCCTCTCTTCTCCTCCTTCACCACCATCGTCACGTACCACCTTACCAAAGAGCTCAAGGATGCAG GGGCTGGGCTTCTTGCTGCTGCCATGATTGCTGTAGTTCCTGGATATATCTCCCGATCTGTGGCTGGCTCCTATGATAATGAAGGGATTG CCATCTTTTGCATGCTACTCACCTACTACATGTGGATCAAGGCAGTAAAGACTGGTTCCATCTGTTGGGCAGCTAAGTGTGCCCTTGCTT ATTTCTACATGGTCTCGTCATGGGGAGGTTATGTGTTCCTGATCAACTTAATTCCTCTCCACGTCCTCGTGCTGATGCTCACAGGCCGTT TCTCTCACCGGATCTATGTGGCCTACTGTACTGTTTACTGCCTGGGCACTATACTTTCTATGCAGATCTCCTTTGTGGGTTTCCAGCCTG TCCTTTCATCAGAGCACATGGCAGCCTTTGGGGTCTTTGGTCTCTGCCAGATCCATGCCTTTGTGGATTACCTGCGCAGCAAGTTGAATC CACAACAATTTGAAGTTCTTTTCCGGAGCGTCATCTCTCTGGTAGGCTTTGTCCTTCTCACCGTGGGAGCTCTCCTCATGCTGACAGGAA AAATATCTCCCTGGACGGGGCGTTTCTACTCGCTGCTGGATCCCTCTTATGCTAAGAACAACATCCCCATCATTGCTTCTGTGTCTGAGC ATCAGCCCACAACCTGGTCCTCATACTATTTTGACCTGCAGCTCCTCGTCTTCATGTTTCCAGTTGGCCTCTATTACTGCTTTAGCAACC TGTCTGATGCCCGGATTTTTATCATCATGTATGGTGTGACCAGCATGTACTTTTCAGCTGTAATGGTGCGTCTAATGCTAGTGTTGGCAC CTGTTATGTGCATTCTCTCTGGCATTGGAGTCTCCCAGGTGCTGTCCACATACATGAAGAATCTGGACATAAGTCGTCCAGACAAGAAGA GCAAGAAGCAACAGGATTCCACCTACCCTATTAAGAATGAAGTGGCAAGTGGGATGATACTGGTCATGGCTTTCTTTCTCATCACCTACA CCTTTCATTCAACCTGGGTGACCAGTGAGGCCTACTCTTCTCCGTCCATTGTACTATCTGCCCGTGGTGGGGATGGCAGTAGGATCATAT TTGATGACTTCCGAGAAGCATATTATTGGCTTCGTCATAATACTCCAGAGGATGCGAAGGTCATGTCCTGGTGGGATTATGGCTATCAGA TTACAGCTATGGCAAACCGAACAATTTTAGTGGACAATAACACATGGAATAATACCCATATTTCTCGAGTAGGGCAGGCAATGGCGTCCA CAGAGGAAAAAGCCTATGAGATCATGAGGGAGCTCGATGTCAGCTATGTGCTGGTCATTTTTGGAGGCCTCACTGGGTATTCCTCTGATG CGTGAAGGTTCTCCAGTGCTGCTCAACTGCCTCATGTACAAGATGTGTTACTATCGCTTTGGACAGGTTTACACAGAAGCCAGCGGTGTG GACACGGCTGCGGTGGGCGGGGTCTTCGACGTCTCCAACGCTGACCGCCTGGGCTTCTCAGAGGTGGAGCTGGTGCAGATGGTGGTGGAC GGAGTGAAGCTGCTCATCGAGATGGAGCAGCGGCTGGAGCAGGGCCAGGCCATCGACGACCTCATGCCTGCCCAGAAATGAAGCCCGGCC CACACCCGACACCAGCCCTGCTGCTTCCTAACTTATTGCCTGGGCAGTGCCCACCATGCACCCCTGATGTTCGCCGTCTGGCGAGCCCTT AGCCTTGCTGTAGAGACTTCCGTCACCCTTGGTAGAGTTTATTTTTTTGATGGCTAAGATACTGCTGATGCTGAAATAAACTAGGGTTTT >87746_87746_2_STT3A-CKB_STT3A_chr11_125488374_ENST00000529196_CKB_chr14_103986376_ENST00000348956_length(amino acids)=606AA_BP= MQTDRRVLPPIHVKMTKFGFLRLSYEKQDTLLKLLILSMAAVLSFSTRLFAVLRFESVIHEFDPYFNYRTTRFLAEEGFYKFHNWFDDRA WYPLGRIIGGTIYPGLMITSAAIYHVLHFFHITIDIRNVCVFLAPLFSSFTTIVTYHLTKELKDAGAGLLAAAMIAVVPGYISRSVAGSY DNEGIAIFCMLLTYYMWIKAVKTGSICWAAKCALAYFYMVSSWGGYVFLINLIPLHVLVLMLTGRFSHRIYVAYCTVYCLGTILSMQISF VGFQPVLSSEHMAAFGVFGLCQIHAFVDYLRSKLNPQQFEVLFRSVISLVGFVLLTVGALLMLTGKISPWTGRFYSLLDPSYAKNNIPII ASVSEHQPTTWSSYYFDLQLLVFMFPVGLYYCFSNLSDARIFIIMYGVTSMYFSAVMVRLMLVLAPVMCILSGIGVSQVLSTYMKNLDIS RPDKKSKKQQDSTYPIKNEVASGMILVMAFFLITYTFHSTWVTSEAYSSPSIVLSARGGDGSRIIFDDFREAYYWLRHNTPEDAKVMSWW -------------------------------------------------------------- >87746_87746_3_STT3A-CKB_STT3A_chr11_125488374_ENST00000531491_CKB_chr14_103986376_ENST00000348956_length(transcript)=2204nt_BP=1820nt GCTGAACGGATGGCTGAGGGAGCCCCGCGGATCGTTTAGGAAAGCCGGCCAGCCTTCTCCACTCGTCTGTTTGCTGTCCTGAGATTTGAA AGTGTTATCCATGAGTTTGATCCGTACTTTAATTATCGGACTACCAGGTTCCTGGCTGAGGAGGGGTTTTATAAATTCCATAACTGGTTT GATGACCGAGCCTGGTACCCTTTGGGACGAATCATTGGAGGAACAATTTACCCAGGTTTAATGATCACCTCTGCTGCAATCTACCATGTA CTCCATTTTTTCCACATCACCATCGACATTCGGAATGTCTGTGTGTTCCTGGCCCCTCTCTTCTCCTCCTTCACCACCATCGTCACGTAC CACCTTACCAAAGAGCTCAAGGATGCAGGGGCTGGGCTTCTTGCTGCTGCCATGATTGCTGTAGTTCCTGGATATATCTCCCGATCTGTG GCTGGCTCCTATGATAATGAAGGGATTGCCATCTTTTGCATGCTACTCACCTACTACATGTGGATCAAGGCAGTAAAGACTGGTTCCATC TGTTGGGCAGCTAAGTGTGCCCTTGCTTATTTCTACATGGTCTCGTCATGGGGAGGTTATGTGTTCCTGATCAACTTAATTCCTCTCCAC GTCCTCGTGCTGATGCTCACAGGCCGTTTCTCTCACCGGATCTATGTGGCCTACTGTACTGTTTACTGCCTGGGCACTATACTTTCTATG CAGATCTCCTTTGTGGGTTTCCAGCCTGTCCTTTCATCAGAGCACATGGCAGCCTTTGGGGTCTTTGGTCTCTGCCAGATCCATGCCTTT GTGGATTACCTGCGCAGCAAGTTGAATCCACAACAATTTGAAGTTCTTTTCCGGAGCGTCATCTCTCTGGTAGGCTTTGTCCTTCTCACC GTGGGAGCTCTCCTCATGCTGACAGGAAAAATATCTCCCTGGACGGGGCGTTTCTACTCGCTGCTGGATCCCTCTTATGCTAAGAACAAC ATCCCCATCATTGCTTCTGTGTCTGAGCATCAGCCCACAACCTGGTCCTCATACTATTTTGACCTGCAGCTCCTCGTCTTCATGTTTCCA GTTGGCCTCTATTACTGCTTTAGCAACCTGTCTGATGCCCGGATTTTTATCATCATGTATGGTGTGACCAGCATGTACTTTTCAGCTGTA ATGGTGCGTCTAATGCTAGTGTTGGCACCTGTTATGTGCATTCTCTCTGGCATTGGAGTCTCCCAGGTGCTGTCCACATACATGAAGAAT CTGGACATAAGTCGTCCAGACAAGAAGAGCAAGAAGCAACAGGATTCCACCTACCCTATTAAGAATGAAGTGGCAAGTGGGATGATACTG GTCATGGCTTTCTTTCTCATCACCTACACCTTTCATTCAACCTGGGTGACCAGTGAGGCCTACTCTTCTCCGTCCATTGTACTATCTGCC CGTGGTGGGGATGGCAGTAGGATCATATTTGATGACTTCCGAGAAGCATATTATTGGCTTCGTCATAATACTCCAGAGGATGCGAAGGTC ATGTCCTGGTGGGATTATGGCTATCAGATTACAGCTATGGCAAACCGAACAATTTTAGTGGACAATAACACATGGAATAATACCCATATT TCTCGAGTAGGGCAGGCAATGGCGTCCACAGAGGAAAAAGCCTATGAGATCATGAGGGAGCTCGATGTCAGCTATGTGCTGGTCATTTTT GGAGGCCTCACTGGGTATTCCTCTGATGCGTGAAGGTTCTCCAGTGCTGCTCAACTGCCTCATGTACAAGATGTGTTACTATCGCTTTGG ACAGGTTTACACAGAAGCCAGCGGTGTGGACACGGCTGCGGTGGGCGGGGTCTTCGACGTCTCCAACGCTGACCGCCTGGGCTTCTCAGA GGTGGAGCTGGTGCAGATGGTGGTGGACGGAGTGAAGCTGCTCATCGAGATGGAGCAGCGGCTGGAGCAGGGCCAGGCCATCGACGACCT CATGCCTGCCCAGAAATGAAGCCCGGCCCACACCCGACACCAGCCCTGCTGCTTCCTAACTTATTGCCTGGGCAGTGCCCACCATGCACC CCTGATGTTCGCCGTCTGGCGAGCCCTTAGCCTTGCTGTAGAGACTTCCGTCACCCTTGGTAGAGTTTATTTTTTTGATGGCTAAGATAC >87746_87746_3_STT3A-CKB_STT3A_chr11_125488374_ENST00000531491_CKB_chr14_103986376_ENST00000348956_length(amino acids)=558AA_BP= MFAVLRFESVIHEFDPYFNYRTTRFLAEEGFYKFHNWFDDRAWYPLGRIIGGTIYPGLMITSAAIYHVLHFFHITIDIRNVCVFLAPLFS SFTTIVTYHLTKELKDAGAGLLAAAMIAVVPGYISRSVAGSYDNEGIAIFCMLLTYYMWIKAVKTGSICWAAKCALAYFYMVSSWGGYVF LINLIPLHVLVLMLTGRFSHRIYVAYCTVYCLGTILSMQISFVGFQPVLSSEHMAAFGVFGLCQIHAFVDYLRSKLNPQQFEVLFRSVIS LVGFVLLTVGALLMLTGKISPWTGRFYSLLDPSYAKNNIPIIASVSEHQPTTWSSYYFDLQLLVFMFPVGLYYCFSNLSDARIFIIMYGV TSMYFSAVMVRLMLVLAPVMCILSGIGVSQVLSTYMKNLDISRPDKKSKKQQDSTYPIKNEVASGMILVMAFFLITYTFHSTWVTSEAYS SPSIVLSARGGDGSRIIFDDFREAYYWLRHNTPEDAKVMSWWDYGYQITAMANRTILVDNNTWNNTHISRVGQAMASTEEKAYEIMRELD -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for STT3A-CKB |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for STT3A-CKB |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for STT3A-CKB |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
