
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TARDBP-LRCH3 (FusionGDB2 ID:89102) |
Fusion Gene Summary for TARDBP-LRCH3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TARDBP-LRCH3 | Fusion gene ID: 89102 | Hgene | Tgene | Gene symbol | TARDBP | LRCH3 | Gene ID | 23435 | 84859 |
| Gene name | TAR DNA binding protein | leucine rich repeats and calponin homology domain containing 3 | |
| Synonyms | ALS10|TDP-43 | - | |
| Cytomap | 1p36.22 | 3q29 | |
| Type of gene | protein-coding | protein-coding | |
| Description | TAR DNA-binding protein 43TAR DNA-binding protein-43 | DISP complex protein LRCH3leucine-rich repeat and calponin homology domain-containing protein 3leucine-rich repeats and calponin homology (CH) domain containing 3 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | Q96II8 | |
| Ensembl transtripts involved in fusion gene | ENST00000240185, ENST00000315091, ENST00000439080, ENST00000480464, | ENST00000536618, ENST00000334859, ENST00000414675, ENST00000425562, ENST00000441090, ENST00000493726, ENST00000438796, | |
| Fusion gene scores | * DoF score | 8 X 8 X 4=256 | 11 X 12 X 5=660 |
| # samples | 8 | 13 | |
| ** MAII score | log2(8/256*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/660*10)=-2.34395440121736 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TARDBP [Title/Abstract] AND LRCH3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TARDBP(11073830)-LRCH3(197562617), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TARDBP | GO:0008380 | RNA splicing | 11285240 |
| Hgene | TARDBP | GO:0043922 | negative regulation by host of viral transcription | 7745706 |
| Hgene | TARDBP | GO:0061158 | 3'-UTR-mediated mRNA destabilization | 28335005 |
| Hgene | TARDBP | GO:0070935 | 3'-UTR-mediated mRNA stabilization | 17481916 |
 Fusion gene breakpoints across TARDBP (5'-gene) Fusion gene breakpoints across TARDBP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
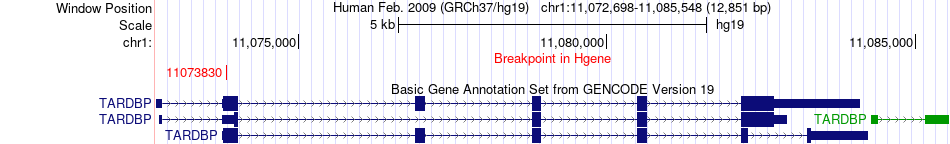 |
 Fusion gene breakpoints across LRCH3 (3'-gene) Fusion gene breakpoints across LRCH3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
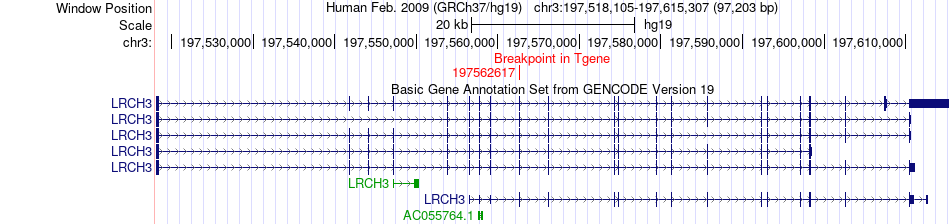 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | DA652289 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
Top |
Fusion Gene ORF analysis for TARDBP-LRCH3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000240185 | ENST00000536618 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-5UTR | ENST00000315091 | ENST00000536618 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000240185 | ENST00000334859 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000240185 | ENST00000414675 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000240185 | ENST00000425562 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000240185 | ENST00000441090 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000240185 | ENST00000493726 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000315091 | ENST00000334859 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000315091 | ENST00000414675 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000315091 | ENST00000425562 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000315091 | ENST00000441090 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5CDS-intron | ENST00000315091 | ENST00000493726 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-3CDS | ENST00000439080 | ENST00000438796 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-5UTR | ENST00000439080 | ENST00000536618 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-intron | ENST00000439080 | ENST00000334859 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-intron | ENST00000439080 | ENST00000414675 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-intron | ENST00000439080 | ENST00000425562 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-intron | ENST00000439080 | ENST00000441090 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| 5UTR-intron | ENST00000439080 | ENST00000493726 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| In-frame | ENST00000240185 | ENST00000438796 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| In-frame | ENST00000315091 | ENST00000438796 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-3CDS | ENST00000480464 | ENST00000438796 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-5UTR | ENST00000480464 | ENST00000536618 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-intron | ENST00000480464 | ENST00000334859 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-intron | ENST00000480464 | ENST00000414675 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-intron | ENST00000480464 | ENST00000425562 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-intron | ENST00000480464 | ENST00000441090 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
| intron-intron | ENST00000480464 | ENST00000493726 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000240185 | TARDBP | chr1 | 11073830 | + | ENST00000438796 | LRCH3 | chr3 | 197562617 | + | 6469 | 294 | 256 | 1530 | 424 |
| ENST00000315091 | TARDBP | chr1 | 11073830 | + | ENST00000438796 | LRCH3 | chr3 | 197562617 | + | 6367 | 192 | 154 | 1428 | 424 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000240185 | ENST00000438796 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + | 0.003370716 | 0.9966293 |
| ENST00000315091 | ENST00000438796 | TARDBP | chr1 | 11073830 | + | LRCH3 | chr3 | 197562617 | + | 0.003215696 | 0.9967843 |
Top |
Fusion Genomic Features for TARDBP-LRCH3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
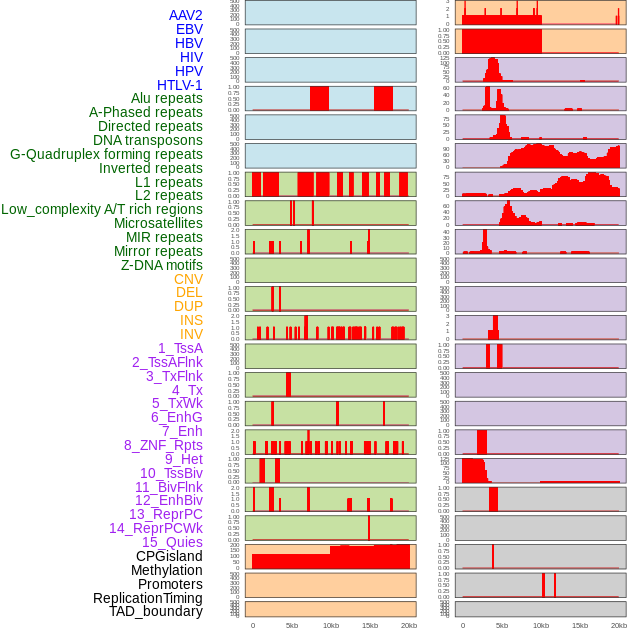 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TARDBP-LRCH3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:11073830/chr3:197562617) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LRCH3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: As part of the DISP complex, may regulate the association of septins with actin and thereby regulate the actin cytoskeleton. {ECO:0000269|PubMed:29467281}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 384_389 | 0 | 713.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 384_389 | 0 | 778.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 384_389 | 0 | 2334.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 652_765 | 0 | 713.0 | Domain | Calponin-homology (CH) | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 652_765 | 0 | 778.0 | Domain | Calponin-homology (CH) | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 652_765 | 0 | 2334.0 | Domain | Calponin-homology (CH) | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 105_127 | 0 | 713.0 | Repeat | LRR 3 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 128_150 | 0 | 713.0 | Repeat | LRR 4 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 152_172 | 0 | 713.0 | Repeat | LRR 5 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 173_195 | 0 | 713.0 | Repeat | LRR 6 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 196_218 | 0 | 713.0 | Repeat | LRR 7 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 220_239 | 0 | 713.0 | Repeat | LRR 8 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 240_264 | 0 | 713.0 | Repeat | LRR 9 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 266_290 | 0 | 713.0 | Repeat | LRR 10 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 56_79 | 0 | 713.0 | Repeat | LRR 1 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000334859 | 0 | 19 | 81_104 | 0 | 713.0 | Repeat | LRR 2 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 105_127 | 0 | 778.0 | Repeat | LRR 3 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 128_150 | 0 | 778.0 | Repeat | LRR 4 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 152_172 | 0 | 778.0 | Repeat | LRR 5 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 173_195 | 0 | 778.0 | Repeat | LRR 6 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 196_218 | 0 | 778.0 | Repeat | LRR 7 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 220_239 | 0 | 778.0 | Repeat | LRR 8 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 240_264 | 0 | 778.0 | Repeat | LRR 9 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 266_290 | 0 | 778.0 | Repeat | LRR 10 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 56_79 | 0 | 778.0 | Repeat | LRR 1 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000425562 | 0 | 21 | 81_104 | 0 | 778.0 | Repeat | LRR 2 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 105_127 | 0 | 2334.0 | Repeat | LRR 3 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 128_150 | 0 | 2334.0 | Repeat | LRR 4 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 152_172 | 0 | 2334.0 | Repeat | LRR 5 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 173_195 | 0 | 2334.0 | Repeat | LRR 6 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 196_218 | 0 | 2334.0 | Repeat | LRR 7 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 220_239 | 0 | 2334.0 | Repeat | LRR 8 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 240_264 | 0 | 2334.0 | Repeat | LRR 9 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 266_290 | 0 | 2334.0 | Repeat | LRR 10 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 56_79 | 0 | 2334.0 | Repeat | LRR 1 | |
| Tgene | LRCH3 | chr1:11073830 | chr3:197562617 | ENST00000438796 | 0 | 22 | 81_104 | 0 | 2334.0 | Repeat | LRR 2 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TARDBP | chr1:11073830 | chr3:197562617 | ENST00000240185 | + | 1 | 6 | 274_413 | 0 | 415.0 | Compositional bias | Note=Gly-rich |
| Hgene | TARDBP | chr1:11073830 | chr3:197562617 | ENST00000240185 | + | 1 | 6 | 104_200 | 0 | 415.0 | Domain | RRM 1 |
| Hgene | TARDBP | chr1:11073830 | chr3:197562617 | ENST00000240185 | + | 1 | 6 | 191_262 | 0 | 415.0 | Domain | RRM 2 |
| Hgene | TARDBP | chr1:11073830 | chr3:197562617 | ENST00000240185 | + | 1 | 6 | 239_250 | 0 | 415.0 | Motif | Nuclear export signal |
| Hgene | TARDBP | chr1:11073830 | chr3:197562617 | ENST00000240185 | + | 1 | 6 | 82_98 | 0 | 415.0 | Motif | Nuclear localization signal |
Top |
Fusion Gene Sequence for TARDBP-LRCH3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >89102_89102_1_TARDBP-LRCH3_TARDBP_chr1_11073830_ENST00000240185_LRCH3_chr3_197562617_ENST00000438796_length(transcript)=6469nt_BP=294nt GGCCCTAGCGCCATTTTGTGGGAGCGAAGCGGTGGCTGGGCTGCGCTTGGGTCCGTCGCTGCTTCGGTGTCCCTGTCGGGCTTCCCAGCA GCGGCCTAGCGGTTGAAATACCATCGGAAGACGATGGGACGGTGCTGCTCTCCACGGTTACAGCCCAGTTTCCAGGGGCGTGTGGGCTTC GCTACAGGAATCCAGTGTCTCAGTGTATGAGAGGTGTCCGGCTGGTAGAAGGAATTCTGCATGCCCCAGATGCTGGCTGGGGAAATCTGG TGTATGTTGTCAACTATCCAAAAGGCCCAAGGGACCAGACCCAGACAGCCTTAGTTCACAGTTTATGGCGTATATTGAACAGCGGCGAAT CTCTCATGAGGGTTCACCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGAGAAGATATTTACATCAAAACAGGGT TCCAGCTGAGCCATCTTCCCTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAGCA GTTAGTAGAGCGCACTCGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGATGC TGTCCTGGACTTTGTCAAACAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTCCC ATCCAGAAGGTCTCAGCACACTGATGATAGTGCCTTGTGCATGTCGCTGTCAGGGTTGAATCAAGTGGGCTGTGCTGCTACCCTGCCTCA TTCTTCTGCCTTCACGCCTCTTAAGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAACAGAAACAGTTCATCATTCCCCTGC ATATTCTTTTCCTGCTGCTATCCAGAGAAATCAGCCTCAGCGCCCTGAAAGCTTCCTTTTCCGAGCAGGTGTCAGGGCAGAAACCAACAA AGGTCATGCTTCACCCCTTCCTCCATCTGCTGCACCTACCACTGATTCTACAGATTCCATAACAGGACAGAATTCAAGACAGAGAGAAGA AGAGCTGGAATTAATAGACCAACTGCGTAAACATATTGAGTACCGGTTGAAAGTGTCTCTACCTTGTGATCTCGGAGCAGCTCTAACTGA CGGTGTTGTTCTTTGCCATTTGGCCAATCATGTGCGACCTCGATCTGTCCCAAGCATTCATGTTCCCTCACCAGCTGTACCTAAATTAAC AATGGCGAAATGCAGGCGAAATGTGGAAAATTTCCTAGAAGCTTGCAGAAAAATTGGTGTACCTCAGGACAATCTTTGTTCCCCTTCCGA CATCCTTCAGCTAAACCTCAGCGTTAAAAGAACTGTTGAAACGCTCCTTTCTCTTGGGGCACACTCAGAAGAATCCAGTTTTGTCTGTCT CTCTCTGCAGCTTCTGGGTTTTGTGGCATTTTACTGTACTGTGATGTTAACTCTCTGTGTGCTTTATTACTGGCTCTTCCCCGCTCGCTG AAAGATTGCACTCCGGTGACTTTCTGCCTCATTCCTGTGCTTGGAGCAATTATGTTTGCCTCTCCACATTTTAGAAGAGAAAGGTTTGAG CCAGGTTGCAGTGACGGTCCAGGCTCTCCTGGAACTTGCCCCACCCAAGCAACAGCAGCACCAGTTATCTGCTGTTTGAGGATCCCCAGG ACGGTGGGCACTGGCCTGGCCAAAACAAGGAACAGGACACCGTGATTGCTGCTGCCAGCTGTCTGCTTAAACAAAGCTCTTGTGTGTTCT CAGAAGGGACCGTTCCCATGATTCCTAACAGGAATATTTTGCTTCATTTCTCCATTTTAGGGGAGGTTATTTCCATTTCCATTGAATTTT TTTACGAAGACACCATTGGTTTTCTCAGATGAAACTTCTCTTACTGAAAACCCAGCACCTAACTTGGCTGTGTTTTCCTGGAGAGCAGGC GTGTTTCACGTGCCAGAAAGCACATCTTCTGTGTTAGGCACAAGCATGCTGTGCCATTGTTTTTACCTGGTCTCAAGACAACTGGTCTGC TGGTTTCTAAACTTGGTTGTTCATTAGACTCTCCTGGAGTATTTGTTAAAATACAGATTCACATTCAGTAGAGCTGGGGTAAGGCCTAGG AATCTGCATTTTCAACAACCATTGCACATGTTTTTGTTTTTGTTGGAGAAGGAGTTCTGCTCAGTCGCCCAGGCTAGAGTGCAGTGGCAT AATCTCAGCTCACTGCAACCTCCGCCTCCCGGGCTCAAGCGATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACAACAGGTGCCCGCCA CCATGCCTGGCTAATTTTCTTGTATTTTTAGTAGAGATGGGATTTTGTCATGTTGGCCAGGCTGGTCTCAAACTCCTGACTTCAAGTGAT CCACCTGCCTCGGCCTCCCAAAATGCTGGGATTACAGGCGTGAGCCACTGTGCCCGGCCTCCCAGAGTGCTGGGATGACAGGCGTGAGCC CCCGCGCCCGGCCTCCGAGAGTGCTGGGATTACAGGCGTGAGCCACTGTGCCCGGCCTCTGAGGGTGCTGGGATTACAGGCGTGAGCCAC TGCGCCCGGCGCCCGGCCCATTGCACGTGTTTTTCATGTAGTGGAACCACTGTTCAAAGCTAGATATTTGTATGTTTGCGGGGGAGGGTA ATGGCCCCCTGCATTTTTCACCCACATTTAAAATCAAAAGCTATATTATGAGGAGTTGGACTGTTCTGTCAAAGGCAAAACAAAAATAAT ATAGAAGGCCTAGGTTTTGCATAGTCTTCTTTTTTTTAAGAGTTGGGGTCTTGCCGTATTGCCCAGGCTGGTCTGAAACTCCTGGGCTCA ACTGATCCTCCCGCCTCAGCCTCCCAAAGTGCTGGGATTACTGGAGTGAGCCATCGGGCCTGGCCCAGATTTTGCATAGTAAAACAGTGA AACAGATAGAGTTGTATTTAAGGTCAGTATTCTAGGCCTCCCTTCAACTGCAGTCATACCATCATAGAAGATAGTTCTATGTCATCCTGT TATCTGGACATTATAATAAACAACAAGCCCTCTATACACAGGTTCCAAACCCCGTTATCTAGACATTATCAACAACAAGCCCTCTCTCTA TACAGAGTTTCCAAATTCTGAAGCTGATAGCGCACTTGGTCTCACTGAGCCTCCCTCCCCGCCCTCCTGCTGCTCCTCAGACATCTGGGC CTGCAGACACATTGCAGTTACAGCACATACACTTCTAGCTCGGAAAAGCCTAGTTTCTTGTATTTCTTTCAGGTAACTGCCAGTTTCTGT GGCTGAATTTAAGCACTTACAGCATCTTCCTAGCTGGGTGTGGTGGCTCACACCTGTAATCTCAGCACTTTGGGAGGCTGAGGCGGGCAG ATCACCTGAAGTTGGGAGTTTGAGACCAGCCTGACCAACATGGAGAAACCCCGTCTCTACTAAAAATACAAAATTAGCTGGGCGTGGTGG TGCATGCCTGTAATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATTGCTTGAACCCAGGAGGCGGAGGTTGTGGTGAGCCAAGATCGTG CCATTGCACCCCAGCCTGGGCAACAAAAGCGAAATTCCATCTCAAAAAAAGAAAACGCGCCCCATCTCGCTCCACCACAAGCGCCTTTAT ATACTCAATGATGGGTGGAGGAAAAGGGCCCAAGGGCTCACACCGCACAGGAGCCTTTACTCAAGTTTCAGCTAGTTTTCCTTCCCCTGC ATTTTGAACTCCTTTTAGTTGTTGGTGATATCCTTTGGTGTTCACTGCCTTTGTAAAACAAGTAAATAAAGTAAGCGGAAGATATCTCTT TTGTTTCTGTAGTTGTTAACTGTTCTGGATTTTCTTTACAATGCATCCATTTGGGCATAATGTTATCCCTCTTTTTATCAGGTAAATGTA GCATCTAAAAGTAGCTTTGGATCAAATCATGTTGAATACCTGGTCAAAGTAATGTTTTCTTCCTTGTTCTTGCAGAGAAACAAATTCATC AGAACATTACCACTTGACCAGGTTTAAAATTGTATAACAGGCTGGGCATGGTGGCTCATGCCTATAATCCCAGCACTTTGGGAGGCCGAG GCAAGAGGATCACTTGAGCCCAGGAGTTTGAGAGCAGCCTGGGCAACATAGCAAGACCCTGTCTCCCAAAAATTAAAAAAAAAAAAAAAA AACCAGGTGTGGTGGCACCTGCCTGTACTCCCAGCTACTCAGGAGGCTGGGGTGGGAGGATCACTTGAGCCTGGGAGGTTGAGGCTACAG TGAGCCATGGCTGCACCACTGCACTCCAGCCTGGGCAACAGAGTGAGACCCCATCTCTAAAAATAAATTGTACAACATCTAACTTCAATG CCCAATCTTATAGAACAAAAGTAAAATGTCTTTATGTAAGACCCCATTATATTTACAGTTCCCAACAGACCCACCAACTCTTATATACCC TTATATAACTTGTTTAGAATTTAGCCATGTGCATTTTTAAACTGTGTTTGTCAACATTAAGCCTCAGTACAGTCTCCAAATTATATGCCA CCAGAAATGAAATTCAGCAAAGGACAGGATTGTTTTTAACATACGCAAGCCAGCCTGGCCAACATGGTGAAACCTTGTCTCTACTAAAAA TACAAAAATTAGCTGGGCGTGGTGGTGGGCACCTGTAATCCCAGCTACTTGGGTGGCTGAGGCACAAGAATCGCTTCAACCTGGAAGCCG AAGGTTGCAGTGGGCTGAGATTGTGCCACTGCACTCCAGCCTGGGCAACAGAGCGAGACTCTGTCTCAAAAAAAAAAAAAAAAAAAAACC CACCAAACCAAAAATATACTATGCAAGGAATAATTCTAGAAGACATTACATAATGCAATACTTGCAACATTTGCAACTAATTTTCCCATA GGGATAAATGGAAATTTCAACTTATTTCAAATTTTGCACATATTATGAAACCTTATTAATGTATTTTTATCAAACTAAATCAGATTTGTA TTTGAATTGTTAGGAAAAACCATGTGCAGTTTTGGCTGATAATTGAAGGAAAAATATCAAATGCTTTGAATTTTTTTTCTCTTTTTTCAA ACCCTCTGCAGAGGTAGGAAGGTATGAATTTCTTTTTTATGTCAAGATGCAAAAACAAATCATGATGCTTTTGTTGGGAGAATTTTTGTA TTCAGTATTTTGTATGTACCTTTTTTTTTTTAAATTGGAAAGCACAATTCGGTTTAACATTTAGCTTTGCTTGACTCCAGTGTAAGATGA AGATGACCTTGTCACAGCTCCCCTGACCTGAAGCAGAGCCCTTCCCATCACTGACAGTGTTGGGGGTTGAGAGCCCCCCAGCAGAGCCCT TCCCATCACTGACAGTGTGTCGGCAGCTGAGAGCTCCCCCAGCAGAGCCCTTCCCATCACTGAACAGTGTTGGCGGCTGAGAGCCCCCCA GCAGAGCCCTTCCCATCACTGACAGTGTTGGGGGCTGAGAGCCCCCCAGCAGAGCCCTTCCCATCACTGAACAGTGTTGGCGGCTGAGAG CCCCCCAGCAGAGCCCTTCCCATCACTGACAGTGTTGGGGGCTGAGAGCCCCCCTAGCAGAGCCCTTCCCATCACTGACAGTGTCTGGGG CTGAGAGCCCCCCCTAGCAGAGCCCTTCCCATCACTGACAGTGTTGGGGGCTGAGAGCCCCCCAGCAGAGCCCTTCCCATCACTGACAGT GTCTGGGGCTGAGAGCCCCCCAGCAGAGCCCTTCCCATCACTGACAGTGTCTGGGGCTGAGAGCCCCCCCTAGCAGAGCCCTTCCCATCA CTGACAGTGTCGGGTTGAGAGCCACCCCCAGCAGAGCCCTTGCCACGTACAGAGATTTTCTGTCAGCAGTTTCCCAGTATTTCAGACTCA TAATCTGTGTTTAGCACTCCAGTGTTCTCTTGTAATAGCCAGGTCTGGAAACGACCCATGGAAAGAGGGGCCTTTGTATTCTGCTAGAAC ACAGAGCATAGTAGTAGATCACTTTTCTGAAGAGCTTTTTAAAATACAGCTAATCAAGGAGTTTGCGATTTCTCTTACTTTCTACAGCTC AGCAGTGACTAATGATGTGTGACTATGCGAATGAGTTTGATGAATTCGTAAAAACTAAGCGCTTAAATGGTTATAGAAATGAAAACATGG ATATTTAGAAGCCTTTCTATATACACAGTGCAGTTGAGATGGTTTACAGCAAATTTCGTTTTATTTCTGAAAATGTTGTAAACATGTAAT TAGTGAAAGATTTTGTAAAACATTGTCCTGTATTTTTGTCTGTAAATATATTGCATAAGTTCTAAGTATAAATATGTCAGTATCATGTAT >89102_89102_1_TARDBP-LRCH3_TARDBP_chr1_11073830_ENST00000240185_LRCH3_chr3_197562617_ENST00000438796_length(amino acids)=424AA_BP=13 MGKSGVCCQLSKRPKGPDPDSLSSQFMAYIEQRRISHEGSPVKPVAIREFQKTEDMRRYLHQNRVPAEPSSLLSLSASHNQLSHTDLELH QRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGECPFPSRRSQHTDDSALCMSLSGLNQVGCA ATLPHSSAFTPLKSDDRPNALLSSPATETVHHSPAYSFPAAIQRNQPQRPESFLFRAGVRAETNKGHASPLPPSAAPTTDSTDSITGQNS RQREEELELIDQLRKHIEYRLKVSLPCDLGAALTDGVVLCHLANHVRPRSVPSIHVPSPAVPKLTMAKCRRNVENFLEACRKIGVPQDNL -------------------------------------------------------------- >89102_89102_2_TARDBP-LRCH3_TARDBP_chr1_11073830_ENST00000315091_LRCH3_chr3_197562617_ENST00000438796_length(transcript)=6367nt_BP=192nt TTGAAATACCATCGGAAGACGATGGGACGGTGCTGCTCTCCACGGTTACAGCCCAGTTTCCAGGGGCGTGTGGGCTTCGCTACAGGAATC CAGTGTCTCAGTGTATGAGAGGTGTCCGGCTGGTAGAAGGAATTCTGCATGCCCCAGATGCTGGCTGGGGAAATCTGGTGTATGTTGTCA ACTATCCAAAAGGCCCAAGGGACCAGACCCAGACAGCCTTAGTTCACAGTTTATGGCGTATATTGAACAGCGGCGAATCTCTCATGAGGG TTCACCAGTAAAGCCAGTAGCCATTAGGGAGTTTCAAAAAACAGAAGATATGAGAAGATATTTACATCAAAACAGGGTTCCAGCTGAGCC ATCTTCCCTCCTGTCACTATCAGCAAGTCACAATCAGCTGTCACACACAGACCTGGAACTTCATCAGAGAAGGGAGCAGTTAGTAGAGCG CACTCGGAGAGAGGCTCAGCTTGCTGCCCTGCAGTATGAGGAGGAGAAAATAAGGACCAAGCAGATCCAGAGAGATGCTGTCCTGGACTT TGTCAAACAAAAAGCATCACAAAGTCCACAAAAACAGCACCCGCTCCTAGATGGCGTAGATGGTGAGTGCCCCTTCCCATCCAGAAGGTC TCAGCACACTGATGATAGTGCCTTGTGCATGTCGCTGTCAGGGTTGAATCAAGTGGGCTGTGCTGCTACCCTGCCTCATTCTTCTGCCTT CACGCCTCTTAAGAGTGATGACAGACCTAATGCTCTATTAAGTTCACCTGCAACAGAAACAGTTCATCATTCCCCTGCATATTCTTTTCC TGCTGCTATCCAGAGAAATCAGCCTCAGCGCCCTGAAAGCTTCCTTTTCCGAGCAGGTGTCAGGGCAGAAACCAACAAAGGTCATGCTTC ACCCCTTCCTCCATCTGCTGCACCTACCACTGATTCTACAGATTCCATAACAGGACAGAATTCAAGACAGAGAGAAGAAGAGCTGGAATT AATAGACCAACTGCGTAAACATATTGAGTACCGGTTGAAAGTGTCTCTACCTTGTGATCTCGGAGCAGCTCTAACTGACGGTGTTGTTCT TTGCCATTTGGCCAATCATGTGCGACCTCGATCTGTCCCAAGCATTCATGTTCCCTCACCAGCTGTACCTAAATTAACAATGGCGAAATG CAGGCGAAATGTGGAAAATTTCCTAGAAGCTTGCAGAAAAATTGGTGTACCTCAGGACAATCTTTGTTCCCCTTCCGACATCCTTCAGCT AAACCTCAGCGTTAAAAGAACTGTTGAAACGCTCCTTTCTCTTGGGGCACACTCAGAAGAATCCAGTTTTGTCTGTCTCTCTCTGCAGCT TCTGGGTTTTGTGGCATTTTACTGTACTGTGATGTTAACTCTCTGTGTGCTTTATTACTGGCTCTTCCCCGCTCGCTGAAAGATTGCACT CCGGTGACTTTCTGCCTCATTCCTGTGCTTGGAGCAATTATGTTTGCCTCTCCACATTTTAGAAGAGAAAGGTTTGAGCCAGGTTGCAGT GACGGTCCAGGCTCTCCTGGAACTTGCCCCACCCAAGCAACAGCAGCACCAGTTATCTGCTGTTTGAGGATCCCCAGGACGGTGGGCACT GGCCTGGCCAAAACAAGGAACAGGACACCGTGATTGCTGCTGCCAGCTGTCTGCTTAAACAAAGCTCTTGTGTGTTCTCAGAAGGGACCG TTCCCATGATTCCTAACAGGAATATTTTGCTTCATTTCTCCATTTTAGGGGAGGTTATTTCCATTTCCATTGAATTTTTTTACGAAGACA CCATTGGTTTTCTCAGATGAAACTTCTCTTACTGAAAACCCAGCACCTAACTTGGCTGTGTTTTCCTGGAGAGCAGGCGTGTTTCACGTG CCAGAAAGCACATCTTCTGTGTTAGGCACAAGCATGCTGTGCCATTGTTTTTACCTGGTCTCAAGACAACTGGTCTGCTGGTTTCTAAAC TTGGTTGTTCATTAGACTCTCCTGGAGTATTTGTTAAAATACAGATTCACATTCAGTAGAGCTGGGGTAAGGCCTAGGAATCTGCATTTT CAACAACCATTGCACATGTTTTTGTTTTTGTTGGAGAAGGAGTTCTGCTCAGTCGCCCAGGCTAGAGTGCAGTGGCATAATCTCAGCTCA CTGCAACCTCCGCCTCCCGGGCTCAAGCGATTCTCCTGCCTCAGCCTCCCGAGTAGCTGGGACAACAGGTGCCCGCCACCATGCCTGGCT AATTTTCTTGTATTTTTAGTAGAGATGGGATTTTGTCATGTTGGCCAGGCTGGTCTCAAACTCCTGACTTCAAGTGATCCACCTGCCTCG GCCTCCCAAAATGCTGGGATTACAGGCGTGAGCCACTGTGCCCGGCCTCCCAGAGTGCTGGGATGACAGGCGTGAGCCCCCGCGCCCGGC CTCCGAGAGTGCTGGGATTACAGGCGTGAGCCACTGTGCCCGGCCTCTGAGGGTGCTGGGATTACAGGCGTGAGCCACTGCGCCCGGCGC CCGGCCCATTGCACGTGTTTTTCATGTAGTGGAACCACTGTTCAAAGCTAGATATTTGTATGTTTGCGGGGGAGGGTAATGGCCCCCTGC ATTTTTCACCCACATTTAAAATCAAAAGCTATATTATGAGGAGTTGGACTGTTCTGTCAAAGGCAAAACAAAAATAATATAGAAGGCCTA GGTTTTGCATAGTCTTCTTTTTTTTAAGAGTTGGGGTCTTGCCGTATTGCCCAGGCTGGTCTGAAACTCCTGGGCTCAACTGATCCTCCC GCCTCAGCCTCCCAAAGTGCTGGGATTACTGGAGTGAGCCATCGGGCCTGGCCCAGATTTTGCATAGTAAAACAGTGAAACAGATAGAGT TGTATTTAAGGTCAGTATTCTAGGCCTCCCTTCAACTGCAGTCATACCATCATAGAAGATAGTTCTATGTCATCCTGTTATCTGGACATT ATAATAAACAACAAGCCCTCTATACACAGGTTCCAAACCCCGTTATCTAGACATTATCAACAACAAGCCCTCTCTCTATACAGAGTTTCC AAATTCTGAAGCTGATAGCGCACTTGGTCTCACTGAGCCTCCCTCCCCGCCCTCCTGCTGCTCCTCAGACATCTGGGCCTGCAGACACAT TGCAGTTACAGCACATACACTTCTAGCTCGGAAAAGCCTAGTTTCTTGTATTTCTTTCAGGTAACTGCCAGTTTCTGTGGCTGAATTTAA GCACTTACAGCATCTTCCTAGCTGGGTGTGGTGGCTCACACCTGTAATCTCAGCACTTTGGGAGGCTGAGGCGGGCAGATCACCTGAAGT TGGGAGTTTGAGACCAGCCTGACCAACATGGAGAAACCCCGTCTCTACTAAAAATACAAAATTAGCTGGGCGTGGTGGTGCATGCCTGTA ATCCCAGCTACTCGGGAGGCTGAGGCAGGAGAATTGCTTGAACCCAGGAGGCGGAGGTTGTGGTGAGCCAAGATCGTGCCATTGCACCCC AGCCTGGGCAACAAAAGCGAAATTCCATCTCAAAAAAAGAAAACGCGCCCCATCTCGCTCCACCACAAGCGCCTTTATATACTCAATGAT GGGTGGAGGAAAAGGGCCCAAGGGCTCACACCGCACAGGAGCCTTTACTCAAGTTTCAGCTAGTTTTCCTTCCCCTGCATTTTGAACTCC TTTTAGTTGTTGGTGATATCCTTTGGTGTTCACTGCCTTTGTAAAACAAGTAAATAAAGTAAGCGGAAGATATCTCTTTTGTTTCTGTAG TTGTTAACTGTTCTGGATTTTCTTTACAATGCATCCATTTGGGCATAATGTTATCCCTCTTTTTATCAGGTAAATGTAGCATCTAAAAGT AGCTTTGGATCAAATCATGTTGAATACCTGGTCAAAGTAATGTTTTCTTCCTTGTTCTTGCAGAGAAACAAATTCATCAGAACATTACCA CTTGACCAGGTTTAAAATTGTATAACAGGCTGGGCATGGTGGCTCATGCCTATAATCCCAGCACTTTGGGAGGCCGAGGCAAGAGGATCA CTTGAGCCCAGGAGTTTGAGAGCAGCCTGGGCAACATAGCAAGACCCTGTCTCCCAAAAATTAAAAAAAAAAAAAAAAAACCAGGTGTGG TGGCACCTGCCTGTACTCCCAGCTACTCAGGAGGCTGGGGTGGGAGGATCACTTGAGCCTGGGAGGTTGAGGCTACAGTGAGCCATGGCT GCACCACTGCACTCCAGCCTGGGCAACAGAGTGAGACCCCATCTCTAAAAATAAATTGTACAACATCTAACTTCAATGCCCAATCTTATA GAACAAAAGTAAAATGTCTTTATGTAAGACCCCATTATATTTACAGTTCCCAACAGACCCACCAACTCTTATATACCCTTATATAACTTG TTTAGAATTTAGCCATGTGCATTTTTAAACTGTGTTTGTCAACATTAAGCCTCAGTACAGTCTCCAAATTATATGCCACCAGAAATGAAA TTCAGCAAAGGACAGGATTGTTTTTAACATACGCAAGCCAGCCTGGCCAACATGGTGAAACCTTGTCTCTACTAAAAATACAAAAATTAG CTGGGCGTGGTGGTGGGCACCTGTAATCCCAGCTACTTGGGTGGCTGAGGCACAAGAATCGCTTCAACCTGGAAGCCGAAGGTTGCAGTG GGCTGAGATTGTGCCACTGCACTCCAGCCTGGGCAACAGAGCGAGACTCTGTCTCAAAAAAAAAAAAAAAAAAAAACCCACCAAACCAAA AATATACTATGCAAGGAATAATTCTAGAAGACATTACATAATGCAATACTTGCAACATTTGCAACTAATTTTCCCATAGGGATAAATGGA AATTTCAACTTATTTCAAATTTTGCACATATTATGAAACCTTATTAATGTATTTTTATCAAACTAAATCAGATTTGTATTTGAATTGTTA GGAAAAACCATGTGCAGTTTTGGCTGATAATTGAAGGAAAAATATCAAATGCTTTGAATTTTTTTTCTCTTTTTTCAAACCCTCTGCAGA GGTAGGAAGGTATGAATTTCTTTTTTATGTCAAGATGCAAAAACAAATCATGATGCTTTTGTTGGGAGAATTTTTGTATTCAGTATTTTG TATGTACCTTTTTTTTTTTAAATTGGAAAGCACAATTCGGTTTAACATTTAGCTTTGCTTGACTCCAGTGTAAGATGAAGATGACCTTGT CACAGCTCCCCTGACCTGAAGCAGAGCCCTTCCCATCACTGACAGTGTTGGGGGTTGAGAGCCCCCCAGCAGAGCCCTTCCCATCACTGA CAGTGTGTCGGCAGCTGAGAGCTCCCCCAGCAGAGCCCTTCCCATCACTGAACAGTGTTGGCGGCTGAGAGCCCCCCAGCAGAGCCCTTC CCATCACTGACAGTGTTGGGGGCTGAGAGCCCCCCAGCAGAGCCCTTCCCATCACTGAACAGTGTTGGCGGCTGAGAGCCCCCCAGCAGA GCCCTTCCCATCACTGACAGTGTTGGGGGCTGAGAGCCCCCCTAGCAGAGCCCTTCCCATCACTGACAGTGTCTGGGGCTGAGAGCCCCC CCTAGCAGAGCCCTTCCCATCACTGACAGTGTTGGGGGCTGAGAGCCCCCCAGCAGAGCCCTTCCCATCACTGACAGTGTCTGGGGCTGA GAGCCCCCCAGCAGAGCCCTTCCCATCACTGACAGTGTCTGGGGCTGAGAGCCCCCCCTAGCAGAGCCCTTCCCATCACTGACAGTGTCG GGTTGAGAGCCACCCCCAGCAGAGCCCTTGCCACGTACAGAGATTTTCTGTCAGCAGTTTCCCAGTATTTCAGACTCATAATCTGTGTTT AGCACTCCAGTGTTCTCTTGTAATAGCCAGGTCTGGAAACGACCCATGGAAAGAGGGGCCTTTGTATTCTGCTAGAACACAGAGCATAGT AGTAGATCACTTTTCTGAAGAGCTTTTTAAAATACAGCTAATCAAGGAGTTTGCGATTTCTCTTACTTTCTACAGCTCAGCAGTGACTAA TGATGTGTGACTATGCGAATGAGTTTGATGAATTCGTAAAAACTAAGCGCTTAAATGGTTATAGAAATGAAAACATGGATATTTAGAAGC CTTTCTATATACACAGTGCAGTTGAGATGGTTTACAGCAAATTTCGTTTTATTTCTGAAAATGTTGTAAACATGTAATTAGTGAAAGATT TTGTAAAACATTGTCCTGTATTTTTGTCTGTAAATATATTGCATAAGTTCTAAGTATAAATATGTCAGTATCATGTATGTTATTTTAAAT >89102_89102_2_TARDBP-LRCH3_TARDBP_chr1_11073830_ENST00000315091_LRCH3_chr3_197562617_ENST00000438796_length(amino acids)=424AA_BP=13 MGKSGVCCQLSKRPKGPDPDSLSSQFMAYIEQRRISHEGSPVKPVAIREFQKTEDMRRYLHQNRVPAEPSSLLSLSASHNQLSHTDLELH QRREQLVERTRREAQLAALQYEEEKIRTKQIQRDAVLDFVKQKASQSPQKQHPLLDGVDGECPFPSRRSQHTDDSALCMSLSGLNQVGCA ATLPHSSAFTPLKSDDRPNALLSSPATETVHHSPAYSFPAAIQRNQPQRPESFLFRAGVRAETNKGHASPLPPSAAPTTDSTDSITGQNS RQREEELELIDQLRKHIEYRLKVSLPCDLGAALTDGVVLCHLANHVRPRSVPSIHVPSPAVPKLTMAKCRRNVENFLEACRKIGVPQDNL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TARDBP-LRCH3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | TARDBP | chr1:11073830 | chr3:197562617 | ENST00000240185 | + | 1 | 6 | 216_414 | 0 | 415.0 | UBQLN2 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TARDBP-LRCH3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TARDBP-LRCH3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
