
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TBC1D22A-AP3D1 (FusionGDB2 ID:89282) |
Fusion Gene Summary for TBC1D22A-AP3D1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TBC1D22A-AP3D1 | Fusion gene ID: 89282 | Hgene | Tgene | Gene symbol | TBC1D22A | AP3D1 | Gene ID | 25771 | 8943 |
| Gene name | TBC1 domain family member 22A | adaptor related protein complex 3 subunit delta 1 | |
| Synonyms | C22orf4|HSC79E021 | ADTD|HPS10|hBLVR | |
| Cytomap | 22q13.31 | 19p13.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | TBC1 domain family member 22Aputative GTPase activator | AP-3 complex subunit delta-1AP-3 complex delta subunit, partial CDSadapter-related protein complex 3 subunit delta-1adaptor related protein complex 3 delta 1 subunitdelta adaptinsubunit of putative vesicle coat adaptor complex AP-3 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O14617 | |
| Ensembl transtripts involved in fusion gene | ENST00000337137, ENST00000355704, ENST00000406733, ENST00000407381, ENST00000380995, ENST00000472791, | ENST00000590683, ENST00000345016, ENST00000355272, ENST00000356926, ENST00000350812, | |
| Fusion gene scores | * DoF score | 34 X 18 X 14=8568 | 12 X 12 X 8=1152 |
| # samples | 37 | 13 | |
| ** MAII score | log2(37/8568*10)=-4.53336130423394 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(13/1152*10)=-3.14755718841386 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TBC1D22A [Title/Abstract] AND AP3D1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TBC1D22A(47433094)-AP3D1(2102267), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | TBC1D22A-AP3D1 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. TBC1D22A-AP3D1 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across TBC1D22A (5'-gene) Fusion gene breakpoints across TBC1D22A (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
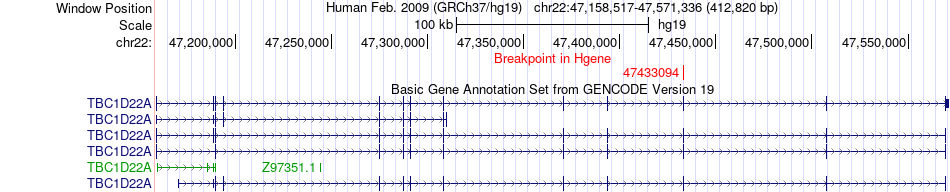 |
 Fusion gene breakpoints across AP3D1 (3'-gene) Fusion gene breakpoints across AP3D1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
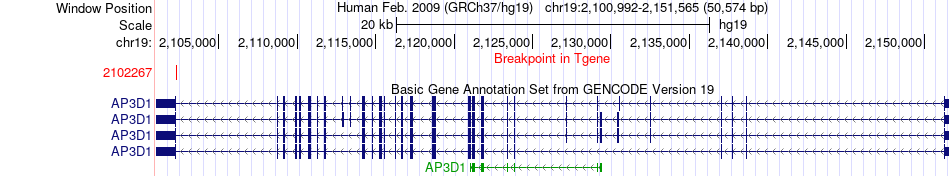 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCS | TCGA-NA-A4R1-01A | TBC1D22A | chr22 | 47433094 | - | AP3D1 | chr19 | 2102267 | - |
| ChimerDB4 | UCS | TCGA-NA-A4R1-01A | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| ChimerDB4 | UCS | TCGA-NA-A4R1 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
Top |
Fusion Gene ORF analysis for TBC1D22A-AP3D1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000337137 | ENST00000590683 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| 5CDS-intron | ENST00000355704 | ENST00000590683 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| 5CDS-intron | ENST00000406733 | ENST00000590683 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| 5CDS-intron | ENST00000407381 | ENST00000590683 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000337137 | ENST00000345016 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000337137 | ENST00000355272 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000337137 | ENST00000356926 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000355704 | ENST00000345016 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000355704 | ENST00000355272 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000355704 | ENST00000356926 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000406733 | ENST00000345016 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000406733 | ENST00000355272 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000406733 | ENST00000356926 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000407381 | ENST00000345016 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000407381 | ENST00000355272 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| Frame-shift | ENST00000407381 | ENST00000356926 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| In-frame | ENST00000337137 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| In-frame | ENST00000355704 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| In-frame | ENST00000406733 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| In-frame | ENST00000407381 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000380995 | ENST00000345016 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000380995 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000380995 | ENST00000355272 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000380995 | ENST00000356926 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000472791 | ENST00000345016 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000472791 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000472791 | ENST00000355272 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-3CDS | ENST00000472791 | ENST00000356926 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-intron | ENST00000380995 | ENST00000590683 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
| intron-intron | ENST00000472791 | ENST00000590683 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000337137 | TBC1D22A | chr22 | 47433094 | + | ENST00000350812 | AP3D1 | chr19 | 2102267 | - | 2768 | 1495 | 166 | 1590 | 474 |
| ENST00000407381 | TBC1D22A | chr22 | 47433094 | + | ENST00000350812 | AP3D1 | chr19 | 2102267 | - | 2540 | 1267 | 115 | 1362 | 415 |
| ENST00000355704 | TBC1D22A | chr22 | 47433094 | + | ENST00000350812 | AP3D1 | chr19 | 2102267 | - | 2474 | 1201 | 106 | 1296 | 396 |
| ENST00000406733 | TBC1D22A | chr22 | 47433094 | + | ENST00000350812 | AP3D1 | chr19 | 2102267 | - | 2849 | 1576 | 241 | 1671 | 476 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000337137 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - | 0.005616967 | 0.994383 |
| ENST00000407381 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - | 0.002512413 | 0.99748755 |
| ENST00000355704 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - | 0.003906109 | 0.9960939 |
| ENST00000406733 | ENST00000350812 | TBC1D22A | chr22 | 47433094 | + | AP3D1 | chr19 | 2102267 | - | 0.006690538 | 0.9933095 |
Top |
Fusion Genomic Features for TBC1D22A-AP3D1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
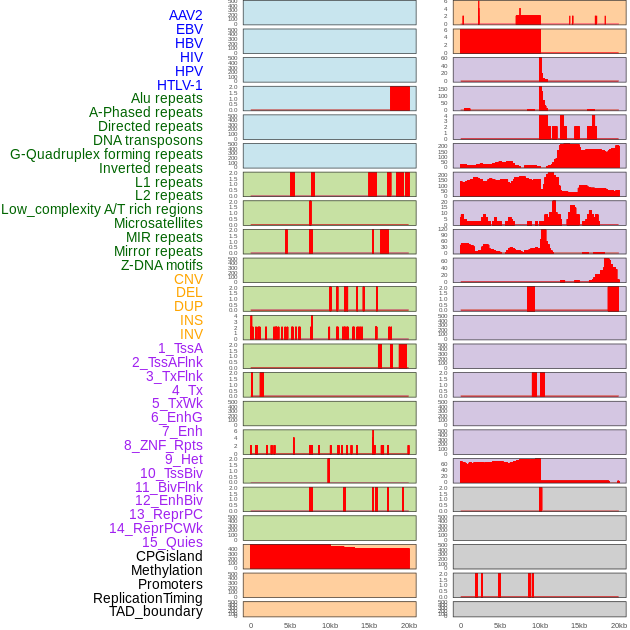 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TBC1D22A-AP3D1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:47433094/chr19:2102267) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | AP3D1 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Part of the AP-3 complex, an adaptor-related complex which is not clathrin-associated. The complex is associated with the Golgi region as well as more peripheral structures. It facilitates the budding of vesicles from the Golgi membrane and may be directly involved in trafficking to lysosomes. Involved in process of CD8+ T-cell and NK cell degranulation (PubMed:26744459). In concert with the BLOC-1 complex, AP-3 is required to target cargos into vesicles assembled at cell bodies for delivery into neurites and nerve terminals (By similarity). {ECO:0000250|UniProtKB:O54774, ECO:0000269|PubMed:26744459}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TBC1D22A | chr22:47433094 | chr19:2102267 | ENST00000337137 | + | 11 | 13 | 222_446 | 443 | 518.0 | Domain | Rab-GAP TBC |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TBC1D22A | chr22:47433094 | chr19:2102267 | ENST00000355704 | + | 9 | 11 | 222_446 | 365 | 440.0 | Domain | Rab-GAP TBC |
| Hgene | TBC1D22A | chr22:47433094 | chr19:2102267 | ENST00000380995 | + | 1 | 9 | 222_446 | 0 | 325.0 | Domain | Rab-GAP TBC |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 659_679 | 1122 | 1154.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 725_756 | 1122 | 1154.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 845_869 | 1122 | 1154.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 659_679 | 1184 | 1216.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 725_756 | 1184 | 1216.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 845_869 | 1184 | 1216.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 828_893 | 1122 | 1154.0 | Compositional bias | Note=Lys-rich | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 828_893 | 1184 | 1216.0 | Compositional bias | Note=Lys-rich | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 142_179 | 1122 | 1154.0 | Repeat | Note=HEAT 3 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 180_216 | 1122 | 1154.0 | Repeat | Note=HEAT 4 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 254_292 | 1122 | 1154.0 | Repeat | Note=HEAT 5 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 299_336 | 1122 | 1154.0 | Repeat | Note=HEAT 6 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 338_373 | 1122 | 1154.0 | Repeat | Note=HEAT 7 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 34_71 | 1122 | 1154.0 | Repeat | Note=HEAT 1 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 375_409 | 1122 | 1154.0 | Repeat | Note=HEAT 8 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 431_468 | 1122 | 1154.0 | Repeat | Note=HEAT 9 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 497_535 | 1122 | 1154.0 | Repeat | Note=HEAT 10 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 548_585 | 1122 | 1154.0 | Repeat | Note=HEAT 11 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000345016 | 28 | 30 | 77_114 | 1122 | 1154.0 | Repeat | Note=HEAT 2 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 142_179 | 1184 | 1216.0 | Repeat | Note=HEAT 3 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 180_216 | 1184 | 1216.0 | Repeat | Note=HEAT 4 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 254_292 | 1184 | 1216.0 | Repeat | Note=HEAT 5 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 299_336 | 1184 | 1216.0 | Repeat | Note=HEAT 6 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 338_373 | 1184 | 1216.0 | Repeat | Note=HEAT 7 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 34_71 | 1184 | 1216.0 | Repeat | Note=HEAT 1 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 375_409 | 1184 | 1216.0 | Repeat | Note=HEAT 8 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 431_468 | 1184 | 1216.0 | Repeat | Note=HEAT 9 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 497_535 | 1184 | 1216.0 | Repeat | Note=HEAT 10 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 548_585 | 1184 | 1216.0 | Repeat | Note=HEAT 11 | |
| Tgene | AP3D1 | chr22:47433094 | chr19:2102267 | ENST00000355272 | 30 | 32 | 77_114 | 1184 | 1216.0 | Repeat | Note=HEAT 2 |
Top |
Fusion Gene Sequence for TBC1D22A-AP3D1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >89282_89282_1_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000337137_AP3D1_chr19_2102267_ENST00000350812_length(transcript)=2768nt_BP=1495nt GCGCGTAGGCACAACTTCCGGAAGGAGGCGGAAGAGCTTCTCGGCTCTAGGCTCTGGAGTCCCGGGAGCAGTGAGGGGCCACCCGGGGCA CAGGAAAGGGCCGCTAGGGGAGGGCCGGGTGCACTCGGGGTGTCTGGGCCGCGGGTCTGAGGGATGAGGAGGGGCCATGGCCAGCGACGG GGCCAGGAAGCAATTCTGGAAGCGCAGCAACAGCAAGCTCCCGGGCAGCATCCAGCACGTGTATGGTGCCCAGCACCCCCCCTTTGATCC ACTGTTACATGGCACTTTGCTCAGGTCCACGGCCAAGATGCCGACCACACCAGTGAAGGCCAAGAGGGTCAGCACCTTCCAGGAGTTTGA GAGCAATACCAGCGATGCCTGGGACGCTGGGGAGGACGACGATGAGCTCCTGGCCATGGCGGCGGAGAGCCTGAACTCCGAGGTGGTCAT GGAGACGGCCAACCGTGTGCTGCGTAACCACAGCCAGCGGCAGGGGCGGCCCACGCTGCAGGAGGGGCCAGGGCTTCAGCAGAAGCCCAG GCCCGAGGCAGAGCCGCCCTCACCCCCCAGCGGCGACCTCCGGCTGGTGAAGTCGGTCAGTGAGAGCCACACGTCCTGTCCTGCAGAAAG TGCCAGCGATGCCGCCCCTCTGCAGAGGTCCCAGTCTCTCCCACACTCGGCCACCGTCACGCTGGGTGGCACATCTGACCCCAGCACTCT CAGCAGCTCAGCGCTGAGCGAAAGAGAGGCCTCCCGGCTCGACAAGTTCAAGCAGCTGCTTGCCGGCCCCAACACGGACCTTGAGGAATT ACGGAGGTTGAGCTGGTCCGGAATCCCTAAGCCAGTGCGTCCAATGACGTGGAAGCTCCTCTCAGGTTACCTTCCCGCCAATGTAGACCG GAGACCAGCCACTCTCCAGAGAAAACAAAAAGAATATTTTGCATTTATTGAGCACTATTACGATTCTAGGAACGACGAAGTTCACCAGGA CACATACAGGCAGATCCACATAGACATCCCTCGCATGAGCCCTGAAGCGTTGATCCTGCAGCCCAAGGTGACGGAGATTTTTGAAAGGAT CTTGTTCATATGGGCGATCCGCCACCCAGCCAGTGGATACGTTCAGGGTATAAATGATCTCGTCACTCCTTTCTTTGTGGTCTTCATTTG TGAATACATAGAGGCAGAGGAGGTGGACACGGTGGACGTCTCCGGCGTGCCCGCAGAGGTGCTGTGCAACATCGAGGCCGACACCTACTG GTGCATGAGCAAGCTGCTGGATGGCATTCAGGACAACTACACCTTTGCCCAACCTGGGATTCAAATGAAAGTGAAAATGTTAGAAGAACT CGTGAGCCGGATTGATGAGCAAGTGCACCGGCACCTGGACCAACACGAAGTGAGATACCTGCAGTTTGCCTTCCGCTGGATGAACAACCT GCTGATGAGGGAGGTGCCCCTGCGTTGTACCATCCGCCTGTGGGACACCTACCAGGGTGAGAACTCTGTCTCAGTCGACGGGAAGTGCAG TGACTCCACGCTACTGAGCAACTTGTTAGAAGAGATGAAGGCGACGCTGGCCAAGTGTTGAGAGCTGCCTGCGAGCCCCGCACCACCCCG CGGAGCACGTACCCAGGGACCGCAGCCCTGACGTGTCTCGCCTCTCCTCAGTCGTGTGTACTGTACCCAAGCCTGAGTGTTAATTTAACT CTATGTTGTCCGCCGTGTAGACATCCGAGGTCATTTGTTGCGTTGAATTATCTGACCATCCTTTTTTACTGTGACTCTTCCCATTCTCTT TGGCAAGAAGTCCCCTTCTCGCCCCCAAACCAGCAAGGGACTCCCCCACCTGGGTCTGTGCCCTGCCCCGCGCTGGGGGCCGAGTCCTTG AATGTGGCTTCAGGGGCTCCTGTCCTGGGCCAGGGCCTGATGGGCACCACGTGAGGGGCACTTGGTGGACAGGGCGGGGCTGACGTGGCC TCCTCTGGGGTCGCCTGCTTTTGACCCAAAGGTCCTGACGGTTGCGTCCGGGGGAGGGGAAGGAAGGGCCGCTGTCGCCAAGGTTTTCTC TCCCAGAACCCACAGTGGGAAAGCGGTCTTGCCAGGCGTTGTCCATTGTCAGTGTGCTCGTGGGCTGGTGACTGGGTCTTGGGATCCCAG GCCACGCGCCAGCCAGGCTGTGGGCAGGGCGGGGCCAGGGACGCCAAAGAGAGGTTGCAGTCAGAACCGTGGACGGGGTGGGTTGAGGCC TCTCTGCCACCCGTCTTCCTGGTCAGCAGAAGTGCATCTCGGCTTGGGTTTGGGGTGGTCCGCATCCCCTGCTTGCCACTATGCGCACCA AGGTTTCCCCACATCCTTCCCAGCACCCTTAGGAAGGCCCAGGCAGGGCCTGGAAGCAGCGGACCTGGGCTGTTCTGTGTTGAAGGAGTG TGCCCAGTGCCCTTGGGCAGGACCTGTGAGAGCCACCTCACAGGCAGAGCCCCCACCAGGCAGGGCAAGGAGACTCCGCTCACTCCCCAC GGCCAGCGTGGGCACAGGACTGACCCTTCTTCAGAGATAATGACATTTTATCTTCTCCTTTTGATGAAAACTGTCACTTTAGCATGTAAT CCATTACAGAATCCCATGCAGTGATTCCAGGATTTGAAATTGTATGATGTGTTACATAAGAATTTATTTGCTATCGACATTCCCGTATAA >89282_89282_1_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000337137_AP3D1_chr19_2102267_ENST00000350812_length(amino acids)=474AA_BP=442 MASDGARKQFWKRSNSKLPGSIQHVYGAQHPPFDPLLHGTLLRSTAKMPTTPVKAKRVSTFQEFESNTSDAWDAGEDDDELLAMAAESLN SEVVMETANRVLRNHSQRQGRPTLQEGPGLQQKPRPEAEPPSPPSGDLRLVKSVSESHTSCPAESASDAAPLQRSQSLPHSATVTLGGTS DPSTLSSSALSEREASRLDKFKQLLAGPNTDLEELRRLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRKQKEYFAFIEHYYDSRND EVHQDTYRQIHIDIPRMSPEALILQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPFFVVFICEYIEAEEVDTVDVSGVPAEVLCNIE ADTYWCMSKLLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRWMNNLLMREVPLRCTIRLWDTYQGENSVSV -------------------------------------------------------------- >89282_89282_2_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000355704_AP3D1_chr19_2102267_ENST00000350812_length(transcript)=2474nt_BP=1201nt CCCGGGAGCAGTGAGGGGCCACCCGGGGCACAGGAAAGGGCCGCTAGGGGAGGGCCGGGTGCACTCGGGGTGTCTGGGCCGCGGGTCTGA GGGATGAGGAGGGGCCATGGCCAGCGACGGGGCCAGGAAGCAATTCTGGAAGCGCAGCAACAGCAAGCTCCCGGGCAGTTTGCTCAGGTC CACGGCCAAGATGCCGACCACACCAGTGAAGGCCAAGAGGGTCAGCACCTTCCAGGAGTTTGAGAGCAATACCAGCGATGCCTGGGACGC TGGGGAGGACGACGATGAGCTCCTGGCCATGGCGGCGGAGAGCCTGAACTCCGAGGTGGTCATGGAGACGGCCAACCGTGTGCTGCGTAA CCACAGCCAGCGGCAGGGGCGGCCCACGCTGCAGGAGGGGCCAGGGCTTCAGCAGAAGCCCAGGCCCGAGGCAGAGCCGCCCTCACCCCC CAGCGGCGACCTCCGGCTGGTGAAGTCGGTCAGTGAGAGCCACACGTCCTGTCCTGCAGAGGAATTACGGAGGTTGAGCTGGTCCGGAAT CCCTAAGCCAGTGCGTCCAATGACGTGGAAGCTCCTCTCAGGTTACCTTCCCGCCAATGTAGACCGGAGACCAGCCACTCTCCAGAGAAA ACAAAAAGAATATTTTGCATTTATTGAGCACTATTACGATTCTAGGAACGACGAAGTTCACCAGGACACATACAGGCAGATCCACATAGA CATCCCTCGCATGAGCCCTGAAGCGTTGATCCTGCAGCCCAAGGTGACGGAGATTTTTGAAAGGATCTTGTTCATATGGGCGATCCGCCA CCCAGCCAGTGGATACGTTCAGGGTATAAATGATCTCGTCACTCCTTTCTTTGTGGTCTTCATTTGTGAATACATAGAGGCAGAGGAGGT GGACACGGTGGACGTCTCCGGCGTGCCCGCAGAGGTGCTGTGCAACATCGAGGCCGACACCTACTGGTGCATGAGCAAGCTGCTGGATGG CATTCAGGACAACTACACCTTTGCCCAACCTGGGATTCAAATGAAAGTGAAAATGTTAGAAGAACTCGTGAGCCGGATTGATGAGCAAGT GCACCGGCACCTGGACCAACACGAAGTGAGATACCTGCAGTTTGCCTTCCGCTGGATGAACAACCTGCTGATGAGGGAGGTGCCCCTGCG TTGTACCATCCGCCTGTGGGACACCTACCAGGGTGAGAACTCTGTCTCAGTCGACGGGAAGTGCAGTGACTCCACGCTACTGAGCAACTT GTTAGAAGAGATGAAGGCGACGCTGGCCAAGTGTTGAGAGCTGCCTGCGAGCCCCGCACCACCCCGCGGAGCACGTACCCAGGGACCGCA GCCCTGACGTGTCTCGCCTCTCCTCAGTCGTGTGTACTGTACCCAAGCCTGAGTGTTAATTTAACTCTATGTTGTCCGCCGTGTAGACAT CCGAGGTCATTTGTTGCGTTGAATTATCTGACCATCCTTTTTTACTGTGACTCTTCCCATTCTCTTTGGCAAGAAGTCCCCTTCTCGCCC CCAAACCAGCAAGGGACTCCCCCACCTGGGTCTGTGCCCTGCCCCGCGCTGGGGGCCGAGTCCTTGAATGTGGCTTCAGGGGCTCCTGTC CTGGGCCAGGGCCTGATGGGCACCACGTGAGGGGCACTTGGTGGACAGGGCGGGGCTGACGTGGCCTCCTCTGGGGTCGCCTGCTTTTGA CCCAAAGGTCCTGACGGTTGCGTCCGGGGGAGGGGAAGGAAGGGCCGCTGTCGCCAAGGTTTTCTCTCCCAGAACCCACAGTGGGAAAGC GGTCTTGCCAGGCGTTGTCCATTGTCAGTGTGCTCGTGGGCTGGTGACTGGGTCTTGGGATCCCAGGCCACGCGCCAGCCAGGCTGTGGG CAGGGCGGGGCCAGGGACGCCAAAGAGAGGTTGCAGTCAGAACCGTGGACGGGGTGGGTTGAGGCCTCTCTGCCACCCGTCTTCCTGGTC AGCAGAAGTGCATCTCGGCTTGGGTTTGGGGTGGTCCGCATCCCCTGCTTGCCACTATGCGCACCAAGGTTTCCCCACATCCTTCCCAGC ACCCTTAGGAAGGCCCAGGCAGGGCCTGGAAGCAGCGGACCTGGGCTGTTCTGTGTTGAAGGAGTGTGCCCAGTGCCCTTGGGCAGGACC TGTGAGAGCCACCTCACAGGCAGAGCCCCCACCAGGCAGGGCAAGGAGACTCCGCTCACTCCCCACGGCCAGCGTGGGCACAGGACTGAC CCTTCTTCAGAGATAATGACATTTTATCTTCTCCTTTTGATGAAAACTGTCACTTTAGCATGTAATCCATTACAGAATCCCATGCAGTGA TTCCAGGATTTGAAATTGTATGATGTGTTACATAAGAATTTATTTGCTATCGACATTCCCGTATAAAGAGAGAGACATATCACGCTGCTG >89282_89282_2_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000355704_AP3D1_chr19_2102267_ENST00000350812_length(amino acids)=396AA_BP=364 MASDGARKQFWKRSNSKLPGSLLRSTAKMPTTPVKAKRVSTFQEFESNTSDAWDAGEDDDELLAMAAESLNSEVVMETANRVLRNHSQRQ GRPTLQEGPGLQQKPRPEAEPPSPPSGDLRLVKSVSESHTSCPAEELRRLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRKQKEYF AFIEHYYDSRNDEVHQDTYRQIHIDIPRMSPEALILQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPFFVVFICEYIEAEEVDTVDV SGVPAEVLCNIEADTYWCMSKLLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRWMNNLLMREVPLRCTIRL -------------------------------------------------------------- >89282_89282_3_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000406733_AP3D1_chr19_2102267_ENST00000350812_length(transcript)=2849nt_BP=1576nt AGAAGTCTGTGCCCTTACCCATCACAGCCTCATCCTTATCTCCTCCATTCCCTAGGGGACTTAACAGGTGTTGAAATTATTACAGAGAAA GCTGACTCACTCACCAGGAATCTGATCCTGCTGTGGGCTGGCTTGGGTGGAGGTGTTCCTCCCGCCCCCGCACCCATCCTCCTGTTTGAA CTCAGGCTGCTGCCTGCTGGGCCTGCCTGCCCTTGGAGCCCTGCTGAGCTCAGCCTGAGGCCTGGCTCCTCCAGGCTGGGGGAAAACCAG GCTTGCTGTGCTCGGCAGCAGAGATTCTTCTGGAGTGAGCATCCAGCACGTGTATGGTGCCCAGCACCCCCCCTTTGATCCACTGTTACA TGGCACTTTGCTCAGGTCCACGGCCAAGATGCCGACCACACCAGTGAAGGCCAAGAGGGTCAGCACCTTCCAGGAGTTTGAGAGCAATAC CAGCGATGCCTGGGACGCTGGGGAGGACGACGATGAGCTCCTGGCCATGGCGGCGGAGAGCCTGAACTCCGAGGTGGTCATGGAGACGGC CAACCGTGTGCTGCGTAACCACAGCCAGCGGCAGGGGCGGCCCACGCTGCAGGAGGGGCCAGGGCTTCAGCAGAAGCCCAGGCCCGAGGC AGAGCCGCCCTCACCCCCCAGCGGCGACCTCCGGCTGGTGAAGTCGGTCAGTGAGAGCCACACGTCCTGTCCTGCAGAAAGTGCCAGCGA TGCCGCCCCTCTGCAGAGGTCCCAGTCTCTCCCACACTCGGCCACCGTCACGCTGGGTGGCACATCTGACCCCAGCACTCTCAGCAGCTC AGCGCTGAGCGAAAGAGAGGCCTCCCGGCTCGACAAGTTCAAGCAGCTGCTTGCCGGCCCCAACACGGACCTTGAGGAATTACGGAGGTT GAGCTGGTCCGGAATCCCTAAGCCAGTGCGTCCAATGACGTGGAAGCTCCTCTCAGGTTACCTTCCCGCCAATGTAGACCGGAGACCAGC CACTCTCCAGAGAAAACAAAAAGAATATTTTGCATTTATTGAGCACTATTACGATTCTAGGAACGACGAAGTTCACCAGGACACATACAG GCAGATCCACATAGACATCCCTCGCATGAGCCCTGAAGCGTTGATCCTGCAGCCCAAGGTGACGGAGATTTTTGAAAGGATCTTGTTCAT ATGGGCGATCCGCCACCCAGCCAGTGGATACGTTCAGGGTATAAATGATCTCGTCACTCCTTTCTTTGTGGTCTTCATTTGTGAATACAT AGAGGCAGAGGAGGTGGACACGGTGGACGTCTCCGGCGTGCCCGCAGAGGTGCTGTGCAACATCGAGGCCGACACCTACTGGTGCATGAG CAAGCTGCTGGATGGCATTCAGGACAACTACACCTTTGCCCAACCTGGGATTCAAATGAAAGTGAAAATGTTAGAAGAACTCGTGAGCCG GATTGATGAGCAAGTGCACCGGCACCTGGACCAACACGAAGTGAGATACCTGCAGTTTGCCTTCCGCTGGATGAACAACCTGCTGATGAG GGAGGTGCCCCTGCGTTGTACCATCCGCCTGTGGGACACCTACCAGGGTGAGAACTCTGTCTCAGTCGACGGGAAGTGCAGTGACTCCAC GCTACTGAGCAACTTGTTAGAAGAGATGAAGGCGACGCTGGCCAAGTGTTGAGAGCTGCCTGCGAGCCCCGCACCACCCCGCGGAGCACG TACCCAGGGACCGCAGCCCTGACGTGTCTCGCCTCTCCTCAGTCGTGTGTACTGTACCCAAGCCTGAGTGTTAATTTAACTCTATGTTGT CCGCCGTGTAGACATCCGAGGTCATTTGTTGCGTTGAATTATCTGACCATCCTTTTTTACTGTGACTCTTCCCATTCTCTTTGGCAAGAA GTCCCCTTCTCGCCCCCAAACCAGCAAGGGACTCCCCCACCTGGGTCTGTGCCCTGCCCCGCGCTGGGGGCCGAGTCCTTGAATGTGGCT TCAGGGGCTCCTGTCCTGGGCCAGGGCCTGATGGGCACCACGTGAGGGGCACTTGGTGGACAGGGCGGGGCTGACGTGGCCTCCTCTGGG GTCGCCTGCTTTTGACCCAAAGGTCCTGACGGTTGCGTCCGGGGGAGGGGAAGGAAGGGCCGCTGTCGCCAAGGTTTTCTCTCCCAGAAC CCACAGTGGGAAAGCGGTCTTGCCAGGCGTTGTCCATTGTCAGTGTGCTCGTGGGCTGGTGACTGGGTCTTGGGATCCCAGGCCACGCGC CAGCCAGGCTGTGGGCAGGGCGGGGCCAGGGACGCCAAAGAGAGGTTGCAGTCAGAACCGTGGACGGGGTGGGTTGAGGCCTCTCTGCCA CCCGTCTTCCTGGTCAGCAGAAGTGCATCTCGGCTTGGGTTTGGGGTGGTCCGCATCCCCTGCTTGCCACTATGCGCACCAAGGTTTCCC CACATCCTTCCCAGCACCCTTAGGAAGGCCCAGGCAGGGCCTGGAAGCAGCGGACCTGGGCTGTTCTGTGTTGAAGGAGTGTGCCCAGTG CCCTTGGGCAGGACCTGTGAGAGCCACCTCACAGGCAGAGCCCCCACCAGGCAGGGCAAGGAGACTCCGCTCACTCCCCACGGCCAGCGT GGGCACAGGACTGACCCTTCTTCAGAGATAATGACATTTTATCTTCTCCTTTTGATGAAAACTGTCACTTTAGCATGTAATCCATTACAG AATCCCATGCAGTGATTCCAGGATTTGAAATTGTATGATGTGTTACATAAGAATTTATTTGCTATCGACATTCCCGTATAAAGAGAGAGA >89282_89282_3_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000406733_AP3D1_chr19_2102267_ENST00000350812_length(amino acids)=476AA_BP=444 MAPPGWGKTRLAVLGSRDSSGVSIQHVYGAQHPPFDPLLHGTLLRSTAKMPTTPVKAKRVSTFQEFESNTSDAWDAGEDDDELLAMAAES LNSEVVMETANRVLRNHSQRQGRPTLQEGPGLQQKPRPEAEPPSPPSGDLRLVKSVSESHTSCPAESASDAAPLQRSQSLPHSATVTLGG TSDPSTLSSSALSEREASRLDKFKQLLAGPNTDLEELRRLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRKQKEYFAFIEHYYDSR NDEVHQDTYRQIHIDIPRMSPEALILQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPFFVVFICEYIEAEEVDTVDVSGVPAEVLCN IEADTYWCMSKLLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRWMNNLLMREVPLRCTIRLWDTYQGENSV -------------------------------------------------------------- >89282_89282_4_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000407381_AP3D1_chr19_2102267_ENST00000350812_length(transcript)=2540nt_BP=1267nt CTCTGGAGTCCCGGGAGCAGTGAGGGGCCACCCGGGGCACAGGAAAGGGCCGCTAGGGGAGGGCCGGGTGCACTCGGGGTGTCTGGGCCG CGGGTCTGAGGGATGAGGAGGGGCCATGGCCAGCGACGGGGCCAGGAAGCAATTCTGGAAGCGCAGCAACAGCAAGCTCCCGGGCAGCAT CCAGCACGTGTATGGTGCCCAGCACCCCCCCTTTGATCCACTGTTACATGGCACTTTGCTCAGGTCCACGGCCAAGATGCCGACCACACC AGTGAAGGCCAAGAGGGTCAGCACCTTCCAGGAGTTTGAGAGCAATACCAGCGATGCCTGGGACGCTGGGGAGGACGACGATGAGCTCCT GGCCATGGCGGCGGAGAGCCTGAACTCCGAGGTGGTCATGGAGACGGCCAACCGTGTGCTGCGTAACCACAGCCAGCGGCAGGGGCGGCC CACGCTGCAGGAGGGGCCAGGGCTTCAGCAGAAGCCCAGGCCCGAGGCAGAGCCGCCCTCACCCCCCAGCGGCGACCTCCGGCTGGTGAA GTCGGTCAGTGAGAGCCACACGTCCTGTCCTGCAGAGGAATTACGGAGGTTGAGCTGGTCCGGAATCCCTAAGCCAGTGCGTCCAATGAC GTGGAAGCTCCTCTCAGGTTACCTTCCCGCCAATGTAGACCGGAGACCAGCCACTCTCCAGAGAAAACAAAAAGAATATTTTGCATTTAT TGAGCACTATTACGATTCTAGGAACGACGAAGTTCACCAGGACACATACAGGCAGATCCACATAGACATCCCTCGCATGAGCCCTGAAGC GTTGATCCTGCAGCCCAAGGTGACGGAGATTTTTGAAAGGATCTTGTTCATATGGGCGATCCGCCACCCAGCCAGTGGATACGTTCAGGG TATAAATGATCTCGTCACTCCTTTCTTTGTGGTCTTCATTTGTGAATACATAGAGGCAGAGGAGGTGGACACGGTGGACGTCTCCGGCGT GCCCGCAGAGGTGCTGTGCAACATCGAGGCCGACACCTACTGGTGCATGAGCAAGCTGCTGGATGGCATTCAGGACAACTACACCTTTGC CCAACCTGGGATTCAAATGAAAGTGAAAATGTTAGAAGAACTCGTGAGCCGGATTGATGAGCAAGTGCACCGGCACCTGGACCAACACGA AGTGAGATACCTGCAGTTTGCCTTCCGCTGGATGAACAACCTGCTGATGAGGGAGGTGCCCCTGCGTTGTACCATCCGCCTGTGGGACAC CTACCAGGGTGAGAACTCTGTCTCAGTCGACGGGAAGTGCAGTGACTCCACGCTACTGAGCAACTTGTTAGAAGAGATGAAGGCGACGCT GGCCAAGTGTTGAGAGCTGCCTGCGAGCCCCGCACCACCCCGCGGAGCACGTACCCAGGGACCGCAGCCCTGACGTGTCTCGCCTCTCCT CAGTCGTGTGTACTGTACCCAAGCCTGAGTGTTAATTTAACTCTATGTTGTCCGCCGTGTAGACATCCGAGGTCATTTGTTGCGTTGAAT TATCTGACCATCCTTTTTTACTGTGACTCTTCCCATTCTCTTTGGCAAGAAGTCCCCTTCTCGCCCCCAAACCAGCAAGGGACTCCCCCA CCTGGGTCTGTGCCCTGCCCCGCGCTGGGGGCCGAGTCCTTGAATGTGGCTTCAGGGGCTCCTGTCCTGGGCCAGGGCCTGATGGGCACC ACGTGAGGGGCACTTGGTGGACAGGGCGGGGCTGACGTGGCCTCCTCTGGGGTCGCCTGCTTTTGACCCAAAGGTCCTGACGGTTGCGTC CGGGGGAGGGGAAGGAAGGGCCGCTGTCGCCAAGGTTTTCTCTCCCAGAACCCACAGTGGGAAAGCGGTCTTGCCAGGCGTTGTCCATTG TCAGTGTGCTCGTGGGCTGGTGACTGGGTCTTGGGATCCCAGGCCACGCGCCAGCCAGGCTGTGGGCAGGGCGGGGCCAGGGACGCCAAA GAGAGGTTGCAGTCAGAACCGTGGACGGGGTGGGTTGAGGCCTCTCTGCCACCCGTCTTCCTGGTCAGCAGAAGTGCATCTCGGCTTGGG TTTGGGGTGGTCCGCATCCCCTGCTTGCCACTATGCGCACCAAGGTTTCCCCACATCCTTCCCAGCACCCTTAGGAAGGCCCAGGCAGGG CCTGGAAGCAGCGGACCTGGGCTGTTCTGTGTTGAAGGAGTGTGCCCAGTGCCCTTGGGCAGGACCTGTGAGAGCCACCTCACAGGCAGA GCCCCCACCAGGCAGGGCAAGGAGACTCCGCTCACTCCCCACGGCCAGCGTGGGCACAGGACTGACCCTTCTTCAGAGATAATGACATTT TATCTTCTCCTTTTGATGAAAACTGTCACTTTAGCATGTAATCCATTACAGAATCCCATGCAGTGATTCCAGGATTTGAAATTGTATGAT GTGTTACATAAGAATTTATTTGCTATCGACATTCCCGTATAAAGAGAGAGACATATCACGCTGCTGTCATGATTTTGTGTCAAGATGATC >89282_89282_4_TBC1D22A-AP3D1_TBC1D22A_chr22_47433094_ENST00000407381_AP3D1_chr19_2102267_ENST00000350812_length(amino acids)=415AA_BP=383 MASDGARKQFWKRSNSKLPGSIQHVYGAQHPPFDPLLHGTLLRSTAKMPTTPVKAKRVSTFQEFESNTSDAWDAGEDDDELLAMAAESLN SEVVMETANRVLRNHSQRQGRPTLQEGPGLQQKPRPEAEPPSPPSGDLRLVKSVSESHTSCPAEELRRLSWSGIPKPVRPMTWKLLSGYL PANVDRRPATLQRKQKEYFAFIEHYYDSRNDEVHQDTYRQIHIDIPRMSPEALILQPKVTEIFERILFIWAIRHPASGYVQGINDLVTPF FVVFICEYIEAEEVDTVDVSGVPAEVLCNIEADTYWCMSKLLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAF -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TBC1D22A-AP3D1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TBC1D22A-AP3D1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TBC1D22A-AP3D1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
