
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TBC1D2-GABBR2 (FusionGDB2 ID:89350) |
Fusion Gene Summary for TBC1D2-GABBR2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TBC1D2-GABBR2 | Fusion gene ID: 89350 | Hgene | Tgene | Gene symbol | TBC1D2 | GABBR2 | Gene ID | 55357 | 9568 |
| Gene name | TBC1 domain family member 2 | gamma-aminobutyric acid type B receptor subunit 2 | |
| Synonyms | PARIS-1|PARIS1|TBC1D2A | EIEE59|GABABR2|GPR51|GPRC3B|HG20|HRIHFB2099|NDPLHS | |
| Cytomap | 9q22.33 | 9q22.33 | |
| Type of gene | protein-coding | protein-coding | |
| Description | TBC1 domain family member 2ATBC1 domain family, member 2Aarmusprostate antigen recognized and identified by SEREX 1 | gamma-aminobutyric acid type B receptor subunit 2G-protein coupled receptor 51GABA-B receptor 2GABA-B receptor, R2 subunitGABA-B-R2GABA-BR2gamma-aminobutyric acid (GABA) B receptor, 2gamma-aminobutyric acid B receptor 2gb2 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O75899 | |
| Ensembl transtripts involved in fusion gene | ENST00000342112, ENST00000375064, ENST00000375066, ENST00000493589, ENST00000375063, | ENST00000477471, ENST00000259455, | |
| Fusion gene scores | * DoF score | 3 X 3 X 2=18 | 8 X 8 X 5=320 |
| # samples | 3 | 8 | |
| ** MAII score | log2(3/18*10)=0.736965594166206 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(8/320*10)=-2 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TBC1D2 [Title/Abstract] AND GABBR2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TBC1D2(100991234)-GABBR2(101258796), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | TBC1D2-GABBR2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. TBC1D2-GABBR2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. TBC1D2-GABBR2 seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TBC1D2 | GO:0043547 | positive regulation of GTPase activity | 20116244 |
| Tgene | GABBR2 | GO:0007214 | gamma-aminobutyric acid signaling pathway | 9872316 |
 Fusion gene breakpoints across TBC1D2 (5'-gene) Fusion gene breakpoints across TBC1D2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
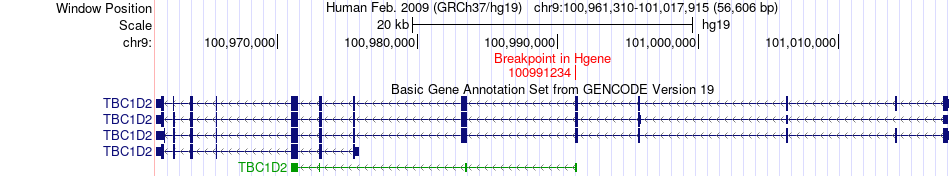 |
 Fusion gene breakpoints across GABBR2 (3'-gene) Fusion gene breakpoints across GABBR2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
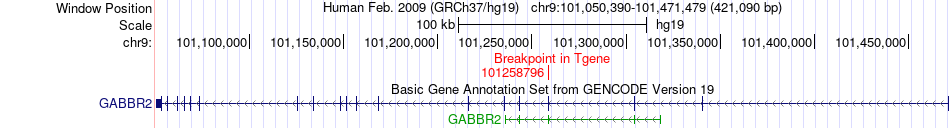 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | HNSC | TCGA-CN-4739-01A | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| ChimerDB4 | HNSC | TCGA-CN-4739 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
Top |
Fusion Gene ORF analysis for TBC1D2-GABBR2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-5UTR | ENST00000342112 | ENST00000477471 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| 5CDS-5UTR | ENST00000375064 | ENST00000477471 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| 5CDS-5UTR | ENST00000375066 | ENST00000477471 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| 5UTR-3CDS | ENST00000493589 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| 5UTR-5UTR | ENST00000493589 | ENST00000477471 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| Frame-shift | ENST00000342112 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| In-frame | ENST00000375064 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| In-frame | ENST00000375066 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| intron-3CDS | ENST00000375063 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
| intron-5UTR | ENST00000375063 | ENST00000477471 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000375064 | TBC1D2 | chr9 | 100991234 | - | ENST00000259455 | GABBR2 | chr9 | 101258796 | - | 5688 | 1017 | 30 | 3212 | 1060 |
| ENST00000375066 | TBC1D2 | chr9 | 100991234 | - | ENST00000259455 | GABBR2 | chr9 | 101258796 | - | 5741 | 1070 | 14 | 3265 | 1083 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000375064 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - | 0.001445734 | 0.9985543 |
| ENST00000375066 | ENST00000259455 | TBC1D2 | chr9 | 100991234 | - | GABBR2 | chr9 | 101258796 | - | 0.001458124 | 0.99854195 |
Top |
Fusion Genomic Features for TBC1D2-GABBR2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
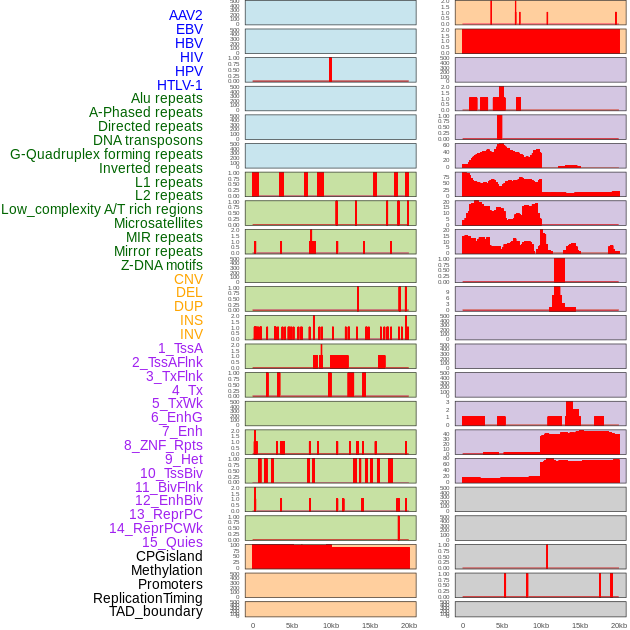 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TBC1D2-GABBR2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:100991234/chr9:101258796) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | GABBR2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Component of a heterodimeric G-protein coupled receptor for GABA, formed by GABBR1 and GABBR2 (PubMed:9872316, PubMed:9872744, PubMed:15617512, PubMed:18165688, PubMed:22660477, PubMed:24305054). Within the heterodimeric GABA receptor, only GABBR1 seems to bind agonists, while GABBR2 mediates coupling to G proteins (PubMed:18165688). Ligand binding causes a conformation change that triggers signaling via guanine nucleotide-binding proteins (G proteins) and modulates the activity of down-stream effectors, such as adenylate cyclase (PubMed:10075644, PubMed:10773016, PubMed:24305054). Signaling inhibits adenylate cyclase, stimulates phospholipase A2, activates potassium channels, inactivates voltage-dependent calcium-channels and modulates inositol phospholipid hydrolysis (PubMed:10075644, PubMed:9872744, PubMed:10906333, PubMed:10773016). Plays a critical role in the fine-tuning of inhibitory synaptic transmission (PubMed:9872744, PubMed:22660477). Pre-synaptic GABA receptor inhibits neurotransmitter release by down-regulating high-voltage activated calcium channels, whereas postsynaptic GABA receptor decreases neuronal excitability by activating a prominent inwardly rectifying potassium (Kir) conductance that underlies the late inhibitory postsynaptic potentials (PubMed:9872316, PubMed:10075644, PubMed:9872744, PubMed:22660477). Not only implicated in synaptic inhibition but also in hippocampal long-term potentiation, slow wave sleep, muscle relaxation and antinociception (Probable). {ECO:0000269|PubMed:10075644, ECO:0000269|PubMed:10328880, ECO:0000269|PubMed:15617512, ECO:0000269|PubMed:18165688, ECO:0000269|PubMed:22660477, ECO:0000269|PubMed:24305054, ECO:0000269|PubMed:9872316, ECO:0000269|PubMed:9872744, ECO:0000305}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 11_14 | 108 | 711.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 11_14 | 326 | 861.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 11_14 | 326 | 918.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 45_142 | 326 | 861.0 | Domain | PH |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 45_142 | 326 | 918.0 | Domain | PH |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 781_819 | 210 | 942.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 505_522 | 210 | 942.0 | Topological domain | Cytoplasmic | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 544_551 | 210 | 942.0 | Topological domain | Extracellular | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 573_597 | 210 | 942.0 | Topological domain | Cytoplasmic | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 619_654 | 210 | 942.0 | Topological domain | Extracellular | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 676_691 | 210 | 942.0 | Topological domain | Cytoplasmic | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 713_720 | 210 | 942.0 | Topological domain | Extracellular | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 742_941 | 210 | 942.0 | Topological domain | Cytoplasmic | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 484_504 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D1 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 523_543 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D2 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 552_572 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D3 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 598_618 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D4 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 655_675 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D5 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 692_712 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D6 | |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 721_741 | 210 | 942.0 | Transmembrane | Helical%3B Name%3D7 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 298_416 | 108 | 711.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 875_913 | 108 | 711.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 298_416 | 0 | 469.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 875_913 | 0 | 469.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 298_416 | 326 | 861.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 875_913 | 326 | 861.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 298_416 | 326 | 918.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 875_913 | 326 | 918.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 11_14 | 0 | 469.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 45_142 | 108 | 711.0 | Domain | PH |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 625_817 | 108 | 711.0 | Domain | Rab-GAP TBC |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 45_142 | 0 | 469.0 | Domain | PH |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 625_817 | 0 | 469.0 | Domain | Rab-GAP TBC |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 625_817 | 326 | 861.0 | Domain | Rab-GAP TBC |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 625_817 | 326 | 918.0 | Domain | Rab-GAP TBC |
| Tgene | GABBR2 | chr9:100991234 | chr9:101258796 | ENST00000259455 | 2 | 19 | 42_483 | 210 | 942.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for TBC1D2-GABBR2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >89350_89350_1_TBC1D2-GABBR2_TBC1D2_chr9_100991234_ENST00000375064_GABBR2_chr9_101258796_ENST00000259455_length(transcript)=5688nt_BP=1017nt GCGGACCCCTCGTCCCTCCGCAGTCTCCGGCTGGCAGCGATGGAGGGCGCTGGGGAGAACGCCCCGGAGTCCAGCTCCTCTGCCCCTGGG TCCGAAGAGTCTGCCAGGGATCCACAGGTGCCGCCTCCGGAGGAAGAATCGGGGGACTGCGCCCGGTCCCTGGAGGCGGTCCCCAAGAAA CTCTGTGGGTATTTAAGTAAGTTCGGCGGCAAAGGGCCCATCCGGGGCTGGAAATCCCGCTGGTTCTTCTACGACGAAAGGAAATGTCAG CTGTATTACTCGCGGACCGCTCAGGATGCCAATCCCTTGGACAGCATCGACCTCTCCAGTGCAGTGTTTGACTGTAAGGCGGACGCTGAG GAGGGGATCTTCGAAATCAAGACTCCCAGCCGGGTTATTACCCTGAAGGCCGCCACCAAGCAAGCGATGCTGTACTGGCTGCAGCAGCTG CAGATGAAGCGCTGGGAATTCCACAACAGCCCGCCGGCACCTCCTGCCACCCCTGATGCCGCCCTGGCTGGGAATGGGCCCGTCCTGCAC CTCGAGCTAGGGCAAGAAGAGGCAGAGCTGGAGGAGTTCCTGTGCCCTGTGAAAACACCCCCTGGGCTAGTGGGCGTGGCAGCTGCCTTG CAGCCCTTCCCTGCCCTTCAGAATATTTCCCTCAAGCACCTGGGGACTGAAATACAGAACACAATGCACAACATCCGTGGCAACAAGCAG GCCCAGGGAACAGGCCATGAACCTCCAGGGGAAGATTCTCCACAGAGTGGGGAGCCTCAGAGGGAGGAGCAGCCCTTGGCCTCTGACGCC AGCACCCCAGGGAGAGAGCCAGAGGATTCTCCAAAGCCTGCACCCAAGCCTTCTCTGACCATCAGTTTCGCTCAGAAAGCCAAGCGCCAG AACAACACCTTCCCATTCTTTTCTGAAGGAATCACACGGAACCGAACTGCCCAGGAGAAAGTGGCAGCCTTGGAGCAACAGGTTCTGATG CTCACCAAGGAGTTAAAGTCTCAGAAGGTGCGGAATGACCTGACTGGAGTTCTGTATGGCGAGGACATTGAGATTTCAGACACCGAGAGC TTCTCCAACGATCCCTGTACCAGTGTCAAAAAGCTGAAGGGGAATGATGTGCGGATCATCCTTGGCCAGTTTGACCAGAATATGGCAGCA AAAGTGTTCTGTTGTGCATACGAGGAGAACATGTATGGTAGTAAATATCAGTGGATCATTCCGGGCTGGTACGAGCCTTCTTGGTGGGAG CAGGTGCACACGGAAGCCAACTCATCCCGCTGCCTCCGGAAGAATCTGCTTGCTGCCATGGAGGGCTACATTGGCGTGGATTTCGAGCCC CTGAGCTCCAAGCAGATCAAGACCATCTCAGGAAAGACTCCACAGCAGTATGAGAGAGAGTACAACAACAAGCGGTCAGGCGTGGGGCCC AGCAAGTTCCACGGGTACGCCTACGATGGCATCTGGGTCATCGCCAAGACACTGCAGAGGGCCATGGAGACACTGCATGCCAGCAGCCGG CACCAGCGGATCCAGGACTTCAACTACACGGACCACACGCTGGGCAGGATCATCCTCAATGCCATGAACGAGACCAACTTCTTCGGGGTC ACGGGTCAAGTTGTATTCCGGAATGGGGAGAGAATGGGGACCATTAAATTTACTCAATTTCAAGACAGCAGGGAGGTGAAGGTGGGAGAG TACAACGCTGTGGCCGACACACTGGAGATCATCAATGACACCATCAGGTTCCAAGGATCCGAACCACCAAAAGACAAGACCATCATCCTG GAGCAGCTGCGGAAGATCTCCCTACCTCTCTACAGCATCCTCTCTGCCCTCACCATCCTCGGGATGATCATGGCCAGTGCTTTTCTCTTC TTCAACATCAAGAACCGGAATCAGAAGCTCATAAAGATGTCGAGTCCATACATGAACAACCTTATCATCCTTGGAGGGATGCTCTCCTAT GCTTCCATATTTCTCTTTGGCCTTGATGGATCCTTTGTCTCTGAAAAGACCTTTGAAACACTTTGCACCGTCAGGACCTGGATTCTCACC GTGGGCTACACGACCGCTTTTGGGGCCATGTTTGCAAAGACCTGGAGAGTCCACGCCATCTTCAAAAATGTGAAAATGAAGAAGAAGATC ATCAAGGACCAGAAACTGCTTGTGATCGTGGGGGGCATGCTGCTGATCGACCTGTGTATCCTGATCTGCTGGCAGGCTGTGGACCCCCTG CGAAGGACAGTGGAGAAGTACAGCATGGAGCCGGACCCAGCAGGACGGGATATCTCCATCCGCCCTCTCCTGGAGCACTGTGAGAACACC CATATGACCATCTGGCTTGGCATCGTCTATGCCTACAAGGGACTTCTCATGTTGTTCGGTTGTTTCTTAGCTTGGGAGACCCGCAACGTC AGCATCCCCGCACTCAACGACAGCAAGTACATCGGGATGAGTGTCTACAACGTGGGGATCATGTGCATCATCGGGGCCGCTGTCTCCTTC CTGACCCGGGACCAGCCCAATGTGCAGTTCTGCATCGTGGCTCTGGTCATCATCTTCTGCAGCACCATCACCCTCTGCCTGGTATTCGTG CCGAAGCTCATCACCCTGAGAACAAACCCAGATGCAGCAACGCAGAACAGGCGATTCCAGTTCACTCAGAATCAGAAGAAAGAAGATTCT AAAACGTCCACCTCGGTCACCAGTGTGAACCAAGCCAGCACATCCCGCCTGGAGGGCCTACAGTCAGAAAACCATCGCCTGCGAATGAAG ATCACAGAGCTGGATAAAGACTTGGAAGAGGTCACCATGCAGCTGCAGGACACACCAGAAAAGACCACCTACATTAAACAGAACCACTAC CAAGAGCTCAATGACATCCTCAACCTGGGAAACTTCACTGAGAGCACAGATGGAGGAAAGGCCATTTTAAAAAATCACCTCGATCAAAAT CCCCAGCTACAGTGGAACACAACAGAGCCCTCTCGAACATGCAAAGATCCTATAGAAGATATAAACTCTCCAGAACACATCCAGCGTCGG CTGTCCCTCCAGCTCCCCATCCTCCACCACGCCTACCTCCCATCCATCGGAGGCGTGGACGCCAGCTGTGTCAGCCCCTGCGTCAGCCCC ACCGCCAGCCCCCGCCACAGACATGTGCCACCCTCCTTCCGAGTCATGGTCTCGGGCCTGTAAGGGTGGGAGGCCTGGGCCCGGGGCCTC CCCCGTGACAGAACCACACTGGGCAGAGGGGTCTGCTGCAGAAACACTGTCGGCTCTGGCTGCGGAGAAGCTGGGCACCATGGCTGGCCT CTCAGGACCACTCGGATGGCACTCAGGTGGACAGGACGGGGCAGGGGGAGACTTGGCACCTGACCTCGAGCCTTATTTGTGAAGTCCTTA TTTCTTCACAAAGAAGAGGAACGGAAATGGGACGTCTTCCTTAACATCTGCAAACAAGGAGGCGCTGGGATATCAAACTTGCAAAAAAAA AAAAAAAAAAAAAAAAACAAAAAAACTAGACAAGGAGAGAGGCACTAGAACTCCAGCTGGAAGTCACGGAGTGGCTCGAGCAGCCTTGGG AAGAGGCAAGGAGCTTCTGAAGAAACTGCCTCTGCACACACATCACTGGCTGTGACCCCTCAGGCTAGCCCTTCTCCACTCTGGGGGAGG AGGTGGGAAGGGCCACCAGGCCCCCAGCTGCCAGGCCAGCTGACCCCAGCCTTCCTGGAACAGGGAGTCTGCAGGAGCGCAGACAGGCAC AGCCCTGGAGCAGGCAGGCCGAGGGCTGCGGCACTGGAGCAGGCTGACTTACATGCTCCACATGGGACCTGTGTCACCCAATGAGATGTT TGTTACTCTGGTAAATGCCACACGTTAACACAATAACACCCATTCCTGGGACCGTGGGGATTTAGGGCACGTCACTGCAGACACGCTCTG CAGCATTCACCGACAGTCTGTCATGCACCCACCACGTTGGCCATGTCCTTGTGTTCCTATCGGATGCTCCCAGTAACCAGGGGGACCACC CGAGCTAATCATGGAATGTCTGTTCCCAGCAAACACGATAAAGAAAGATTGTGCACTTTAACCTCTCTCATCAGGGCCCAAGGGCTGGCT GGGATTTTTTTTTTTTTTTTCCCACTAACTTTGTTTCTGACCAAAGTGAATTGGAGGCACTCTGCTAAAAGACATCCCCGTAGACATAGG GGAGAGAGTTGCTGGCTGAGGGCTTCCCTTGGCTTCCAGAAGGCAGCCTTCCATCCAGACAAGCCAGTGAGCTCTCCCCTTGGGATCACT GGGGTGATCAGTCAGCAGATTGATTCTCATTCATAAGATCATTCCTCCCTTTAAATTGAGCCCCTAAGAGCACTGGCCTGGGAGTCAGAC AGACCTGGGTTCAAGTCCTCAGTCCCCTGCCCACTCCCTACGTGACTTTGATCAGGTCACTAGTGTCTCTCTGAGCCTCAGTTTCCCCTC TGTAACTTGGGGTTGAACTAAAACACCTGTCCTGCCTACCTCACAAGGTCACTCTGAGGATTGAAACTTGATCTTGTCCAGGAAAGCTTT GTACCAAACAGTGAAGCCGCCCTGATCCGTGAGGTATGAGTATGACTCTGACCTTCAGCCCTCCCTACAGCCGGGGGTGTGGCCCAGAGA AGCTTCCAGCACAGCCCTCTACCCAGAACATCCGGGCTGGAGGGAGGCTCCCAGTGACTTTTCTGACATTCCTAGACAGGTTCATTCTTT GCTCAAGAAAGGCCTGAATGACAATGTCCAGGATGTCTGCACAACTGAGCAGCTCGCTCACTCCCTAAAGAAACCTATTGGCAGCTTCAA CAGGCAGGCAATAATCTCTTCCCAGAACCACTGCAGTCAGGAATAAACTGTTTTCTCCACCAGGCTTTGACAAAAGGGCCCACAGGAATC TTACCAATGCCAACATTTCAAAGCACCCTATTTCACGTAGCATAGCTTTCTGCTCCCCTTCCCCAAAGAGAGGTTATGGAGGTACTGTAG CTTTTAGGGAAAAAAAAATGTTAACACATCACAGGTCAAGTTGAAGTCATTCTCTGTTTAGGCACTAAAAATCGGTGTTGTCACTCACTG TGTATTACCAGTATTTACTTGCTTTCTTGATTTCACCAAAACCAAATTTAATTTAAAGGACCACATTAATTTTTCAAAGGGAAAGAGACA ATTAATTGTACATAATGTATACACACACACAAAAAAAAATACCTGTAGAAATATTATTCCAGCATAGCAGGAAAACAAACAAAAGTATTG GACTGTCGGAGGTGAGCCTGTGCGTCTGTAACCCTTTGTGACTCCTGAGCGTGCGCTGTCTTCTAGGTTAACTCACGAAGTACATTCTCT GTCTTACTGATACTGTAGGTTCACCCATTTTTTTTTAATTTCCTCGCAAATAACAAGACCCACAGAAGTGACTCTAGCTACTTAATGGTT CTGTTCTTTTATATGCAGCAAACACACCGTCCATTTCTGAAGAGGCTTCGGCCTGAAGGCATTTTCCAATGATGTTAGTGCACAAAACGC TTTAAATTAGACTGGAACTGCCAGAATCAAATGTAAATGAGGAATTTCTCGTACCCCTACTGCATGGTATCGATTTTTAATAAATTGTTG >89350_89350_1_TBC1D2-GABBR2_TBC1D2_chr9_100991234_ENST00000375064_GABBR2_chr9_101258796_ENST00000259455_length(amino acids)=1060AA_BP=329 MAAMEGAGENAPESSSSAPGSEESARDPQVPPPEEESGDCARSLEAVPKKLCGYLSKFGGKGPIRGWKSRWFFYDERKCQLYYSRTAQDA NPLDSIDLSSAVFDCKADAEEGIFEIKTPSRVITLKAATKQAMLYWLQQLQMKRWEFHNSPPAPPATPDAALAGNGPVLHLELGQEEAEL EEFLCPVKTPPGLVGVAAALQPFPALQNISLKHLGTEIQNTMHNIRGNKQAQGTGHEPPGEDSPQSGEPQREEQPLASDASTPGREPEDS PKPAPKPSLTISFAQKAKRQNNTFPFFSEGITRNRTAQEKVAALEQQVLMLTKELKSQKVRNDLTGVLYGEDIEISDTESFSNDPCTSVK KLKGNDVRIILGQFDQNMAAKVFCCAYEENMYGSKYQWIIPGWYEPSWWEQVHTEANSSRCLRKNLLAAMEGYIGVDFEPLSSKQIKTIS GKTPQQYEREYNNKRSGVGPSKFHGYAYDGIWVIAKTLQRAMETLHASSRHQRIQDFNYTDHTLGRIILNAMNETNFFGVTGQVVFRNGE RMGTIKFTQFQDSREVKVGEYNAVADTLEIINDTIRFQGSEPPKDKTIILEQLRKISLPLYSILSALTILGMIMASAFLFFNIKNRNQKL IKMSSPYMNNLIILGGMLSYASIFLFGLDGSFVSEKTFETLCTVRTWILTVGYTTAFGAMFAKTWRVHAIFKNVKMKKKIIKDQKLLVIV GGMLLIDLCILICWQAVDPLRRTVEKYSMEPDPAGRDISIRPLLEHCENTHMTIWLGIVYAYKGLLMLFGCFLAWETRNVSIPALNDSKY IGMSVYNVGIMCIIGAAVSFLTRDQPNVQFCIVALVIIFCSTITLCLVFVPKLITLRTNPDAATQNRRFQFTQNQKKEDSKTSTSVTSVN QASTSRLEGLQSENHRLRMKITELDKDLEEVTMQLQDTPEKTTYIKQNHYQELNDILNLGNFTESTDGGKAILKNHLDQNPQLQWNTTEP -------------------------------------------------------------- >89350_89350_2_TBC1D2-GABBR2_TBC1D2_chr9_100991234_ENST00000375066_GABBR2_chr9_101258796_ENST00000259455_length(transcript)=5741nt_BP=1070nt CTCCCTTTGGGAAGCTGCCCGCCGAGTCTCCGAGATTTGTCCCTGGTGGTCCCGCGGACCCCTCGTCCCTCCGCAGTCTCCGGCTGGCAG CGATGGAGGGCGCTGGGGAGAACGCCCCGGAGTCCAGCTCCTCTGCCCCTGGGTCCGAAGAGTCTGCCAGGGATCCACAGGTGCCGCCTC CGGAGGAAGAATCGGGGGACTGCGCCCGGTCCCTGGAGGCGGTCCCCAAGAAACTCTGTGGGTATTTAAGTAAGTTCGGCGGCAAAGGGC CCATCCGGGGCTGGAAATCCCGCTGGTTCTTCTACGACGAAAGGAAATGTCAGCTGTATTACTCGCGGACCGCTCAGGATGCCAATCCCT TGGACAGCATCGACCTCTCCAGTGCAGTGTTTGACTGTAAGGCGGACGCTGAGGAGGGGATCTTCGAAATCAAGACTCCCAGCCGGGTTA TTACCCTGAAGGCCGCCACCAAGCAAGCGATGCTGTACTGGCTGCAGCAGCTGCAGATGAAGCGCTGGGAATTCCACAACAGCCCGCCGG CACCTCCTGCCACCCCTGATGCCGCCCTGGCTGGGAATGGGCCCGTCCTGCACCTCGAGCTAGGGCAAGAAGAGGCAGAGCTGGAGGAGT TCCTGTGCCCTGTGAAAACACCCCCTGGGCTAGTGGGCGTGGCAGCTGCCTTGCAGCCCTTCCCTGCCCTTCAGAATATTTCCCTCAAGC ACCTGGGGACTGAAATACAGAACACAATGCACAACATCCGTGGCAACAAGCAGGCCCAGGGAACAGGCCATGAACCTCCAGGGGAAGATT CTCCACAGAGTGGGGAGCCTCAGAGGGAGGAGCAGCCCTTGGCCTCTGACGCCAGCACCCCAGGGAGAGAGCCAGAGGATTCTCCAAAGC CTGCACCCAAGCCTTCTCTGACCATCAGTTTCGCTCAGAAAGCCAAGCGCCAGAACAACACCTTCCCATTCTTTTCTGAAGGAATCACAC GGAACCGAACTGCCCAGGAGAAAGTGGCAGCCTTGGAGCAACAGGTTCTGATGCTCACCAAGGAGTTAAAGTCTCAGAAGGTGCGGAATG ACCTGACTGGAGTTCTGTATGGCGAGGACATTGAGATTTCAGACACCGAGAGCTTCTCCAACGATCCCTGTACCAGTGTCAAAAAGCTGA AGGGGAATGATGTGCGGATCATCCTTGGCCAGTTTGACCAGAATATGGCAGCAAAAGTGTTCTGTTGTGCATACGAGGAGAACATGTATG GTAGTAAATATCAGTGGATCATTCCGGGCTGGTACGAGCCTTCTTGGTGGGAGCAGGTGCACACGGAAGCCAACTCATCCCGCTGCCTCC GGAAGAATCTGCTTGCTGCCATGGAGGGCTACATTGGCGTGGATTTCGAGCCCCTGAGCTCCAAGCAGATCAAGACCATCTCAGGAAAGA CTCCACAGCAGTATGAGAGAGAGTACAACAACAAGCGGTCAGGCGTGGGGCCCAGCAAGTTCCACGGGTACGCCTACGATGGCATCTGGG TCATCGCCAAGACACTGCAGAGGGCCATGGAGACACTGCATGCCAGCAGCCGGCACCAGCGGATCCAGGACTTCAACTACACGGACCACA CGCTGGGCAGGATCATCCTCAATGCCATGAACGAGACCAACTTCTTCGGGGTCACGGGTCAAGTTGTATTCCGGAATGGGGAGAGAATGG GGACCATTAAATTTACTCAATTTCAAGACAGCAGGGAGGTGAAGGTGGGAGAGTACAACGCTGTGGCCGACACACTGGAGATCATCAATG ACACCATCAGGTTCCAAGGATCCGAACCACCAAAAGACAAGACCATCATCCTGGAGCAGCTGCGGAAGATCTCCCTACCTCTCTACAGCA TCCTCTCTGCCCTCACCATCCTCGGGATGATCATGGCCAGTGCTTTTCTCTTCTTCAACATCAAGAACCGGAATCAGAAGCTCATAAAGA TGTCGAGTCCATACATGAACAACCTTATCATCCTTGGAGGGATGCTCTCCTATGCTTCCATATTTCTCTTTGGCCTTGATGGATCCTTTG TCTCTGAAAAGACCTTTGAAACACTTTGCACCGTCAGGACCTGGATTCTCACCGTGGGCTACACGACCGCTTTTGGGGCCATGTTTGCAA AGACCTGGAGAGTCCACGCCATCTTCAAAAATGTGAAAATGAAGAAGAAGATCATCAAGGACCAGAAACTGCTTGTGATCGTGGGGGGCA TGCTGCTGATCGACCTGTGTATCCTGATCTGCTGGCAGGCTGTGGACCCCCTGCGAAGGACAGTGGAGAAGTACAGCATGGAGCCGGACC CAGCAGGACGGGATATCTCCATCCGCCCTCTCCTGGAGCACTGTGAGAACACCCATATGACCATCTGGCTTGGCATCGTCTATGCCTACA AGGGACTTCTCATGTTGTTCGGTTGTTTCTTAGCTTGGGAGACCCGCAACGTCAGCATCCCCGCACTCAACGACAGCAAGTACATCGGGA TGAGTGTCTACAACGTGGGGATCATGTGCATCATCGGGGCCGCTGTCTCCTTCCTGACCCGGGACCAGCCCAATGTGCAGTTCTGCATCG TGGCTCTGGTCATCATCTTCTGCAGCACCATCACCCTCTGCCTGGTATTCGTGCCGAAGCTCATCACCCTGAGAACAAACCCAGATGCAG CAACGCAGAACAGGCGATTCCAGTTCACTCAGAATCAGAAGAAAGAAGATTCTAAAACGTCCACCTCGGTCACCAGTGTGAACCAAGCCA GCACATCCCGCCTGGAGGGCCTACAGTCAGAAAACCATCGCCTGCGAATGAAGATCACAGAGCTGGATAAAGACTTGGAAGAGGTCACCA TGCAGCTGCAGGACACACCAGAAAAGACCACCTACATTAAACAGAACCACTACCAAGAGCTCAATGACATCCTCAACCTGGGAAACTTCA CTGAGAGCACAGATGGAGGAAAGGCCATTTTAAAAAATCACCTCGATCAAAATCCCCAGCTACAGTGGAACACAACAGAGCCCTCTCGAA CATGCAAAGATCCTATAGAAGATATAAACTCTCCAGAACACATCCAGCGTCGGCTGTCCCTCCAGCTCCCCATCCTCCACCACGCCTACC TCCCATCCATCGGAGGCGTGGACGCCAGCTGTGTCAGCCCCTGCGTCAGCCCCACCGCCAGCCCCCGCCACAGACATGTGCCACCCTCCT TCCGAGTCATGGTCTCGGGCCTGTAAGGGTGGGAGGCCTGGGCCCGGGGCCTCCCCCGTGACAGAACCACACTGGGCAGAGGGGTCTGCT GCAGAAACACTGTCGGCTCTGGCTGCGGAGAAGCTGGGCACCATGGCTGGCCTCTCAGGACCACTCGGATGGCACTCAGGTGGACAGGAC GGGGCAGGGGGAGACTTGGCACCTGACCTCGAGCCTTATTTGTGAAGTCCTTATTTCTTCACAAAGAAGAGGAACGGAAATGGGACGTCT TCCTTAACATCTGCAAACAAGGAGGCGCTGGGATATCAAACTTGCAAAAAAAAAAAAAAAAAAAAAAAAACAAAAAAACTAGACAAGGAG AGAGGCACTAGAACTCCAGCTGGAAGTCACGGAGTGGCTCGAGCAGCCTTGGGAAGAGGCAAGGAGCTTCTGAAGAAACTGCCTCTGCAC ACACATCACTGGCTGTGACCCCTCAGGCTAGCCCTTCTCCACTCTGGGGGAGGAGGTGGGAAGGGCCACCAGGCCCCCAGCTGCCAGGCC AGCTGACCCCAGCCTTCCTGGAACAGGGAGTCTGCAGGAGCGCAGACAGGCACAGCCCTGGAGCAGGCAGGCCGAGGGCTGCGGCACTGG AGCAGGCTGACTTACATGCTCCACATGGGACCTGTGTCACCCAATGAGATGTTTGTTACTCTGGTAAATGCCACACGTTAACACAATAAC ACCCATTCCTGGGACCGTGGGGATTTAGGGCACGTCACTGCAGACACGCTCTGCAGCATTCACCGACAGTCTGTCATGCACCCACCACGT TGGCCATGTCCTTGTGTTCCTATCGGATGCTCCCAGTAACCAGGGGGACCACCCGAGCTAATCATGGAATGTCTGTTCCCAGCAAACACG ATAAAGAAAGATTGTGCACTTTAACCTCTCTCATCAGGGCCCAAGGGCTGGCTGGGATTTTTTTTTTTTTTTTCCCACTAACTTTGTTTC TGACCAAAGTGAATTGGAGGCACTCTGCTAAAAGACATCCCCGTAGACATAGGGGAGAGAGTTGCTGGCTGAGGGCTTCCCTTGGCTTCC AGAAGGCAGCCTTCCATCCAGACAAGCCAGTGAGCTCTCCCCTTGGGATCACTGGGGTGATCAGTCAGCAGATTGATTCTCATTCATAAG ATCATTCCTCCCTTTAAATTGAGCCCCTAAGAGCACTGGCCTGGGAGTCAGACAGACCTGGGTTCAAGTCCTCAGTCCCCTGCCCACTCC CTACGTGACTTTGATCAGGTCACTAGTGTCTCTCTGAGCCTCAGTTTCCCCTCTGTAACTTGGGGTTGAACTAAAACACCTGTCCTGCCT ACCTCACAAGGTCACTCTGAGGATTGAAACTTGATCTTGTCCAGGAAAGCTTTGTACCAAACAGTGAAGCCGCCCTGATCCGTGAGGTAT GAGTATGACTCTGACCTTCAGCCCTCCCTACAGCCGGGGGTGTGGCCCAGAGAAGCTTCCAGCACAGCCCTCTACCCAGAACATCCGGGC TGGAGGGAGGCTCCCAGTGACTTTTCTGACATTCCTAGACAGGTTCATTCTTTGCTCAAGAAAGGCCTGAATGACAATGTCCAGGATGTC TGCACAACTGAGCAGCTCGCTCACTCCCTAAAGAAACCTATTGGCAGCTTCAACAGGCAGGCAATAATCTCTTCCCAGAACCACTGCAGT CAGGAATAAACTGTTTTCTCCACCAGGCTTTGACAAAAGGGCCCACAGGAATCTTACCAATGCCAACATTTCAAAGCACCCTATTTCACG TAGCATAGCTTTCTGCTCCCCTTCCCCAAAGAGAGGTTATGGAGGTACTGTAGCTTTTAGGGAAAAAAAAATGTTAACACATCACAGGTC AAGTTGAAGTCATTCTCTGTTTAGGCACTAAAAATCGGTGTTGTCACTCACTGTGTATTACCAGTATTTACTTGCTTTCTTGATTTCACC AAAACCAAATTTAATTTAAAGGACCACATTAATTTTTCAAAGGGAAAGAGACAATTAATTGTACATAATGTATACACACACACAAAAAAA AATACCTGTAGAAATATTATTCCAGCATAGCAGGAAAACAAACAAAAGTATTGGACTGTCGGAGGTGAGCCTGTGCGTCTGTAACCCTTT GTGACTCCTGAGCGTGCGCTGTCTTCTAGGTTAACTCACGAAGTACATTCTCTGTCTTACTGATACTGTAGGTTCACCCATTTTTTTTTA ATTTCCTCGCAAATAACAAGACCCACAGAAGTGACTCTAGCTACTTAATGGTTCTGTTCTTTTATATGCAGCAAACACACCGTCCATTTC TGAAGAGGCTTCGGCCTGAAGGCATTTTCCAATGATGTTAGTGCACAAAACGCTTTAAATTAGACTGGAACTGCCAGAATCAAATGTAAA >89350_89350_2_TBC1D2-GABBR2_TBC1D2_chr9_100991234_ENST00000375066_GABBR2_chr9_101258796_ENST00000259455_length(amino acids)=1083AA_BP=352 MPAESPRFVPGGPADPSSLRSLRLAAMEGAGENAPESSSSAPGSEESARDPQVPPPEEESGDCARSLEAVPKKLCGYLSKFGGKGPIRGW KSRWFFYDERKCQLYYSRTAQDANPLDSIDLSSAVFDCKADAEEGIFEIKTPSRVITLKAATKQAMLYWLQQLQMKRWEFHNSPPAPPAT PDAALAGNGPVLHLELGQEEAELEEFLCPVKTPPGLVGVAAALQPFPALQNISLKHLGTEIQNTMHNIRGNKQAQGTGHEPPGEDSPQSG EPQREEQPLASDASTPGREPEDSPKPAPKPSLTISFAQKAKRQNNTFPFFSEGITRNRTAQEKVAALEQQVLMLTKELKSQKVRNDLTGV LYGEDIEISDTESFSNDPCTSVKKLKGNDVRIILGQFDQNMAAKVFCCAYEENMYGSKYQWIIPGWYEPSWWEQVHTEANSSRCLRKNLL AAMEGYIGVDFEPLSSKQIKTISGKTPQQYEREYNNKRSGVGPSKFHGYAYDGIWVIAKTLQRAMETLHASSRHQRIQDFNYTDHTLGRI ILNAMNETNFFGVTGQVVFRNGERMGTIKFTQFQDSREVKVGEYNAVADTLEIINDTIRFQGSEPPKDKTIILEQLRKISLPLYSILSAL TILGMIMASAFLFFNIKNRNQKLIKMSSPYMNNLIILGGMLSYASIFLFGLDGSFVSEKTFETLCTVRTWILTVGYTTAFGAMFAKTWRV HAIFKNVKMKKKIIKDQKLLVIVGGMLLIDLCILICWQAVDPLRRTVEKYSMEPDPAGRDISIRPLLEHCENTHMTIWLGIVYAYKGLLM LFGCFLAWETRNVSIPALNDSKYIGMSVYNVGIMCIIGAAVSFLTRDQPNVQFCIVALVIIFCSTITLCLVFVPKLITLRTNPDAATQNR RFQFTQNQKKEDSKTSTSVTSVNQASTSRLEGLQSENHRLRMKITELDKDLEEVTMQLQDTPEKTTYIKQNHYQELNDILNLGNFTESTD GGKAILKNHLDQNPQLQWNTTEPSRTCKDPIEDINSPEHIQRRLSLQLPILHHAYLPSIGGVDASCVSPCVSPTASPRHRHVPPSFRVMV -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TBC1D2-GABBR2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 1_169 | 326.0 | 861.0 | CADH1 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 1_169 | 326.0 | 918.0 | CADH1 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 1_169 | 108.0 | 711.0 | CADH1 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 1_169 | 0 | 469.0 | CADH1 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000342112 | - | 4 | 12 | 295_433 | 108.0 | 711.0 | RAC1 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375063 | - | 1 | 7 | 295_433 | 0 | 469.0 | RAC1 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375064 | - | 5 | 13 | 295_433 | 326.0 | 861.0 | RAC1 |
| Hgene | TBC1D2 | chr9:100991234 | chr9:101258796 | ENST00000375066 | - | 5 | 13 | 295_433 | 326.0 | 918.0 | RAC1 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TBC1D2-GABBR2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TBC1D2-GABBR2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
