
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TCTN1-GORASP2 (FusionGDB2 ID:89898) |
Fusion Gene Summary for TCTN1-GORASP2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TCTN1-GORASP2 | Fusion gene ID: 89898 | Hgene | Tgene | Gene symbol | TCTN1 | GORASP2 | Gene ID | 79600 | 26003 |
| Gene name | tectonic family member 1 | golgi reassembly stacking protein 2 | |
| Synonyms | JBTS13|TECT1 | GOLPH6|GRASP55|GRS2|p59 | |
| Cytomap | 12q24.11 | 2q31.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tectonic-1 | Golgi reassembly-stacking protein 2golgi phosphoprotein 6golgi reassembly stacking protein 2, 55kDa | |
| Modification date | 20200313 | 20200327 | |
| UniProtAcc | . | Q9H8Y8 | |
| Ensembl transtripts involved in fusion gene | ENST00000551555, ENST00000377654, ENST00000397655, ENST00000397659, ENST00000550703, ENST00000551590, ENST00000471804, | ENST00000234160, ENST00000452526, ENST00000493692, | |
| Fusion gene scores | * DoF score | 4 X 4 X 3=48 | 9 X 6 X 3=162 |
| # samples | 4 | 7 | |
| ** MAII score | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/162*10)=-1.21056698593966 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TCTN1 [Title/Abstract] AND GORASP2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TCTN1(111066723)-GORASP2(171806048), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TCTN1-GORASP2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | GORASP2 | GO:0034976 | response to endoplasmic reticulum stress | 21884936|27062250|28067262 |
| Tgene | GORASP2 | GO:0061951 | establishment of protein localization to plasma membrane | 28067262 |
 Fusion gene breakpoints across TCTN1 (5'-gene) Fusion gene breakpoints across TCTN1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
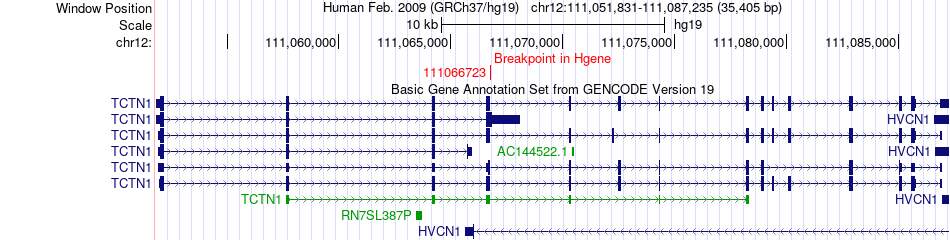 |
 Fusion gene breakpoints across GORASP2 (3'-gene) Fusion gene breakpoints across GORASP2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
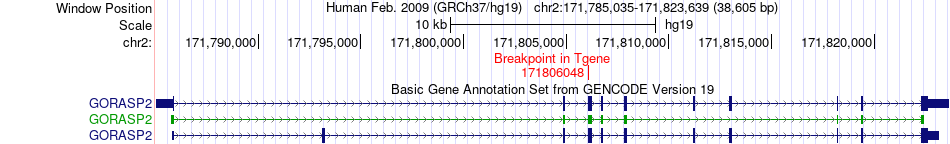 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | ESCA | TCGA-LN-A4A4 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
Top |
Fusion Gene ORF analysis for TCTN1-GORASP2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000551555 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 3UTR-3UTR | ENST00000551555 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 3UTR-3UTR | ENST00000551555 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000377654 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000377654 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000397655 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000397655 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000397659 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000397659 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000550703 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000550703 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000551590 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| 5CDS-3UTR | ENST00000551590 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| Frame-shift | ENST00000550703 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| In-frame | ENST00000377654 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| In-frame | ENST00000397655 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| In-frame | ENST00000397659 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| In-frame | ENST00000551590 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| intron-3CDS | ENST00000471804 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| intron-3UTR | ENST00000471804 | ENST00000452526 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
| intron-3UTR | ENST00000471804 | ENST00000493692 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000551590 | TCTN1 | chr12 | 111066723 | + | ENST00000234160 | GORASP2 | chr2 | 171806048 | + | 2994 | 780 | 9 | 1994 | 661 |
| ENST00000397655 | TCTN1 | chr12 | 111066723 | + | ENST00000234160 | GORASP2 | chr2 | 171806048 | + | 2908 | 694 | 4 | 1908 | 634 |
| ENST00000377654 | TCTN1 | chr12 | 111066723 | + | ENST00000234160 | GORASP2 | chr2 | 171806048 | + | 2884 | 670 | 46 | 1884 | 612 |
| ENST00000397659 | TCTN1 | chr12 | 111066723 | + | ENST00000234160 | GORASP2 | chr2 | 171806048 | + | 2857 | 643 | 19 | 1857 | 612 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000551590 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + | 0.001132514 | 0.9988675 |
| ENST00000397655 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + | 0.000937357 | 0.9990626 |
| ENST00000377654 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + | 0.000930346 | 0.99906963 |
| ENST00000397659 | ENST00000234160 | TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + | 0.000962418 | 0.9990376 |
Top |
Fusion Genomic Features for TCTN1-GORASP2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + | 1.20E-06 | 0.9999988 |
| TCTN1 | chr12 | 111066723 | + | GORASP2 | chr2 | 171806048 | + | 1.20E-06 | 0.9999988 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for TCTN1-GORASP2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:111066723/chr2:171806048) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | GORASP2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Plays a role in the assembly and membrane stacking of the Golgi cisternae, and in the process by which Golgi stacks reform after breakdown during mitosis and meiosis (PubMed:10487747, PubMed:21515684, PubMed:22523075). May regulate the intracellular transport and presentation of a defined set of transmembrane proteins, such as transmembrane TGFA (PubMed:11101516). Required for normal acrosome formation during spermiogenesis and normal male fertility, probably by promoting colocalization of JAM2 and JAM3 at contact sites between germ cells and Sertoli cells (By similarity). Mediates ER stress-induced unconventional (ER/Golgi-independent) trafficking of core-glycosylated CFTR to cell membrane (PubMed:21884936, PubMed:27062250, PubMed:28067262). {ECO:0000250|UniProtKB:Q99JX3, ECO:0000269|PubMed:10487747, ECO:0000269|PubMed:11101516, ECO:0000269|PubMed:21515684, ECO:0000269|PubMed:21884936, ECO:0000269|PubMed:22523075, ECO:0000269|PubMed:27062250, ECO:0000269|PubMed:28067262}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TCTN1 | chr12:111066723 | chr2:171806048 | ENST00000397655 | + | 4 | 15 | 78_97 | 208 | 506.0 | Compositional bias | Note=Cys-rich |
| Hgene | TCTN1 | chr12:111066723 | chr2:171806048 | ENST00000397659 | + | 4 | 15 | 78_97 | 208 | 542.0 | Compositional bias | Note=Cys-rich |
| Hgene | TCTN1 | chr12:111066723 | chr2:171806048 | ENST00000551590 | + | 4 | 15 | 78_97 | 208 | 592.0 | Compositional bias | Note=Cys-rich |
| Tgene | GORASP2 | chr12:111066723 | chr2:171806048 | ENST00000234160 | 1 | 10 | 277_372 | 48 | 453.0 | Compositional bias | Note=Pro-rich | |
| Tgene | GORASP2 | chr12:111066723 | chr2:171806048 | ENST00000234160 | 1 | 10 | 111_199 | 48 | 453.0 | Domain | PDZ GRASP-type 2 | |
| Tgene | GORASP2 | chr12:111066723 | chr2:171806048 | ENST00000234160 | 1 | 10 | 194_199 | 48 | 453.0 | Region | Note=Important for membrane binding |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TCTN1 | chr12:111066723 | chr2:171806048 | ENST00000377654 | + | 4 | 13 | 78_97 | 30 | 208.0 | Compositional bias | Note=Cys-rich |
| Tgene | GORASP2 | chr12:111066723 | chr2:171806048 | ENST00000234160 | 1 | 10 | 15_105 | 48 | 453.0 | Domain | PDZ GRASP-type 1 | |
| Tgene | GORASP2 | chr12:111066723 | chr2:171806048 | ENST00000234160 | 1 | 10 | 15_215 | 48 | 453.0 | Region | GRASP |
Top |
Fusion Gene Sequence for TCTN1-GORASP2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >89898_89898_1_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000377654_GORASP2_chr2_171806048_ENST00000234160_length(transcript)=2884nt_BP=670nt GCAACGCGCTGTCCATGTCGCGGGCCTCGCTGGGACTCCCTGGGAGATGAGGCCGCGAGGTCTCCCGCCGCTCCTGGTGGTGCTCCTGGG CTGCTGGGCCTCCGTGAGCGCCCAGACCGATGCCACCCCGGCGGTGACGACAGAGGGCCTCAACTCCACCGAGGCAGCCCTGGCCACCTT CGGAACTTTCCCGTCGACCAGGCCCCCCGGGACTCCCAGGGCTCCAGGGCCCTCCTCCGGCCCCAGGCCTACCCCAGTCACGGACGTTGC TGTTCTCTGTGTCTGTGACTTATCCCCAGCACAGTGTGACATCAACTGCTGCTGTGATCCCGACTGCAGCTCCGTGGATTTCAGTGTCTT TTCTGCCTGCTCAGTTCCAGTTGTCACGGGCGACAGCCAGTTTTGTAGTCAAAAAGCAGTCATCTATTCATTGAATTTTACAGCAAACCC ACCTCAAAGAGTATTTGAACTTGTTGACCAGATTAATCCATCTATTTTCTGCATTCATATTACAAACTATAAACCTGCATTATCCTTTAT TAATCCAGAAGTACCTGATGAAAACAATTTTGATACATTGATGAAAACATCTGATGGTTTTACATTGAATGCTGAATCATATGTTTCCTT CACAACCAAACTGGATATTCCTACTGCTGCTAAATATGAGAATAAAGACAATGACACTCTTAAGGATCTGCTGAAAGCAAACGTTGAAAA GCCTGTAAAGATGCTTATCTATAGCAGCAAAACATTGGAACTGCGAGAGACCTCAGTCACACCAAGTAACCTGTGGGGCGGCCAGGGCTT ATTGGGAGTGAGCATTCGTTTCTGCAGCTTTGATGGGGCAAATGAAAATGTTTGGCATGTGCTGGAGGTGGAATCAAATTCTCCTGCAGC ACTGGCAGGTCTTAGACCACACAGTGATTATATAATTGGAGCAGATACAGTCATGAATGAGTCTGAAGATCTATTCAGCCTTATCGAAAC ACATGAAGCAAAACCATTGAAACTGTATGTGTACAACACAGACACTGATAACTGTCGAGAAGTGATTATTACACCAAATTCTGCATGGGG TGGAGAAGGCAGCCTAGGATGTGGCATTGGATATGGTTATTTGCATCGAATACCTACACGCCCATTTGAGGAAGGAAAGAAAATTTCTCT TCCAGGACAAATGGCTGGTACACCTATTACACCTCTTAAAGATGGGTTTACAGAGGTCCAGCTGTCCTCAGTTAATCCCCCGTCTTTGTC ACCACCAGGAACTACAGGAATTGAACAGAGTCTGACTGGACTTTCTATTAGCTCAACTCCACCAGCTGTCAGTAGTGTTCTCAGTACAGG TGTACCAACAGTACCGTTATTGCCACCACAAGTAAACCAGTCCCTCACTTCTGTGCCACCAATGAATCCAGCTACTACATTACCAGGTCT GATGCCTTTACCAGCAGGACTGCCCAACCTCCCCAACCTCAACCTCAACCTCCCAGCACCACACATCATGCCAGGGGTTGGCTTACCAGA ACTTGTAAACCCAGGTCTGCCACCTCTTCCTTCCATGCCTCCCCGAAACTTACCTGGCATTGCACCTCTCCCCCTGCCATCCGAGTTCCT CCCGTCATTCCCCTTGGTTCCAGAGAGCTCTTCTGCAGCAAGCTCAGGAGAGCTGCTGTCTTCCCTCCCGCCCACCAGCAACGCACCCTC CGACCCTGCCACAACTACTGCAAAGGCAGACGCTGCCTCCTCACTCACTGTGGATGTGACGCCCCCCACTGCCAAGGCCCCCACCACCGT TGAGGACAGAGTCGGCGACTCCACCCCAGTCAGCGAGAAGCCTGTTTCTGCGGCTGTGGATGCCAATGCTTCTGAGTCACCTTAACTTTG AACCATTCTTTGGAATTGGCGTGGTATATTTAACCACGGGAGCGTGTCTGGAAACGCAAACTATCATTAATTTCATACTAGTTTGTACCG TATCTGTAGGCATCCTGTAAATAATTCCAAGGGGAAAACTAAACGAGGACGTGGGTTGTATCCTGCCAGGTTGAGTGGGGCTCACACGCT AGGGTGAGATGTCAGAAAGCGCTTGTATTTTAAACAACCAAAAAGAATTGTAAGGGTGGCTTGCTGCCAGGCTTGCACTGCCGTTCCTGG GGGTGTGCATCTTCGGGAAAGGTGGTGGCGGGGCGTCCACTAGGTTTCCTGTCCCCTGCTGCTCCTTCCGTAAGAAAATGAAATATTCTA TGCCTAATACTCACACGCAACATTTCTTGTACTTTGTAAGTCGTTTGCGAGAATGCAGACCACCTCACTAAACTGTAAACGGTAAAGAGA TTTTTACTTTTGGTCTCCGTGAGTCGCATCTCTACTAAGGTTTACACAGGAATTCCACCTGAAGACTTGTGTTAAAGTTCTACAGCGCGC ACTGTTAACTGAACGTCTTTTTCTTCAGCCTATACGCGGATCCTTGTTTTGAGCTCTCAGAATCACTCAGACAACATTTTGTAACTGCTG CTGTTGCTTTCTACATACACCTTATAAAGTGACATTTCAAAAGAAATAAGGTGCCACAGTTTTAAACCAGAAGGTGGCACTCTGTGGCTC CTTGTAGTATTATAGCTATACTGGGAAAGCATAGATACAGCAATAAAGTACAGTAATTTTACTTTTTTTCTTGTGTTACATCTAAATTAC AACCCTTAATTGCCACGTGTGCACTTACTACTCTCCAGTATGTCTTATTACTCTCCAGTATGTCACGCATCTTTAACTTTTCACGTCCTA TGTTTGCTTTCTCCCATTTTTAAGAGATGGTAAGTTAACTGGAATTGATTTACTGAATGAAATTAAATGCAGATATCCCTGTTTTTGAAA >89898_89898_1_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000377654_GORASP2_chr2_171806048_ENST00000234160_length(amino acids)=612AA_BP=208 MRPRGLPPLLVVLLGCWASVSAQTDATPAVTTEGLNSTEAALATFGTFPSTRPPGTPRAPGPSSGPRPTPVTDVAVLCVCDLSPAQCDIN CCCDPDCSSVDFSVFSACSVPVVTGDSQFCSQKAVIYSLNFTANPPQRVFELVDQINPSIFCIHITNYKPALSFINPEVPDENNFDTLMK TSDGFTLNAESYVSFTTKLDIPTAAKYENKDNDTLKDLLKANVEKPVKMLIYSSKTLELRETSVTPSNLWGGQGLLGVSIRFCSFDGANE NVWHVLEVESNSPAALAGLRPHSDYIIGADTVMNESEDLFSLIETHEAKPLKLYVYNTDTDNCREVIITPNSAWGGEGSLGCGIGYGYLH RIPTRPFEEGKKISLPGQMAGTPITPLKDGFTEVQLSSVNPPSLSPPGTTGIEQSLTGLSISSTPPAVSSVLSTGVPTVPLLPPQVNQSL TSVPPMNPATTLPGLMPLPAGLPNLPNLNLNLPAPHIMPGVGLPELVNPGLPPLPSMPPRNLPGIAPLPLPSEFLPSFPLVPESSSAASS -------------------------------------------------------------- >89898_89898_2_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000397655_GORASP2_chr2_171806048_ENST00000234160_length(transcript)=2908nt_BP=694nt GCGCCTGGCTGTCGCGGTTGCCGGGCAACGCGCTGTCCATGTCGCGGGCCTCGCTGGGACTCCCTGGGAGATGAGGCCGCGAGGTCTCCC GCCGCTCCTGGTGGTGCTCCTGGGCTGCTGGGCCTCCGTGAGCGCCCAGACCGATGCCACCCCGGCGGTGACGACAGAGGGCCTCAACTC CACCGAGGCAGCCCTGGCCACCTTCGGAACTTTCCCGTCGACCAGGCCCCCCGGGACTCCCAGGGCTCCAGGGCCCTCCTCCGGCCCCAG GCCTACCCCAGTCACGGACGTTGCTGTTCTCTGTGTCTGTGACTTATCCCCAGCACAGTGTGACATCAACTGCTGCTGTGATCCCGACTG CAGCTCCGTGGATTTCAGTGTCTTTTCTGCCTGCTCAGTTCCAGTTGTCACGGGCGACAGCCAGTTTTGTAGTCAAAAAGCAGTCATCTA TTCATTGAATTTTACAGCAAACCCACCTCAAAGAGTATTTGAACTTGTTGACCAGATTAATCCATCTATTTTCTGCATTCATATTACAAA CTATAAACCTGCATTATCCTTTATTAATCCAGAAGTACCTGATGAAAACAATTTTGATACATTGATGAAAACATCTGATGGTTTTACATT GAATGCTGAATCATATGTTTCCTTCACAACCAAACTGGATATTCCTACTGCTGCTAAATATGAGAATAAAGACAATGACACTCTTAAGGA TCTGCTGAAAGCAAACGTTGAAAAGCCTGTAAAGATGCTTATCTATAGCAGCAAAACATTGGAACTGCGAGAGACCTCAGTCACACCAAG TAACCTGTGGGGCGGCCAGGGCTTATTGGGAGTGAGCATTCGTTTCTGCAGCTTTGATGGGGCAAATGAAAATGTTTGGCATGTGCTGGA GGTGGAATCAAATTCTCCTGCAGCACTGGCAGGTCTTAGACCACACAGTGATTATATAATTGGAGCAGATACAGTCATGAATGAGTCTGA AGATCTATTCAGCCTTATCGAAACACATGAAGCAAAACCATTGAAACTGTATGTGTACAACACAGACACTGATAACTGTCGAGAAGTGAT TATTACACCAAATTCTGCATGGGGTGGAGAAGGCAGCCTAGGATGTGGCATTGGATATGGTTATTTGCATCGAATACCTACACGCCCATT TGAGGAAGGAAAGAAAATTTCTCTTCCAGGACAAATGGCTGGTACACCTATTACACCTCTTAAAGATGGGTTTACAGAGGTCCAGCTGTC CTCAGTTAATCCCCCGTCTTTGTCACCACCAGGAACTACAGGAATTGAACAGAGTCTGACTGGACTTTCTATTAGCTCAACTCCACCAGC TGTCAGTAGTGTTCTCAGTACAGGTGTACCAACAGTACCGTTATTGCCACCACAAGTAAACCAGTCCCTCACTTCTGTGCCACCAATGAA TCCAGCTACTACATTACCAGGTCTGATGCCTTTACCAGCAGGACTGCCCAACCTCCCCAACCTCAACCTCAACCTCCCAGCACCACACAT CATGCCAGGGGTTGGCTTACCAGAACTTGTAAACCCAGGTCTGCCACCTCTTCCTTCCATGCCTCCCCGAAACTTACCTGGCATTGCACC TCTCCCCCTGCCATCCGAGTTCCTCCCGTCATTCCCCTTGGTTCCAGAGAGCTCTTCTGCAGCAAGCTCAGGAGAGCTGCTGTCTTCCCT CCCGCCCACCAGCAACGCACCCTCCGACCCTGCCACAACTACTGCAAAGGCAGACGCTGCCTCCTCACTCACTGTGGATGTGACGCCCCC CACTGCCAAGGCCCCCACCACCGTTGAGGACAGAGTCGGCGACTCCACCCCAGTCAGCGAGAAGCCTGTTTCTGCGGCTGTGGATGCCAA TGCTTCTGAGTCACCTTAACTTTGAACCATTCTTTGGAATTGGCGTGGTATATTTAACCACGGGAGCGTGTCTGGAAACGCAAACTATCA TTAATTTCATACTAGTTTGTACCGTATCTGTAGGCATCCTGTAAATAATTCCAAGGGGAAAACTAAACGAGGACGTGGGTTGTATCCTGC CAGGTTGAGTGGGGCTCACACGCTAGGGTGAGATGTCAGAAAGCGCTTGTATTTTAAACAACCAAAAAGAATTGTAAGGGTGGCTTGCTG CCAGGCTTGCACTGCCGTTCCTGGGGGTGTGCATCTTCGGGAAAGGTGGTGGCGGGGCGTCCACTAGGTTTCCTGTCCCCTGCTGCTCCT TCCGTAAGAAAATGAAATATTCTATGCCTAATACTCACACGCAACATTTCTTGTACTTTGTAAGTCGTTTGCGAGAATGCAGACCACCTC ACTAAACTGTAAACGGTAAAGAGATTTTTACTTTTGGTCTCCGTGAGTCGCATCTCTACTAAGGTTTACACAGGAATTCCACCTGAAGAC TTGTGTTAAAGTTCTACAGCGCGCACTGTTAACTGAACGTCTTTTTCTTCAGCCTATACGCGGATCCTTGTTTTGAGCTCTCAGAATCAC TCAGACAACATTTTGTAACTGCTGCTGTTGCTTTCTACATACACCTTATAAAGTGACATTTCAAAAGAAATAAGGTGCCACAGTTTTAAA CCAGAAGGTGGCACTCTGTGGCTCCTTGTAGTATTATAGCTATACTGGGAAAGCATAGATACAGCAATAAAGTACAGTAATTTTACTTTT TTTCTTGTGTTACATCTAAATTACAACCCTTAATTGCCACGTGTGCACTTACTACTCTCCAGTATGTCTTATTACTCTCCAGTATGTCAC GCATCTTTAACTTTTCACGTCCTATGTTTGCTTTCTCCCATTTTTAAGAGATGGTAAGTTAACTGGAATTGATTTACTGAATGAAATTAA >89898_89898_2_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000397655_GORASP2_chr2_171806048_ENST00000234160_length(amino acids)=634AA_BP=230 MAVAVAGQRAVHVAGLAGTPWEMRPRGLPPLLVVLLGCWASVSAQTDATPAVTTEGLNSTEAALATFGTFPSTRPPGTPRAPGPSSGPRP TPVTDVAVLCVCDLSPAQCDINCCCDPDCSSVDFSVFSACSVPVVTGDSQFCSQKAVIYSLNFTANPPQRVFELVDQINPSIFCIHITNY KPALSFINPEVPDENNFDTLMKTSDGFTLNAESYVSFTTKLDIPTAAKYENKDNDTLKDLLKANVEKPVKMLIYSSKTLELRETSVTPSN LWGGQGLLGVSIRFCSFDGANENVWHVLEVESNSPAALAGLRPHSDYIIGADTVMNESEDLFSLIETHEAKPLKLYVYNTDTDNCREVII TPNSAWGGEGSLGCGIGYGYLHRIPTRPFEEGKKISLPGQMAGTPITPLKDGFTEVQLSSVNPPSLSPPGTTGIEQSLTGLSISSTPPAV SSVLSTGVPTVPLLPPQVNQSLTSVPPMNPATTLPGLMPLPAGLPNLPNLNLNLPAPHIMPGVGLPELVNPGLPPLPSMPPRNLPGIAPL PLPSEFLPSFPLVPESSSAASSGELLSSLPPTSNAPSDPATTTAKADAASSLTVDVTPPTAKAPTTVEDRVGDSTPVSEKPVSAAVDANA -------------------------------------------------------------- >89898_89898_3_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000397659_GORASP2_chr2_171806048_ENST00000234160_length(transcript)=2857nt_BP=643nt CGCTGGGACTCCCTGGGAGATGAGGCCGCGAGGTCTCCCGCCGCTCCTGGTGGTGCTCCTGGGCTGCTGGGCCTCCGTGAGCGCCCAGAC CGATGCCACCCCGGCGGTGACGACAGAGGGCCTCAACTCCACCGAGGCAGCCCTGGCCACCTTCGGAACTTTCCCGTCGACCAGGCCCCC CGGGACTCCCAGGGCTCCAGGGCCCTCCTCCGGCCCCAGGCCTACCCCAGTCACGGACGTTGCTGTTCTCTGTGTCTGTGACTTATCCCC AGCACAGTGTGACATCAACTGCTGCTGTGATCCCGACTGCAGCTCCGTGGATTTCAGTGTCTTTTCTGCCTGCTCAGTTCCAGTTGTCAC GGGCGACAGCCAGTTTTGTAGTCAAAAAGCAGTCATCTATTCATTGAATTTTACAGCAAACCCACCTCAAAGAGTATTTGAACTTGTTGA CCAGATTAATCCATCTATTTTCTGCATTCATATTACAAACTATAAACCTGCATTATCCTTTATTAATCCAGAAGTACCTGATGAAAACAA TTTTGATACATTGATGAAAACATCTGATGGTTTTACATTGAATGCTGAATCATATGTTTCCTTCACAACCAAACTGGATATTCCTACTGC TGCTAAATATGAGAATAAAGACAATGACACTCTTAAGGATCTGCTGAAAGCAAACGTTGAAAAGCCTGTAAAGATGCTTATCTATAGCAG CAAAACATTGGAACTGCGAGAGACCTCAGTCACACCAAGTAACCTGTGGGGCGGCCAGGGCTTATTGGGAGTGAGCATTCGTTTCTGCAG CTTTGATGGGGCAAATGAAAATGTTTGGCATGTGCTGGAGGTGGAATCAAATTCTCCTGCAGCACTGGCAGGTCTTAGACCACACAGTGA TTATATAATTGGAGCAGATACAGTCATGAATGAGTCTGAAGATCTATTCAGCCTTATCGAAACACATGAAGCAAAACCATTGAAACTGTA TGTGTACAACACAGACACTGATAACTGTCGAGAAGTGATTATTACACCAAATTCTGCATGGGGTGGAGAAGGCAGCCTAGGATGTGGCAT TGGATATGGTTATTTGCATCGAATACCTACACGCCCATTTGAGGAAGGAAAGAAAATTTCTCTTCCAGGACAAATGGCTGGTACACCTAT TACACCTCTTAAAGATGGGTTTACAGAGGTCCAGCTGTCCTCAGTTAATCCCCCGTCTTTGTCACCACCAGGAACTACAGGAATTGAACA GAGTCTGACTGGACTTTCTATTAGCTCAACTCCACCAGCTGTCAGTAGTGTTCTCAGTACAGGTGTACCAACAGTACCGTTATTGCCACC ACAAGTAAACCAGTCCCTCACTTCTGTGCCACCAATGAATCCAGCTACTACATTACCAGGTCTGATGCCTTTACCAGCAGGACTGCCCAA CCTCCCCAACCTCAACCTCAACCTCCCAGCACCACACATCATGCCAGGGGTTGGCTTACCAGAACTTGTAAACCCAGGTCTGCCACCTCT TCCTTCCATGCCTCCCCGAAACTTACCTGGCATTGCACCTCTCCCCCTGCCATCCGAGTTCCTCCCGTCATTCCCCTTGGTTCCAGAGAG CTCTTCTGCAGCAAGCTCAGGAGAGCTGCTGTCTTCCCTCCCGCCCACCAGCAACGCACCCTCCGACCCTGCCACAACTACTGCAAAGGC AGACGCTGCCTCCTCACTCACTGTGGATGTGACGCCCCCCACTGCCAAGGCCCCCACCACCGTTGAGGACAGAGTCGGCGACTCCACCCC AGTCAGCGAGAAGCCTGTTTCTGCGGCTGTGGATGCCAATGCTTCTGAGTCACCTTAACTTTGAACCATTCTTTGGAATTGGCGTGGTAT ATTTAACCACGGGAGCGTGTCTGGAAACGCAAACTATCATTAATTTCATACTAGTTTGTACCGTATCTGTAGGCATCCTGTAAATAATTC CAAGGGGAAAACTAAACGAGGACGTGGGTTGTATCCTGCCAGGTTGAGTGGGGCTCACACGCTAGGGTGAGATGTCAGAAAGCGCTTGTA TTTTAAACAACCAAAAAGAATTGTAAGGGTGGCTTGCTGCCAGGCTTGCACTGCCGTTCCTGGGGGTGTGCATCTTCGGGAAAGGTGGTG GCGGGGCGTCCACTAGGTTTCCTGTCCCCTGCTGCTCCTTCCGTAAGAAAATGAAATATTCTATGCCTAATACTCACACGCAACATTTCT TGTACTTTGTAAGTCGTTTGCGAGAATGCAGACCACCTCACTAAACTGTAAACGGTAAAGAGATTTTTACTTTTGGTCTCCGTGAGTCGC ATCTCTACTAAGGTTTACACAGGAATTCCACCTGAAGACTTGTGTTAAAGTTCTACAGCGCGCACTGTTAACTGAACGTCTTTTTCTTCA GCCTATACGCGGATCCTTGTTTTGAGCTCTCAGAATCACTCAGACAACATTTTGTAACTGCTGCTGTTGCTTTCTACATACACCTTATAA AGTGACATTTCAAAAGAAATAAGGTGCCACAGTTTTAAACCAGAAGGTGGCACTCTGTGGCTCCTTGTAGTATTATAGCTATACTGGGAA AGCATAGATACAGCAATAAAGTACAGTAATTTTACTTTTTTTCTTGTGTTACATCTAAATTACAACCCTTAATTGCCACGTGTGCACTTA CTACTCTCCAGTATGTCTTATTACTCTCCAGTATGTCACGCATCTTTAACTTTTCACGTCCTATGTTTGCTTTCTCCCATTTTTAAGAGA >89898_89898_3_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000397659_GORASP2_chr2_171806048_ENST00000234160_length(amino acids)=612AA_BP=208 MRPRGLPPLLVVLLGCWASVSAQTDATPAVTTEGLNSTEAALATFGTFPSTRPPGTPRAPGPSSGPRPTPVTDVAVLCVCDLSPAQCDIN CCCDPDCSSVDFSVFSACSVPVVTGDSQFCSQKAVIYSLNFTANPPQRVFELVDQINPSIFCIHITNYKPALSFINPEVPDENNFDTLMK TSDGFTLNAESYVSFTTKLDIPTAAKYENKDNDTLKDLLKANVEKPVKMLIYSSKTLELRETSVTPSNLWGGQGLLGVSIRFCSFDGANE NVWHVLEVESNSPAALAGLRPHSDYIIGADTVMNESEDLFSLIETHEAKPLKLYVYNTDTDNCREVIITPNSAWGGEGSLGCGIGYGYLH RIPTRPFEEGKKISLPGQMAGTPITPLKDGFTEVQLSSVNPPSLSPPGTTGIEQSLTGLSISSTPPAVSSVLSTGVPTVPLLPPQVNQSL TSVPPMNPATTLPGLMPLPAGLPNLPNLNLNLPAPHIMPGVGLPELVNPGLPPLPSMPPRNLPGIAPLPLPSEFLPSFPLVPESSSAASS -------------------------------------------------------------- >89898_89898_4_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000551590_GORASP2_chr2_171806048_ENST00000234160_length(transcript)=2994nt_BP=780nt ATCCGCCCCCTGCCTCATCCAGGCTTCGCCCCAGTCAGTCTCCGGCAGCTTCTCGCGGCTTCGCAAGCCCCTTCCCGTGATGCCCCGCGC CTGGCTGTCGCGGTTGCCGGGCAACGCGCTGTCCATGTCGCGGGCCTCGCTGGGACTCCCTGGGAGATGAGGCCGCGAGGTCTCCCGCCG CTCCTGGTGGTGCTCCTGGGCTGCTGGGCCTCCGTGAGCGCCCAGACCGATGCCACCCCGGCGGTGACGACAGAGGGCCTCAACTCCACC GAGGCAGCCCTGGCCACCTTCGGAACTTTCCCGTCGACCAGGCCCCCCGGGACTCCCAGGGCTCCAGGGCCCTCCTCCGGCCCCAGGCCT ACCCCAGTCACGGACGTTGCTGTTCTCTGTGTCTGTGACTTATCCCCAGCACAGTGTGACATCAACTGCTGCTGTGATCCCGACTGCAGC TCCGTGGATTTCAGTGTCTTTTCTGCCTGCTCAGTTCCAGTTGTCACGGGCGACAGCCAGTTTTGTAGTCAAAAAGCAGTCATCTATTCA TTGAATTTTACAGCAAACCCACCTCAAAGAGTATTTGAACTTGTTGACCAGATTAATCCATCTATTTTCTGCATTCATATTACAAACTAT AAACCTGCATTATCCTTTATTAATCCAGAAGTACCTGATGAAAACAATTTTGATACATTGATGAAAACATCTGATGGTTTTACATTGAAT GCTGAATCATATGTTTCCTTCACAACCAAACTGGATATTCCTACTGCTGCTAAATATGAGAATAAAGACAATGACACTCTTAAGGATCTG CTGAAAGCAAACGTTGAAAAGCCTGTAAAGATGCTTATCTATAGCAGCAAAACATTGGAACTGCGAGAGACCTCAGTCACACCAAGTAAC CTGTGGGGCGGCCAGGGCTTATTGGGAGTGAGCATTCGTTTCTGCAGCTTTGATGGGGCAAATGAAAATGTTTGGCATGTGCTGGAGGTG GAATCAAATTCTCCTGCAGCACTGGCAGGTCTTAGACCACACAGTGATTATATAATTGGAGCAGATACAGTCATGAATGAGTCTGAAGAT CTATTCAGCCTTATCGAAACACATGAAGCAAAACCATTGAAACTGTATGTGTACAACACAGACACTGATAACTGTCGAGAAGTGATTATT ACACCAAATTCTGCATGGGGTGGAGAAGGCAGCCTAGGATGTGGCATTGGATATGGTTATTTGCATCGAATACCTACACGCCCATTTGAG GAAGGAAAGAAAATTTCTCTTCCAGGACAAATGGCTGGTACACCTATTACACCTCTTAAAGATGGGTTTACAGAGGTCCAGCTGTCCTCA GTTAATCCCCCGTCTTTGTCACCACCAGGAACTACAGGAATTGAACAGAGTCTGACTGGACTTTCTATTAGCTCAACTCCACCAGCTGTC AGTAGTGTTCTCAGTACAGGTGTACCAACAGTACCGTTATTGCCACCACAAGTAAACCAGTCCCTCACTTCTGTGCCACCAATGAATCCA GCTACTACATTACCAGGTCTGATGCCTTTACCAGCAGGACTGCCCAACCTCCCCAACCTCAACCTCAACCTCCCAGCACCACACATCATG CCAGGGGTTGGCTTACCAGAACTTGTAAACCCAGGTCTGCCACCTCTTCCTTCCATGCCTCCCCGAAACTTACCTGGCATTGCACCTCTC CCCCTGCCATCCGAGTTCCTCCCGTCATTCCCCTTGGTTCCAGAGAGCTCTTCTGCAGCAAGCTCAGGAGAGCTGCTGTCTTCCCTCCCG CCCACCAGCAACGCACCCTCCGACCCTGCCACAACTACTGCAAAGGCAGACGCTGCCTCCTCACTCACTGTGGATGTGACGCCCCCCACT GCCAAGGCCCCCACCACCGTTGAGGACAGAGTCGGCGACTCCACCCCAGTCAGCGAGAAGCCTGTTTCTGCGGCTGTGGATGCCAATGCT TCTGAGTCACCTTAACTTTGAACCATTCTTTGGAATTGGCGTGGTATATTTAACCACGGGAGCGTGTCTGGAAACGCAAACTATCATTAA TTTCATACTAGTTTGTACCGTATCTGTAGGCATCCTGTAAATAATTCCAAGGGGAAAACTAAACGAGGACGTGGGTTGTATCCTGCCAGG TTGAGTGGGGCTCACACGCTAGGGTGAGATGTCAGAAAGCGCTTGTATTTTAAACAACCAAAAAGAATTGTAAGGGTGGCTTGCTGCCAG GCTTGCACTGCCGTTCCTGGGGGTGTGCATCTTCGGGAAAGGTGGTGGCGGGGCGTCCACTAGGTTTCCTGTCCCCTGCTGCTCCTTCCG TAAGAAAATGAAATATTCTATGCCTAATACTCACACGCAACATTTCTTGTACTTTGTAAGTCGTTTGCGAGAATGCAGACCACCTCACTA AACTGTAAACGGTAAAGAGATTTTTACTTTTGGTCTCCGTGAGTCGCATCTCTACTAAGGTTTACACAGGAATTCCACCTGAAGACTTGT GTTAAAGTTCTACAGCGCGCACTGTTAACTGAACGTCTTTTTCTTCAGCCTATACGCGGATCCTTGTTTTGAGCTCTCAGAATCACTCAG ACAACATTTTGTAACTGCTGCTGTTGCTTTCTACATACACCTTATAAAGTGACATTTCAAAAGAAATAAGGTGCCACAGTTTTAAACCAG AAGGTGGCACTCTGTGGCTCCTTGTAGTATTATAGCTATACTGGGAAAGCATAGATACAGCAATAAAGTACAGTAATTTTACTTTTTTTC TTGTGTTACATCTAAATTACAACCCTTAATTGCCACGTGTGCACTTACTACTCTCCAGTATGTCTTATTACTCTCCAGTATGTCACGCAT CTTTAACTTTTCACGTCCTATGTTTGCTTTCTCCCATTTTTAAGAGATGGTAAGTTAACTGGAATTGATTTACTGAATGAAATTAAATGC >89898_89898_4_TCTN1-GORASP2_TCTN1_chr12_111066723_ENST00000551590_GORASP2_chr2_171806048_ENST00000234160_length(amino acids)=661AA_BP=257 MPHPGFAPVSLRQLLAASQAPSRDAPRLAVAVAGQRAVHVAGLAGTPWEMRPRGLPPLLVVLLGCWASVSAQTDATPAVTTEGLNSTEAA LATFGTFPSTRPPGTPRAPGPSSGPRPTPVTDVAVLCVCDLSPAQCDINCCCDPDCSSVDFSVFSACSVPVVTGDSQFCSQKAVIYSLNF TANPPQRVFELVDQINPSIFCIHITNYKPALSFINPEVPDENNFDTLMKTSDGFTLNAESYVSFTTKLDIPTAAKYENKDNDTLKDLLKA NVEKPVKMLIYSSKTLELRETSVTPSNLWGGQGLLGVSIRFCSFDGANENVWHVLEVESNSPAALAGLRPHSDYIIGADTVMNESEDLFS LIETHEAKPLKLYVYNTDTDNCREVIITPNSAWGGEGSLGCGIGYGYLHRIPTRPFEEGKKISLPGQMAGTPITPLKDGFTEVQLSSVNP PSLSPPGTTGIEQSLTGLSISSTPPAVSSVLSTGVPTVPLLPPQVNQSLTSVPPMNPATTLPGLMPLPAGLPNLPNLNLNLPAPHIMPGV GLPELVNPGLPPLPSMPPRNLPGIAPLPLPSEFLPSFPLVPESSSAASSGELLSSLPPTSNAPSDPATTTAKADAASSLTVDVTPPTAKA -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TCTN1-GORASP2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TCTN1-GORASP2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TCTN1-GORASP2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
