
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TFEB-CADM2 (FusionGDB2 ID:90322) |
Fusion Gene Summary for TFEB-CADM2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TFEB-CADM2 | Fusion gene ID: 90322 | Hgene | Tgene | Gene symbol | TFEB | CADM2 | Gene ID | 7942 | 253559 |
| Gene name | transcription factor EB | cell adhesion molecule 2 | |
| Synonyms | ALPHATFEB|BHLHE35|TCFEB | IGSF4D|NECL3|Necl-3|SynCAM 2|synCAM2 | |
| Cytomap | 6p21.1 | 3p12.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transcription factor EBT-cell transcription factor EBclass E basic helix-loop-helix protein 35 | cell adhesion molecule 2immunoglobulin superfamily member 4Dnectin-like 3nectin-like protein 3synaptic cell adhesion molecule 2 | |
| Modification date | 20200329 | 20200322 | |
| UniProtAcc | . | Q8N3J6 | |
| Ensembl transtripts involved in fusion gene | ENST00000230323, ENST00000358871, ENST00000373033, ENST00000403298, ENST00000420312, ENST00000394283, | ENST00000485126, ENST00000383699, ENST00000405615, ENST00000407528, | |
| Fusion gene scores | * DoF score | 12 X 11 X 8=1056 | 15 X 13 X 8=1560 |
| # samples | 14 | 17 | |
| ** MAII score | log2(14/1056*10)=-2.91511110241349 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/1560*10)=-3.19793937761191 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TFEB [Title/Abstract] AND CADM2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TFEB(41653828)-CADM2(85851197), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TFEB-CADM2 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. TFEB-CADM2 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. TFEB-CADM2 seems lost the major protein functional domain in Hgene partner, which is a transcription factor due to the frame-shifted ORF. TFEB-CADM2 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. TFEB-CADM2 seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TFEB | GO:0010468 | regulation of gene expression | 27278822 |
| Hgene | TFEB | GO:0045893 | positive regulation of transcription, DNA-templated | 27278822|29146937 |
| Hgene | TFEB | GO:0045944 | positive regulation of transcription by RNA polymerase II | 19556463 |
 Fusion gene breakpoints across TFEB (5'-gene) Fusion gene breakpoints across TFEB (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
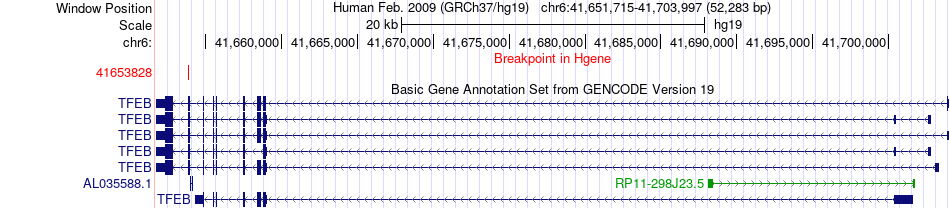 |
 Fusion gene breakpoints across CADM2 (3'-gene) Fusion gene breakpoints across CADM2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
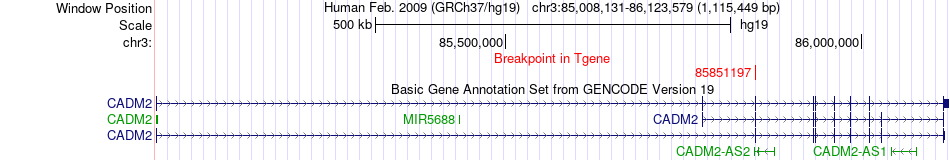 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | KIRP | TCGA-B9-A69E-01A | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
Top |
Fusion Gene ORF analysis for TFEB-CADM2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000230323 | ENST00000485126 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| 5CDS-intron | ENST00000358871 | ENST00000485126 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| 5CDS-intron | ENST00000373033 | ENST00000485126 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| 5CDS-intron | ENST00000403298 | ENST00000485126 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| 5CDS-intron | ENST00000420312 | ENST00000485126 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000230323 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000230323 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000230323 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000358871 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000358871 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000358871 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000403298 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000403298 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000403298 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000420312 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000420312 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| Frame-shift | ENST00000420312 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| In-frame | ENST00000373033 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| In-frame | ENST00000373033 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| In-frame | ENST00000373033 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| intron-3CDS | ENST00000394283 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| intron-3CDS | ENST00000394283 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| intron-3CDS | ENST00000394283 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
| intron-intron | ENST00000394283 | ENST00000485126 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000373033 | TFEB | chr6 | 41653828 | - | ENST00000383699 | CADM2 | chr3 | 85851197 | + | 10005 | 1232 | 1093 | 2358 | 421 |
| ENST00000373033 | TFEB | chr6 | 41653828 | - | ENST00000407528 | CADM2 | chr3 | 85851197 | + | 2531 | 1232 | 1093 | 2478 | 461 |
| ENST00000373033 | TFEB | chr6 | 41653828 | - | ENST00000405615 | CADM2 | chr3 | 85851197 | + | 2479 | 1232 | 1093 | 2478 | 462 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000373033 | ENST00000383699 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + | 0.000147702 | 0.9998523 |
| ENST00000373033 | ENST00000407528 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + | 0.003337619 | 0.9966624 |
| ENST00000373033 | ENST00000405615 | TFEB | chr6 | 41653828 | - | CADM2 | chr3 | 85851197 | + | 0.003246341 | 0.99675363 |
Top |
Fusion Genomic Features for TFEB-CADM2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TFEB | chr6 | 41653827 | - | CADM2 | chr3 | 85851196 | + | 0.000485724 | 0.9995142 |
| TFEB | chr6 | 41653827 | - | CADM2 | chr3 | 85851196 | + | 0.000485724 | 0.9995142 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
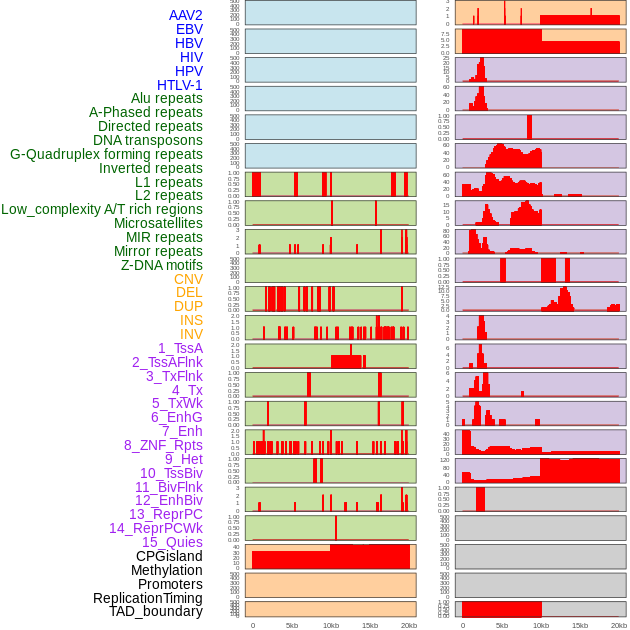 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
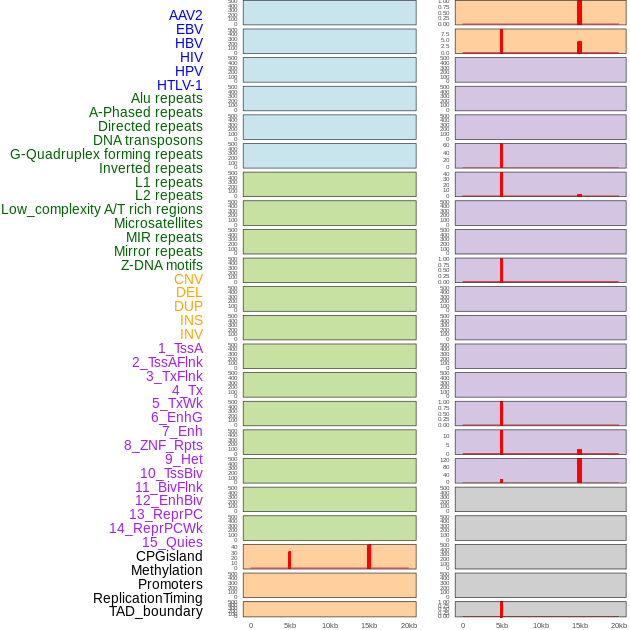 |
Top |
Fusion Protein Features for TFEB-CADM2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr6:41653828/chr3:85851197) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | CADM2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Adhesion molecule that engages in homo- and heterophilic interactions with the other nectin-like family members, leading to cell aggregation. Important for synapse organization, providing regulated trans-synaptic adhesion. Preferentially binds to oligodendrocytes. {ECO:0000269|PubMed:17967169}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 10_44 | 317 | 477.0 | Compositional bias | Note=Gln-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 35_44 | 317 | 477.0 | Compositional bias | Note=Poly-Gln |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 10_44 | 317 | 477.0 | Compositional bias | Note=Gln-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 35_44 | 317 | 477.0 | Compositional bias | Note=Poly-Gln |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 10_44 | 317 | 477.0 | Compositional bias | Note=Gln-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 35_44 | 317 | 477.0 | Compositional bias | Note=Poly-Gln |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 10_44 | 232 | 392.0 | Compositional bias | Note=Gln-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 35_44 | 232 | 392.0 | Compositional bias | Note=Poly-Gln |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 235_288 | 317 | 477.0 | Domain | bHLH |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 235_288 | 317 | 477.0 | Domain | bHLH |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 235_288 | 317 | 477.0 | Domain | bHLH |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 136_153 | 317 | 477.0 | Motif | Nuclear export signal |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 136_153 | 317 | 477.0 | Motif | Nuclear export signal |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 136_153 | 317 | 477.0 | Motif | Nuclear export signal |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 136_153 | 232 | 392.0 | Motif | Nuclear export signal |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 156_165 | 317 | 477.0 | Region | Strong transcription activation domain |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 298_319 | 317 | 477.0 | Region | Note=Leucine-zipper |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 156_165 | 317 | 477.0 | Region | Strong transcription activation domain |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 298_319 | 317 | 477.0 | Region | Note=Leucine-zipper |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 156_165 | 317 | 477.0 | Region | Strong transcription activation domain |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 298_319 | 317 | 477.0 | Region | Note=Leucine-zipper |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 156_165 | 232 | 392.0 | Region | Strong transcription activation domain |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 319_350 | 29 | 405.0 | Compositional bias | Note=Thr-rich | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 319_350 | 22 | 438.0 | Compositional bias | Note=Thr-rich | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 319_350 | 20 | 436.0 | Compositional bias | Note=Thr-rich | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 127_219 | 29 | 405.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 227_312 | 29 | 405.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 27_119 | 29 | 405.0 | Domain | Note=Ig-like V-type | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 127_219 | 22 | 438.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 227_312 | 22 | 438.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 27_119 | 22 | 438.0 | Domain | Note=Ig-like V-type | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 127_219 | 20 | 436.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 227_312 | 20 | 436.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 27_119 | 20 | 436.0 | Domain | Note=Ig-like V-type | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 389_435 | 29 | 405.0 | Topological domain | Cytoplasmic | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 25_367 | 22 | 438.0 | Topological domain | Extracellular | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 389_435 | 22 | 438.0 | Topological domain | Cytoplasmic | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 25_367 | 20 | 436.0 | Topological domain | Extracellular | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 389_435 | 20 | 436.0 | Topological domain | Cytoplasmic | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 368_388 | 29 | 405.0 | Transmembrane | Helical | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000405615 | 0 | 10 | 368_388 | 22 | 438.0 | Transmembrane | Helical | |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000407528 | 0 | 10 | 368_388 | 20 | 436.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000230323 | - | 9 | 10 | 366_414 | 317 | 477.0 | Compositional bias | Note=Pro-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000373033 | - | 8 | 9 | 366_414 | 317 | 477.0 | Compositional bias | Note=Pro-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000403298 | - | 8 | 9 | 366_414 | 317 | 477.0 | Compositional bias | Note=Pro-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 366_414 | 232 | 392.0 | Compositional bias | Note=Pro-rich |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 235_288 | 232 | 392.0 | Domain | bHLH |
| Hgene | TFEB | chr6:41653828 | chr3:85851197 | ENST00000420312 | - | 8 | 9 | 298_319 | 232 | 392.0 | Region | Note=Leucine-zipper |
| Tgene | CADM2 | chr6:41653828 | chr3:85851197 | ENST00000383699 | 1 | 10 | 25_367 | 29 | 405.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for TFEB-CADM2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >90322_90322_1_TFEB-CADM2_TFEB_chr6_41653828_ENST00000373033_CADM2_chr3_85851197_ENST00000383699_length(transcript)=10005nt_BP=1232nt GTGGAGTGACAGCCGGAGCCCGGGCGCCGGGCGCGGGGCTCGGTGACAGCGGAGGCGGCGGCCCGGGCGGGACGCAGGGAGCGGCCGCGG CGCCGACGGCGGCGGGAAGGGCGGACGGGGCGGGCCTCCCTGGTGGCACGGGGCCGGAGCAGGCCGGGAGCAGGTGGCGGGCACAGCTTG CGGGCCAGGCACCCGAACTTGCGACAAGTTGCCGGAGCCGCGGGGCGCGGGCGGCGGACAGATTGACCTTCAGAGCGAGGGAGCCAGCGC CGGCAGCCACCATGGCGTCACGCATAGGGTTGCGCATGCAGCTCATGCGGGAGCAGGCGCAGCAGGAGGAGCAGCGGGAGCGCATGCAGC AACAGGCTGTCATGCATTACATGCAGCAGCAGCAGCAGCAGCAACAGCAGCAGCTCGGAGGGCCGCCCACCCCGGCCATCAATACCCCCG TCCACTTCCAGTCGCCACCACCTGTGCCTGGGGAGGTGTTGAAGGTGCAGTCCTACCTGGAGAATCCCACATCCTACCATCTGCAGCAGT CGCAGCATCAGAAGGTGCGGGAGTACCTGTCCGAGACCTATGGGAACAAGTTTGCTGCCCACATCAGCCCAGCCCAGGGCTCTCCGAAAC CCCCACCAGCCGCCTCCCCAGGGGTGCGAGCTGGACACGTGCTGTCCTCCTCCGCTGGCAACAGTGCTCCCAATAGCCCCATGGCCATGC TGCACATTGGCTCCAACCCTGAGAGGGAGTTGGATGATGTCATTGACAACATTATGCGTCTGGACGATGTCCTTGGCTACATCAATCCTG AAATGCAGATGCCCAACACGCTACCCCTGTCCAGCAGCCACCTGAATGTGTACAGCAGCGACCCCCAGGTCACAGCCTCCCTGGTGGGCG TCACCAGCAGCTCCTGCCCTGCGGACCTGACCCAGAAGCGAGAGCTCACAGATGCTGAGAGCAGGGCCCTGGCCAAGGAGCGGCAGAAGA AAGACAATCACAACTTAATTGAAAGGAGACGAAGGTTCAACATCAATGACCGCATCAAGGAGTTGGGAATGCTGATCCCCAAGGCCAATG ACCTGGACGTGCGCTGGAACAAGGGCACCATCCTCAAGGCCTCTGTGGATTACATCCGGAGGATGCAGAAGGACCTGCAAAAGTCCAGGG AGCTGGAGAACCACTCTCGCCGCCTGGAGATGACCAACAAGCAGCTCTGGCTCCGTATCCAGGCAGCCAAGGGCAGTTTCCACTAACACA GAATGTAACCGTTGTTGAAGGTGGAACTGCAATTTTGACCTGCAGGGTTGATCAAAATGATAACACCTCCCTCCAGTGGTCAAATCCAGC TCAACAGACTCTGTACTTTGACGACAAGAAAGCTTTAAGGGACAATAGGATCGAGCTGGTTCGCGCTTCCTGGCATGAATTGAGTATTAG TGTCAGTGATGTGTCTCTCTCTGATGAAGGACAGTACACCTGTTCTTTATTTACAATGCCTGTCAAAACTTCCAAGGCATATCTCACCGT TCTGGGTGTTCCTGAAAAGCCTCAGATTAGTGGATTCTCATCACCAGTTATGGAGGGTGACTTGATGCAGCTGACTTGCAAAACATCTGG TAGTAAACCTGCAGCTGATATAAGATGGTTCAAAAATGACAAAGAGATTAAAGATGTAAAATATTTAAAAGAAGAGGATGCAAATCGCAA GACATTCACTGTCAGCAGCACACTGGACTTCCGAGTGGACCGGAGTGATGATGGAGTGGCGGTCATCTGCAGAGTAGATCACGAATCCCT CAATGCCACCCCTCAGGTAGCCATGCAGGTGCTAGAAATACACTATACACCATCAGTTAAGATTATACCATCGACTCCTTTTCCACAAGA AGGACAGCCTTTAATTTTGACTTGTGAATCCAAAGGAAAACCACTGCCAGAACCTGTTTTGTGGACAAAGGATGGCGGAGAATTACCAGA TCCTGACCGAATGGTTGTGAGTGGTAGGGAGCTAAACATTCTTTTCCTGAACAAAACGGATAATGGTACATATCGATGTGAAGCCACAAA CACCATTGGCCAAAGCAGTGCGGAATATGTTCTCATTGTGCATGATCCTAATGCTTTGGCTGGCCAGAATGGCCCTGACCATGCTCTCAT AGGAGGAATAGTGGCTGTAGTTGTATTTGTCACGCTGTGTTCTATCTTTCTGCTTGGTCGATATCTGGCAAGGCATAAAGGAACGTATTT AACAAATGAAGCTAAAGGAGCTGAAGATGCACCAGATGCTGATACAGCCATTATCAATGCTGAAGGCAGCCAAGTCAATGCTGAAGAGAA AAAAGAGTATTTCATTTAAGATGCAGGCCAAGATTCTGAGTTTTACTACCAGGCTGAATGCTGGAGAAAACTGGCTATCATCTTTCAGAA GTCATTTCTACCATCGTCTGCTACCCTTATTAACTCCCATACTGTACTGCTATCAGTAGCCAGTGTATACCAACAATCAGCTGTTGAAAG CATCATTCTTTAATTACTGTACCATCCATAATGCAGGACATTTCTTACTGCCTAAATTTCACACCATTGCTCTTTTAACATACAGTGCTT GAATATACAGCCTTAACAATGTTAATCATCTCCTTGGATCATTATATTGAGTGGTTTTTATACATTAAAAAATGTATGCAGAGTTTTTTT CCCCCATTTTTTCCCCTTTAAGTCATAGACCTTATCAGTTTGCCATTGGACAGCTTTCACAAGTAACAGGATTAGGGAAGGACATAATGT ATTTAAGAATAAGTGAAGGTTAGAGGTTTACAATTAATGTTTTCTACATGTCTGTCTTCAAAGACAACATGCTTTGGAAACATATGTCCT AGAAAACATAAAATTAAAAAAAAACACTATAGTGTAGTTTTTGGAATGTTGGAGATTATACTGGTAAAGCTGTATCGATTTGCTATTGAA ATATAGTATTTGATACAGGAGCCATGTGTACGAATTGCAATCATGACATGGTATTTTCTATAATTATAGAGATCTTACTATTAGGATATT GTAGCTTCATTTACCAATAAATTATTCCAAATGATTTACTGCTTTTGGAATCACAAATATTTAATTCATTTCTCTAATTTGATTCTATAA TTTATTTGCTTCATAAAGATTCACCTGCCTAAACAATATTGAAGTATCCATAGGGCACAATTTTCAGACATTTTAGTATCTGTATATAGT GCATATAATAAATCCAATTAAACATTATTTGATACATATTTAACTTTTTTGGCTGTATGTAATTCAGTCATAAGTAAGCATTTTCTAACG TCCATTATATCCCTTTAGGTATTACTTAAACCAATATTATAAGTAGATAACATGTATTAGTATTTTTGTCTGTATGTCCTAGCACTGTTC AACAACAAATTTTTCTAGTTCTTGTTAATTTTTATTTGTTATACAATGGAAGCACAATGTTATAAGGAAAGGTAATTTTAAGCTAACAAC CAGTGCACAGCCTCAGGTTTTAAATTACAACCACAGTTGAATAATAACCTAAAAATAAATTTTAGTTTGCTAAAAATTTACAAATTTTCA TTTTGCAGTTCCAGAGCTGCTTACCTATGTTGGTTTGCCAAAAAGAAGTGTTTAAGATATGTTTTCTGTATAAATGAACTAATTCTTTAT ATTAAATTCCTGTCTATGCATTTTGTGAGGTACAAACTGATGAAACAGTGAGTGATAGAGAATTTCTGCCATGCTTTTAACTCCAAAGTG AAAGTCTCTTTCTCTGGATTATTTTAAAATTTTGGCGTGTTTAACCATTAAGAAATGCTGTATTCTTAAATATGAAACAGTTAATCTAAA GTCTTATTATTTCTTCTAGTAAAATAAAATATTGTTAGATTAGTTCTCATTAAATTTATTATATGCTAAAGAATAAAACTCCAGTTAGAT TTAAAAAAATCCCATTTACAAGCATTGCACCTTTAAGAAGAAAACAAATATGACAGTATGGTAGCTATACTTGTGATGTCTGACGCATGA TGGCCTATGGTTTTAAAAATAGTATTGTGGAATATATTTTTGGGTAATAAGAATGGTTGAGTGCAACATCATGTATTAATATCAAATGAA AGACAAGGGTGCTGTATCTTTGATTATTTATCAAAAAAGTATAAATCTTTTAAGGAAAATGTTAGAATTTTAAAGTTTTTTTTTGATTGT TGAAGCATTTATCTTGTTGATTTCTTACAAAAGAAAAAGGACGATGTCAGTCAAGCAGCACTTTTTCAGAATATACAGAACATAAATAAT ATGATGTGGTTGAGTGTTAACATAATAAATCATATACAGTATGACATTTTAAGGAAATAAGTCTGCATTGATGTGATTGCATGCTTGTTT TTACAGTTCACTCCATACCATCTGTGGAAAAATGCAATAATATGTTAATATAGTATGGTAGTTTGAGAGAATCAGGTAGGTGCTACTAGA CACATTGAAACTAGAATAGGTTTATAAACTGTTCATATCTTTCAATGCATAATCTTTAGAAAGACTCCACAAAGCAAAATTCATCCTGTT AGTACAAAATGGAAAGGTTCTTATTACAGAAGTAAGTTGTACAAACTGCATCATCAATTACTATGTAAAGCCTAATTTCAGGATGCACTT ACAAAATTGCTACTCTTCATCGATGCCGTATTTACATCCCTCTTCATTTCATCTATATTCTACTTGGCTCCAACAACTAAATTGTTGCCT AAATAACTCAATCATTATTTATAGCACTGTAAAATTAGAAATCCAAAATTGGTTGCATTTCATGATTTTTGACTCTTTTAACCCTTCAGA GTAATATAAAATCTCATTAGAACTCTTATAAAACCAGTTAGCTAAAACAACTAGATTATTATACTAGTTATAAATGCTGGTATATTATAA TTTGTGGGATCTAATTAAATGTTTGCCTATTAAATATAGTTTGATTTAAATGAGACACCATGTTCTGAACTGGAGAAAACAATATCAATG AAGTCATATTTTCATACCTTTAAAAATTATTTAATTTTCAACCAAACAAAAAATAAATTGTAAAAATTATGCTCATTTCTATTCCAACAA GTCAGAAAATAATGACATAATATTTCTAAATGAGAATGATGTCAATTTCATTGTCTGGAACATGCACACTTGTGTGCACATAGAGTCAAT TTCTGTCTCTTCTAAGTGAGCACAGTAGGTAGGGAACTCAATGGAAGCAGTGTGCACACACACACACCCTTAGTAGAATATGGAGCATTA TTCTTTACCAAAAGTAGCTATTCTCTGCCAGTGTACTGTTTACAGTGTAAATTGATTGTCCTCCGTCAAAATCAAGTCTAAGCGGCATTT TACTTTTCTTTTCTTTTTTATTTTCCATTTTTGGGTGTTCTAGATAGGATTATTATCAAAAGTAGGAGGAAAAAGAAATGTTTGTGAGTG CAAGATTAAAGACAGAGCGCATGAGCAGAGTACTGATGGTGTGCGTGTTTGTGTGTGTGTATATGTGTGTGTATAAGAGAGAAAGAGAAA TCGAAAAGAGAGAGAAATTTCAACTTGTTTTCTTTTACCAGAGAATCCAAGAAGCAGTTTAAGGAACCTGATATTGGAGTTTAATCTAGA AACTGGATGATCTCATATTGATTCATGATGCTTAATCTTTCATTGAAACTCCACAATTAAAATAACAAATAGAGAAAGAAAACTAACTTT CCTAAATATGGCAAAGAAACTAACACCTCATTTATTTTTATTTCTTTGTAAAAATATAAATATCTCAATCAAAAGTACAGGCTCATTTGT AATTATGTACTCATGATTGTAACAGCATTGAGATTTCATATAGGCACATGTATAGCAGCTGTTTGACAGTGATGGCTCTTCCATGTAACA TAGCAGTTATCGTGTAATTTAGTGCCACTTATTGCAAAGTGTTACCAAGGGGAACATTTCAAATGCCTTTTGTTTTTCTTTGGACATCAA GGTAATTTAGTGGACAACAAATATTAAATTTTACATTATGCTTTTTAATTGTGGAGAAAATGCAAAACCTCATGTCTTACCACAACTTTC CCATGGTATTGCTAATTACTTAGAGTCTCTAAAATTGCCCAATATTATTTCCCTTTGCAAATGGTAATTTCTGATAAAACAATACAAAAC AAAAAAAAAGCAGTAATCACAGCCAAATAAATGAAACACCTAATGAGAATTACTGTTGAGTAATGCTTTTTATTAAGTGGCACAAAGTAC ACACTAAAAACTATGCAAAATATAGGTAACTACAGTGTACGTACATGTAAGTATCTTGTACTTGCTGTGTATGGATGTTGGAAACCCATG TAATTATAATATGCATTTTGAATATTGGGAAAGAGGAAATCTTAGTGTAGATAATAGAATTTGTTTTGAAATATACACTGGCAATATTGA AATATACTCAGTGCAATATCTGCAGTTCTATTGCTGGATACTTAATGCAAAGGGATTTTAAAGAATTCTTCAGCAAGATCCAAGAGGTTG CTAATCTTCACCTCTCACATACTAAAGCATTCATTAAAAATACAACTAATTCAAGGACAAGTTAAGTTTTTCTTATTAATGTGACACAGT AACATTTTTATTTGGATTAGTTTTTGGTGTTATTGCAGAGAAAGAAATAACCTGAGTTTTAAATTTCGATTCATATCTCATTGAATTGGG AATTCTTCAATTTTGTTTGAAATGTGTGCATGTTTTCATAGATTGTAGCAAGTACAAATGTATTCAATGAAAGAACATAGGGTAGTTTTA ATAGTTAAAGTCACTATATCTTAATGGGAATTTTAAAATATCTTTTCTTTTTCTATTCATTTTTTCCCTTTCTCTCTCTATTCTACCCCC AAACAAGCCCTTTGCACTCATACCCTTCCAAAGGCAATGGATTGAAAGTAGCAAGATAATAAAGAGAGTTAAAGGGGACAACATATAACA ATGGGTTCTAAAATCATCAGCATTTTTATGTATTGAGATTATTAATATTGAATGCAATAGTTATTGGATATAGCTGGTAAGTTCTCTGCC ATGCATACTTACAATAATCCTTCTACCTCTCACTTTTAACATGGGCTATGATACTTCAATATAAATAGAGGAAATAATTATTTACCAGAT ATCCCAGGAAGCATTCTTATGGAACTATTATAACAGTGCACTCCCCACCCCCTGGGAAATCTCAAAAGCCAAGTTCTTCAGTGTATTCTA TGGTGTATGAATCTGCATTTGTTTTTCCTTTTCTATCATTAGCTTTTCCTAAAGGCAAAATATTATAACTTTATAAGAATAGTCCTTCTC CTGCCTATTTAATATTTCAATGTTAATATAATTGTATTGGTTTTTAAAAATAATTAAAATGGCAACACTTTGCGATCCAATCTGATATTT AATTTTTAAAATGTAACAATTTCTTATAAATTCATGCTGTGTTTTAAGTAAAAATCATGTATATGAATTATTTTACCAAGTAGTATTATC TGCACGAGACAAATTTAAGTATTTTAAAAATCATCCTTATTAATAAATAATCCTCATATAATGCACAGTATTGCCACAAATGCCCACTTC ATTTTGTCTGCATAGACATATTAAGAATTTTTCAATGATGTAATTTAATTATCCTAAAAGAAATGACAGATTCTTCTGTAGGAAAAAATA AAAACATGAATATCTCAAATTATAATGGTTAATCTCTAAAATATGCTTAAAGTAAGATGTTTCTATGTTATGTGGTTATGTTATCAATAA TATTTGGTTGATCTTCACATATTTTATATGCATATATATATCAAGAAGTTATATATATATAACTCAAGAAACTCAAGTTATATATATATG TCAAGAAATGTATGATTATTTTGGAGAGAATGGGCCCAAATGTGAAAAAGATATAAAAAAAACAAAAAAAACAAAAAAAAAACACTATTT TCCCCTACGAAATATACTGTATATTTCAAAAGAAGAAAAGTTAAAAGATATTTGAAGATTGCAGGGGCAAAACAAAAACCTACCAGGGCT CAGCACTTAAGCACTTTTCCCACATCCAGGGTCAAAGCAGCAGTAACTACATGGACTTTTAAGTGCGGTATGAAATGCAACATTACGCTT ACAAACAGTACTGTCAAACCTCAAATGCTTTCTTTCTTCAGATGCTTTTTCGTGTACATGATACTAGTAGACACTTTTCTCTTTATATTT ACTGATAGTGAAAATCATACGCAATAAAATATTGATGTTTGAAGGCAGTGGTCACCAATTGGTTAAAAAACTATGAAATGTAAACTGAAT TGTTATATCTCTATCCTTTTTGCTTTTCTCTGTGTTTTTAATGTATGGAATAAATCTCATAAATAGAAAGAAAAATAATCTAGAAATTTT TCAAAGCTAGTACTCTTTCTCCTTATAAATGTACACAATTTTAATCTTTTTACAAATTTATTTAACTGTACCTACTGTACTTATTGTAGA TTCAATGACGCAGTTAAGTCATCACCCAAGGATTTATGAATTTGAGATTACTGACCTGTTTTCTTCATATTGCATTCACATCAATATTTG TGAATTTGTTGTTCAGCTTTTCATTCAAACAAAAAATATTCCCTCAAGAAAGCTCCATTTTTATCATAAACATTTCAACATAACCAACAT TAGAACAAGTCTGCCATGTTAAAAATAATTTAAAGACTTATCTCTGAAAACGGTATCCAGAAACGCAGGTGTTCCCAGTAATGTAGCTTC AAAAATAAAATGTGCTATTTATATGACATGAAATTCATAACTTTTGGAAGGGTATATTTATGACAGCATAAAAAATAAATTCTGTGCTAT AAAGAAGATCCAACAAATTAACCATATAAGCACAGAAAATAGAGAAACACAGTTATTGAATCTACTCTTGTCATTAACATTTTCAAAAAA CAAAATGCATATTGTAATATTTGGTACATGACACTTGCATGTTGATATGCCTATATACTTACAAAGTATTCAATGTGTACTTAGCGGCGC TTAAAATATGTCATGTACAACTCTTATAAACATTTTTACAGGGTTCCCATTTGCACTTCATCTTTCAGTAAAGTCTTGTCAGAAAAAAAT TGTCTGATAAATATGGAAAAATAAAATTTGAATTTTAGTTATCTGTTGGACATTACTGAATTGTCTATTCATTTAGTACATTTTCTTAAC CTTGAAAACAGACATTGAAAGATGGAACTTTCTTTTTTCCAGTGACAGGTCATTAAATCGATGGTACAAAACATACTTCTTTATGCCAAA CACATTATTTTGAATACCTTTTTTTCAACTTCATACATTATCCATCCATTTTAGTTTTCCTCGATGATGCCCATTTTCTTTGTAAATTTC TAAGTGGAAGTTTTAAAAATTAAGCATCAGTAAAATAAATGTTATCTTATTTGTACAACATTTAATCGTTATTCCTTGGAATATATGAAC CTTGTAAAGTGATGACTAAGGGTCAAAAAAAATGTTGCTGACTTAGGGAAATTTGCTTGAGAATGCCAAAATTTTAAAACTCTGTAAAGG TAATTTTACATTCACTTATGTGTGTTTACTAATGTTGAGTAGGAACAGAAGCAAAAATACCTAAACCTATTTTTTTTAACCTTCACACTC TAACCAGTATTATATATTTCAGCTACCTAAGATAATGCAATTAATCATATATCCAACCTAAGCTTCTGCCAATAAGGATTTCAGATAAGC TTTTAAATTTATGGAGGATTTTCTTTGTGTCTTTGGAAGTTCACTTACCATAAAGTATTTTATAGTCTACTTTAAATAACAGATTTTGTT ACACAGGGTAAGCCAAAGGCTGTACAAGATTTAGTGTCATTGGGTCTCATATTTGTGTTAGACCATGAAATATTCCATTAAAATTTATCT >90322_90322_1_TFEB-CADM2_TFEB_chr6_41653828_ENST00000373033_CADM2_chr3_85851197_ENST00000383699_length(amino acids)=421AA_BP=46 MEQGHHPQGLCGLHPEDAEGPAKVQGAGEPLSPPGDDQQAALAPYPGSQGQFPLTQNVTVVEGGTAILTCRVDQNDNTSLQWSNPAQQTL YFDDKKALRDNRIELVRASWHELSISVSDVSLSDEGQYTCSLFTMPVKTSKAYLTVLGVPEKPQISGFSSPVMEGDLMQLTCKTSGSKPA ADIRWFKNDKEIKDVKYLKEEDANRKTFTVSSTLDFRVDRSDDGVAVICRVDHESLNATPQVAMQVLEIHYTPSVKIIPSTPFPQEGQPL ILTCESKGKPLPEPVLWTKDGGELPDPDRMVVSGRELNILFLNKTDNGTYRCEATNTIGQSSAEYVLIVHDPNALAGQNGPDHALIGGIV -------------------------------------------------------------- >90322_90322_2_TFEB-CADM2_TFEB_chr6_41653828_ENST00000373033_CADM2_chr3_85851197_ENST00000405615_length(transcript)=2479nt_BP=1232nt GTGGAGTGACAGCCGGAGCCCGGGCGCCGGGCGCGGGGCTCGGTGACAGCGGAGGCGGCGGCCCGGGCGGGACGCAGGGAGCGGCCGCGG CGCCGACGGCGGCGGGAAGGGCGGACGGGGCGGGCCTCCCTGGTGGCACGGGGCCGGAGCAGGCCGGGAGCAGGTGGCGGGCACAGCTTG CGGGCCAGGCACCCGAACTTGCGACAAGTTGCCGGAGCCGCGGGGCGCGGGCGGCGGACAGATTGACCTTCAGAGCGAGGGAGCCAGCGC CGGCAGCCACCATGGCGTCACGCATAGGGTTGCGCATGCAGCTCATGCGGGAGCAGGCGCAGCAGGAGGAGCAGCGGGAGCGCATGCAGC AACAGGCTGTCATGCATTACATGCAGCAGCAGCAGCAGCAGCAACAGCAGCAGCTCGGAGGGCCGCCCACCCCGGCCATCAATACCCCCG TCCACTTCCAGTCGCCACCACCTGTGCCTGGGGAGGTGTTGAAGGTGCAGTCCTACCTGGAGAATCCCACATCCTACCATCTGCAGCAGT CGCAGCATCAGAAGGTGCGGGAGTACCTGTCCGAGACCTATGGGAACAAGTTTGCTGCCCACATCAGCCCAGCCCAGGGCTCTCCGAAAC CCCCACCAGCCGCCTCCCCAGGGGTGCGAGCTGGACACGTGCTGTCCTCCTCCGCTGGCAACAGTGCTCCCAATAGCCCCATGGCCATGC TGCACATTGGCTCCAACCCTGAGAGGGAGTTGGATGATGTCATTGACAACATTATGCGTCTGGACGATGTCCTTGGCTACATCAATCCTG AAATGCAGATGCCCAACACGCTACCCCTGTCCAGCAGCCACCTGAATGTGTACAGCAGCGACCCCCAGGTCACAGCCTCCCTGGTGGGCG TCACCAGCAGCTCCTGCCCTGCGGACCTGACCCAGAAGCGAGAGCTCACAGATGCTGAGAGCAGGGCCCTGGCCAAGGAGCGGCAGAAGA AAGACAATCACAACTTAATTGAAAGGAGACGAAGGTTCAACATCAATGACCGCATCAAGGAGTTGGGAATGCTGATCCCCAAGGCCAATG ACCTGGACGTGCGCTGGAACAAGGGCACCATCCTCAAGGCCTCTGTGGATTACATCCGGAGGATGCAGAAGGACCTGCAAAAGTCCAGGG AGCTGGAGAACCACTCTCGCCGCCTGGAGATGACCAACAAGCAGCTCTGGCTCCGTATCCAGGCAGCCAAGGGCAGTTTCCACTAACACA GAATGTAACCGTTGTTGAAGGTGGAACTGCAATTTTGACCTGCAGGGTTGATCAAAATGATAACACCTCCCTCCAGTGGTCAAATCCAGC TCAACAGACTCTGTACTTTGACGACAAGAAAGCTTTAAGGGACAATAGGATCGAGCTGGTTCGCGCTTCCTGGCATGAATTGAGTATTAG TGTCAGTGATGTGTCTCTCTCTGATGAAGGACAGTACACCTGTTCTTTATTTACAATGCCTGTCAAAACTTCCAAGGCATATCTCACCGT TCTGGGTGTTCCTGAAAAGCCTCAGATTAGTGGATTCTCATCACCAGTTATGGAGGGTGACTTGATGCAGCTGACTTGCAAAACATCTGG TAGTAAACCTGCAGCTGATATAAGATGGTTCAAAAATGACAAAGAGATTAAAGATGTAAAATATTTAAAAGAAGAGGATGCAAATCGCAA GACATTCACTGTCAGCAGCACACTGGACTTCCGAGTGGACCGGAGTGATGATGGAGTGGCGGTCATCTGCAGAGTAGATCACGAATCCCT CAATGCCACCCCTCAGGTAGCCATGCAGGTGCTAGAAATACACTATACACCATCAGTTAAGATTATACCATCGACTCCTTTTCCACAAGA AGGACAGCCTTTAATTTTGACTTGTGAATCCAAAGGAAAACCACTGCCAGAACCTGTTTTGTGGACAAAGGATGGCGGAGAATTACCAGA TCCTGACCGAATGGTTGTGAGTGGTAGGGAGCTAAACATTCTTTTCCTGAACAAAACGGATAATGGTACATATCGATGTGAAGCCACAAA CACCATTGGCCAAAGCAGTGCGGAATATGTTCTCATTGTGCATGATGTTCCCAACACTTTGCTTCCCACTACTATCATCCCCTCCCTTAC CACTGCAACAGTCACAACCACTGTAGCCATAACAACCAGCCCAACCACATCTGCAACAACCAGCAGCATCAGAGATCCTAATGCTTTGGC TGGCCAGAATGGCCCTGACCATGCTCTCATAGGAGGAATAGTGGCTGTAGTTGTATTTGTCACGCTGTGTTCTATCTTTCTGCTTGGTCG ATATCTGGCAAGGCATAAAGGAACGTATTTAACAAATGAAGCTAAAGGAGCTGAAGATGCACCAGATGCTGATACAGCCATTATCAATGC >90322_90322_2_TFEB-CADM2_TFEB_chr6_41653828_ENST00000373033_CADM2_chr3_85851197_ENST00000405615_length(amino acids)=462AA_BP=46 MEQGHHPQGLCGLHPEDAEGPAKVQGAGEPLSPPGDDQQAALAPYPGSQGQFPLTQNVTVVEGGTAILTCRVDQNDNTSLQWSNPAQQTL YFDDKKALRDNRIELVRASWHELSISVSDVSLSDEGQYTCSLFTMPVKTSKAYLTVLGVPEKPQISGFSSPVMEGDLMQLTCKTSGSKPA ADIRWFKNDKEIKDVKYLKEEDANRKTFTVSSTLDFRVDRSDDGVAVICRVDHESLNATPQVAMQVLEIHYTPSVKIIPSTPFPQEGQPL ILTCESKGKPLPEPVLWTKDGGELPDPDRMVVSGRELNILFLNKTDNGTYRCEATNTIGQSSAEYVLIVHDVPNTLLPTTIIPSLTTATV TTTVAITTSPTTSATTSSIRDPNALAGQNGPDHALIGGIVAVVVFVTLCSIFLLGRYLARHKGTYLTNEAKGAEDAPDADTAIINAEGSQ -------------------------------------------------------------- >90322_90322_3_TFEB-CADM2_TFEB_chr6_41653828_ENST00000373033_CADM2_chr3_85851197_ENST00000407528_length(transcript)=2531nt_BP=1232nt GTGGAGTGACAGCCGGAGCCCGGGCGCCGGGCGCGGGGCTCGGTGACAGCGGAGGCGGCGGCCCGGGCGGGACGCAGGGAGCGGCCGCGG CGCCGACGGCGGCGGGAAGGGCGGACGGGGCGGGCCTCCCTGGTGGCACGGGGCCGGAGCAGGCCGGGAGCAGGTGGCGGGCACAGCTTG CGGGCCAGGCACCCGAACTTGCGACAAGTTGCCGGAGCCGCGGGGCGCGGGCGGCGGACAGATTGACCTTCAGAGCGAGGGAGCCAGCGC CGGCAGCCACCATGGCGTCACGCATAGGGTTGCGCATGCAGCTCATGCGGGAGCAGGCGCAGCAGGAGGAGCAGCGGGAGCGCATGCAGC AACAGGCTGTCATGCATTACATGCAGCAGCAGCAGCAGCAGCAACAGCAGCAGCTCGGAGGGCCGCCCACCCCGGCCATCAATACCCCCG TCCACTTCCAGTCGCCACCACCTGTGCCTGGGGAGGTGTTGAAGGTGCAGTCCTACCTGGAGAATCCCACATCCTACCATCTGCAGCAGT CGCAGCATCAGAAGGTGCGGGAGTACCTGTCCGAGACCTATGGGAACAAGTTTGCTGCCCACATCAGCCCAGCCCAGGGCTCTCCGAAAC CCCCACCAGCCGCCTCCCCAGGGGTGCGAGCTGGACACGTGCTGTCCTCCTCCGCTGGCAACAGTGCTCCCAATAGCCCCATGGCCATGC TGCACATTGGCTCCAACCCTGAGAGGGAGTTGGATGATGTCATTGACAACATTATGCGTCTGGACGATGTCCTTGGCTACATCAATCCTG AAATGCAGATGCCCAACACGCTACCCCTGTCCAGCAGCCACCTGAATGTGTACAGCAGCGACCCCCAGGTCACAGCCTCCCTGGTGGGCG TCACCAGCAGCTCCTGCCCTGCGGACCTGACCCAGAAGCGAGAGCTCACAGATGCTGAGAGCAGGGCCCTGGCCAAGGAGCGGCAGAAGA AAGACAATCACAACTTAATTGAAAGGAGACGAAGGTTCAACATCAATGACCGCATCAAGGAGTTGGGAATGCTGATCCCCAAGGCCAATG ACCTGGACGTGCGCTGGAACAAGGGCACCATCCTCAAGGCCTCTGTGGATTACATCCGGAGGATGCAGAAGGACCTGCAAAAGTCCAGGG AGCTGGAGAACCACTCTCGCCGCCTGGAGATGACCAACAAGCAGCTCTGGCTCCGTATCCAGGCAGCCAAGGGCAGTTTCCACTAACACA GAATGTAACCGTTGTTGAAGGTGGAACTGCAATTTTGACCTGCAGGGTTGATCAAAATGATAACACCTCCCTCCAGTGGTCAAATCCAGC TCAACAGACTCTGTACTTTGACGACAAGAAAGCTTTAAGGGACAATAGGATCGAGCTGGTTCGCGCTTCCTGGCATGAATTGAGTATTAG TGTCAGTGATGTGTCTCTCTCTGATGAAGGACAGTACACCTGTTCTTTATTTACAATGCCTGTCAAAACTTCCAAGGCATATCTCACCGT TCTGGGTGTTCCTGAAAAGCCTCAGATTAGTGGATTCTCATCACCAGTTATGGAGGGTGACTTGATGCAGCTGACTTGCAAAACATCTGG TAGTAAACCTGCAGCTGATATAAGATGGTTCAAAAATGACAAAGAGATTAAAGATGTAAAATATTTAAAAGAAGAGGATGCAAATCGCAA GACATTCACTGTCAGCAGCACACTGGACTTCCGAGTGGACCGGAGTGATGATGGAGTGGCGGTCATCTGCAGAGTAGATCACGAATCCCT CAATGCCACCCCTCAGGTAGCCATGCAGGTGCTAGAAATACACTATACACCATCAGTTAAGATTATACCATCGACTCCTTTTCCACAAGA AGGACAGCCTTTAATTTTGACTTGTGAATCCAAAGGAAAACCACTGCCAGAACCTGTTTTGTGGACAAAGGATGGCGGAGAATTACCAGA TCCTGACCGAATGGTTGTGAGTGGTAGGGAGCTAAACATTCTTTTCCTGAACAAAACGGATAATGGTACATATCGATGTGAAGCCACAAA CACCATTGGCCAAAGCAGTGCGGAATATGTTCTCATTGTGCATGATGTTCCCAACACTTTGCTTCCCACTACTATCATCCCCTCCCTTAC CACTGCAACAGTCACAACCACTGTAGCCATAACAACCAGCCCAACCACATCTGCAACAACCAGCAGCATCAGAGATCCTAATGCTTTGGC TGGCCAGAATGGCCCTGACCATGCTCTCATAGGAGGAATAGTGGCTGTAGTTGTATTTGTCACGCTGTGTTCTATCTTTCTGCTTGGTCG ATATCTGGCAAGGCATAAAGGAACGTATTTAACAAATGAAGCTAAAGGAGCTGAAGATGCACCAGATGCTGATACAGCCATTATCAATGC TGAAGGCAGCCAAGTCAATGCTGAAGAGAAAAAAGAGTATTTCATTTAAGATGCAGGCCAAGATTCTGAGTTTTACTACCAGGCTGAATG >90322_90322_3_TFEB-CADM2_TFEB_chr6_41653828_ENST00000373033_CADM2_chr3_85851197_ENST00000407528_length(amino acids)=461AA_BP=46 MEQGHHPQGLCGLHPEDAEGPAKVQGAGEPLSPPGDDQQAALAPYPGSQGQFPLTQNVTVVEGGTAILTCRVDQNDNTSLQWSNPAQQTL YFDDKKALRDNRIELVRASWHELSISVSDVSLSDEGQYTCSLFTMPVKTSKAYLTVLGVPEKPQISGFSSPVMEGDLMQLTCKTSGSKPA ADIRWFKNDKEIKDVKYLKEEDANRKTFTVSSTLDFRVDRSDDGVAVICRVDHESLNATPQVAMQVLEIHYTPSVKIIPSTPFPQEGQPL ILTCESKGKPLPEPVLWTKDGGELPDPDRMVVSGRELNILFLNKTDNGTYRCEATNTIGQSSAEYVLIVHDVPNTLLPTTIIPSLTTATV TTTVAITTSPTTSATTSSIRDPNALAGQNGPDHALIGGIVAVVVFVTLCSIFLLGRYLARHKGTYLTNEAKGAEDAPDADTAIINAEGSQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TFEB-CADM2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TFEB-CADM2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TFEB-CADM2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
