
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TGFBR2-XPC (FusionGDB2 ID:90466) |
Fusion Gene Summary for TGFBR2-XPC |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TGFBR2-XPC | Fusion gene ID: 90466 | Hgene | Tgene | Gene symbol | TGFBR2 | XPC | Gene ID | 7048 | 7508 |
| Gene name | transforming growth factor beta receptor 2 | XPC complex subunit, DNA damage recognition and repair factor | |
| Synonyms | AAT3|FAA3|LDS1B|LDS2|LDS2B|MFS2|RIIC|TAAD2|TBR-ii|TBRII|TGFR-2|TGFbeta-RII | RAD4|XP3|XPCC|p125 | |
| Cytomap | 3p24.1 | 3p25.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | TGF-beta receptor type-2TGF-beta receptor type IIBTGF-beta type II receptortbetaR-IItransforming growth factor beta receptor IItransforming growth factor beta receptor type IICtransforming growth factor, beta receptor II (70/80kDa)transforming grow | DNA repair protein complementing XP-C cellsmutant xeroderma pigmentosum group Cxeroderma pigmentosum, complementation group C | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000295754, ENST00000359013, | ENST00000285021, ENST00000449060, | |
| Fusion gene scores | * DoF score | 9 X 6 X 7=378 | 5 X 4 X 4=80 |
| # samples | 9 | 5 | |
| ** MAII score | log2(9/378*10)=-2.0703893278914 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/80*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TGFBR2 [Title/Abstract] AND XPC [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TGFBR2(30648469)-XPC(14214562), # samples:4 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TGFBR2 | GO:0006468 | protein phosphorylation | 12015308 |
| Hgene | TGFBR2 | GO:0006915 | apoptotic process | 17471240 |
| Hgene | TGFBR2 | GO:0007179 | transforming growth factor beta receptor signaling pathway | 18453574 |
| Hgene | TGFBR2 | GO:0010718 | positive regulation of epithelial to mesenchymal transition | 26459119 |
| Hgene | TGFBR2 | GO:0018105 | peptidyl-serine phosphorylation | 12015308 |
| Hgene | TGFBR2 | GO:0018107 | peptidyl-threonine phosphorylation | 12015308 |
| Hgene | TGFBR2 | GO:0042493 | response to drug | 17878231 |
| Hgene | TGFBR2 | GO:0060389 | pathway-restricted SMAD protein phosphorylation | 18453574 |
| Hgene | TGFBR2 | GO:0070723 | response to cholesterol | 17878231 |
| Hgene | TGFBR2 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation | 17164348 |
| Tgene | XPC | GO:0000715 | nucleotide-excision repair, DNA damage recognition | 10873465|19941824 |
| Tgene | XPC | GO:0006289 | nucleotide-excision repair | 8168482|9734359|11259578 |
| Tgene | XPC | GO:0045893 | positive regulation of transcription, DNA-templated | 29973595|31527837 |
| Tgene | XPC | GO:0070914 | UV-damage excision repair | 8077226 |
 Fusion gene breakpoints across TGFBR2 (5'-gene) Fusion gene breakpoints across TGFBR2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
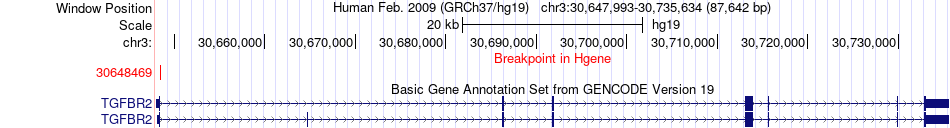 |
 Fusion gene breakpoints across XPC (3'-gene) Fusion gene breakpoints across XPC (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
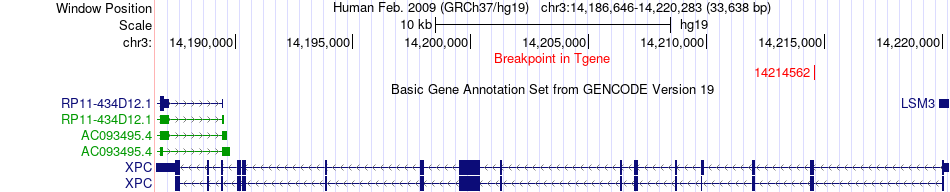 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A1L2-01A | TGFBR2 | chr3 | 30648469 | - | XPC | chr3 | 14214562 | - |
| ChimerDB4 | SARC | TCGA-DX-A1L2-01A | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
| ChimerDB4 | SARC | TCGA-DX-A1L2 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
| ChimerDB4 | SARC | TCGA-DX-A8BJ-01A | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
Top |
Fusion Gene ORF analysis for TGFBR2-XPC |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000295754 | ENST00000285021 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
| In-frame | ENST00000295754 | ENST00000449060 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
| In-frame | ENST00000359013 | ENST00000285021 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
| In-frame | ENST00000359013 | ENST00000449060 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000295754 | TGFBR2 | chr3 | 30648469 | + | ENST00000285021 | XPC | chr3 | 14214562 | - | 3990 | 476 | 382 | 3195 | 937 |
| ENST00000295754 | TGFBR2 | chr3 | 30648469 | + | ENST00000449060 | XPC | chr3 | 14214562 | - | 3085 | 476 | 382 | 3084 | 901 |
| ENST00000359013 | TGFBR2 | chr3 | 30648469 | + | ENST00000285021 | XPC | chr3 | 14214562 | - | 3891 | 377 | 283 | 3096 | 937 |
| ENST00000359013 | TGFBR2 | chr3 | 30648469 | + | ENST00000449060 | XPC | chr3 | 14214562 | - | 2986 | 377 | 283 | 2985 | 901 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000295754 | ENST00000285021 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - | 0.001081785 | 0.9989183 |
| ENST00000295754 | ENST00000449060 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - | 0.002735619 | 0.9972644 |
| ENST00000359013 | ENST00000285021 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - | 0.001120591 | 0.9988794 |
| ENST00000359013 | ENST00000449060 | TGFBR2 | chr3 | 30648469 | + | XPC | chr3 | 14214562 | - | 0.00286506 | 0.9971349 |
Top |
Fusion Genomic Features for TGFBR2-XPC |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
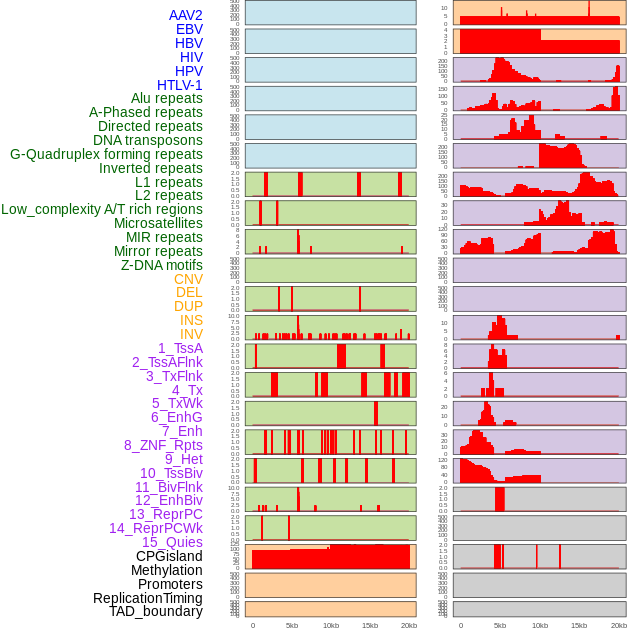 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TGFBR2-XPC |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr3:30648469/chr3:14214562) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 124_130 | 34 | 941.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 359_395 | 34 | 941.0 | Compositional bias | Note=Lys-rich (basic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 408_431 | 34 | 941.0 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 432_461 | 34 | 941.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 466_493 | 34 | 941.0 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 501_507 | 34 | 941.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 124_130 | 34 | 904.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 359_395 | 34 | 904.0 | Compositional bias | Note=Lys-rich (basic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 408_431 | 34 | 904.0 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 432_461 | 34 | 904.0 | Compositional bias | Note=Asp/Glu-rich (acidic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 466_493 | 34 | 904.0 | Compositional bias | Note=Arg/Lys-rich (basic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 501_507 | 34 | 904.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 390_395 | 34 | 941.0 | Motif | Nuclear localization signal | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 390_395 | 34 | 904.0 | Motif | Nuclear localization signal | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 607_741 | 34 | 941.0 | Region | Note=DNA-binding%3B preference for heteroduplex DNA | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 607_766 | 34 | 941.0 | Region | Note=Minimal sensor domain involved in damage recognition | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 767_831 | 34 | 941.0 | Region | Note=DNA-binding%3B preference for single stranded DNA%3B required for formation of stable nucleoprotein complex | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 607_741 | 34 | 904.0 | Region | Note=DNA-binding%3B preference for heteroduplex DNA | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 607_766 | 34 | 904.0 | Region | Note=Minimal sensor domain involved in damage recognition | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 767_831 | 34 | 904.0 | Region | Note=DNA-binding%3B preference for single stranded DNA%3B required for formation of stable nucleoprotein complex |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000295754 | + | 1 | 7 | 244_544 | 31 | 568.0 | Domain | Protein kinase |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000359013 | + | 1 | 8 | 244_544 | 31 | 593.0 | Domain | Protein kinase |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000295754 | + | 1 | 7 | 250_258 | 31 | 568.0 | Nucleotide binding | ATP |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000359013 | + | 1 | 8 | 250_258 | 31 | 593.0 | Nucleotide binding | ATP |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000295754 | + | 1 | 7 | 188_567 | 31 | 568.0 | Topological domain | Cytoplasmic |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000295754 | + | 1 | 7 | 23_166 | 31 | 568.0 | Topological domain | Extracellular |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000359013 | + | 1 | 8 | 188_567 | 31 | 593.0 | Topological domain | Cytoplasmic |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000359013 | + | 1 | 8 | 23_166 | 31 | 593.0 | Topological domain | Extracellular |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000295754 | + | 1 | 7 | 167_187 | 31 | 568.0 | Transmembrane | Helical |
| Hgene | TGFBR2 | chr3:30648469 | chr3:14214562 | ENST00000359013 | + | 1 | 8 | 167_187 | 31 | 593.0 | Transmembrane | Helical |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 30_177 | 34 | 941.0 | Compositional bias | Note=Glu-rich (acidic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 30_34 | 34 | 941.0 | Compositional bias | Note=Poly-Glu | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 30_177 | 34 | 904.0 | Compositional bias | Note=Glu-rich (acidic) | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 30_34 | 34 | 904.0 | Compositional bias | Note=Poly-Glu |
Top |
Fusion Gene Sequence for TGFBR2-XPC |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >90466_90466_1_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000295754_XPC_chr3_14214562_ENST00000285021_length(transcript)=3990nt_BP=476nt GGAGAGGGAGAAGGCTCTCGGGCGGAGAGAGGTCCTGCCCAGCTGTTGGCGAGGAGTTTCCTGTTTCCCCCGCAGCGCTGAGTTGAAGTT GAGTGAGTCACTCGCGCGCACGGAGCGACGACACCCCCGCGCGTGCACCCGCTCGGGACAGGAGCCGGACTCCTGTGCAGCTTCCCTCGG CCGCCGGGGGCCTCCCCGCGCCTCGCCGGCCTCCAGGCCCCCTCCTGGCTGGCGAGCGGGCGCCACATCTGGCCCGCACATCTGCGCTGC CGGCCCGGCGCGGGGTCCGGAGAGGGCGCGGCGCGGAGGCGCAGCCAGGGGTCCGGGAAGGCGCCGTCCGCTGCGCTGGGGGCTCGGTCT ATGACGAGCAGCGGGGTCTGCCATGGGTCGGGGGCTGCTCAGGGGCCTGTGGCCGCTGCACATCGTCCTGTGGACGCGTATCGCCAGCAC GATCCCACCGCACGTTCAGAAGTCGGATGCCTTTGAAGATGAGAAACCCCCAAAGAAGAGCCTTCTCTCCAAAGTTTCACAAGGAAAGAG GAAAAGAGGCTGCAGTCATCCTGGGGGTTCAGCAGATGGTCCAGCAAAAAAGAAAGTGGCCAAGGTGACTGTTAAATCTGAAAACCTCAA GGTTATAAAGGATGAAGCCCTCAGCGATGGGGATGACCTCAGGGACTTTCCAAGTGACCTCAAGAAGGCACACCATCTGAAGAGAGGGGC TACCATGAATGAAGACAGCAATGAAGAAGAGGAAGAAAGTGAAAATGATTGGGAAGAGGTTGAAGAACTTAGTGAGCCTGTGCTGGGTGA CGTGAGAGAAAGTACAGCCTTCTCTCGATCTCTTCTGCCTGTGAAGCCAGTGGAGATAGAGATTGAAACGCCAGAGCAGGCGAAGACAAG AGAAAGAAGTGAAAAGATAAAACTGGAGTTTGAGACATATCTTCGGAGGGCGATGAAACGTTTCAATAAAGGGGTCCATGAGGACACACA CAAGGTTCACCTTCTCTGCCTGCTAGCAAATGGCTTCTATCGAAATAACATCTGCAGCCAGCCAGATCTGCATGCTATTGGCCTGTCCAT CATCCCAGCCCGCTTTACCAGAGTGCTGCCTCGAGATGTGGACACCTACTACCTCTCAAACCTGGTGAAGTGGTTCATTGGAACATTTAC AGTTAATGCAGAACTTTCAGCCAGTGAACAAGATAACCTGCAGACTACATTGGAAAGGAGATTTGCTATTTACTCTGCTCGAGATGATGA GGAATTGGTCCATATATTCTTACTGATTCTCCGGGCTCTGCAGCTCTTGACCCGGCTGGTATTGTCTCTACAGCCAATTCCTCTGAAGTC AGCAACAGCAAAGGGAAAGAAACCTTCCAAGGAAAGATTGACTGCGGATCCAGGAGGCTCCTCAGAAACTTCCAGCCAAGTTCTAGAAAA CCACACCAAACCAAAGACCAGCAAAGGAACCAAACAAGAGGAAACCTTTGCTAAGGGCACCTGCAGGCCAAGTGCCAAAGGGAAGAGGAA CAAGGGAGGCAGAAAGAAACGGAGCAAGCCCTCCTCCAGCGAGGAAGATGAGGGCCCAGGAGACAAGCAGGAGAAGGCAACCCAGCGACG TCCGCATGGCCGGGAGCGGCGGGTGGCCTCCAGGGTGTCTTATAAAGAGGAGAGTGGGAGTGATGAGGCTGGCAGCGGCTCTGATTTTGA GCTCTCCAGTGGAGAAGCCTCTGATCCCTCTGATGAGGATTCCGAACCTGGCCCTCCAAAGCAGAGGAAAGCCCCCGCTCCTCAGAGGAC AAAGGCTGGGTCCAAGAGTGCCTCCAGGACCCATCGTGGGAGCCATCGTAAGGACCCAAGCTTGCCAGCGGCATCCTCAAGCTCTTCAAG CAGTAAAAGAGGCAAGAAAATGTGCAGCGATGGTGAGAAGGCAGAAAAAAGAAGCATAGCTGGTATAGACCAGTGGCTAGAGGTGTTCTG TGAGCAGGAGGAAAAGTGGGTATGTGTAGACTGTGTGCACGGTGTGGTGGGCCAGCCTCTGACCTGTTACAAGTACGCCACCAAGCCCAT GACCTATGTGGTGGGCATTGACAGTGACGGCTGGGTCCGAGATGTCACACAGAGGTACGACCCAGTCTGGATGACAGTGACCCGCAAGTG CCGGGTTGATGCTGAGTGGTGGGCCGAGACCTTGAGACCATACCAGAGCCCATTTATGGACAGGGAGAAGAAAGAAGACTTGGAGTTTCA GGCTAAACACATGGACCAGCCTTTGCCCACTGCCATTGGCTTATATAAGAACCACCCTCTGTATGCCCTGAAGCGGCATCTCCTGAAATA TGAGGCCATCTATCCCGAGACAGCTGCCATCCTTGGGTATTGTCGTGGAGAAGCGGTCTACTCCAGGGATTGTGTGCACACTCTGCATTC CAGGGACACGTGGCTGAAGAAAGCAAGAGTGGTGAGGCTTGGAGAAGTACCCTACAAGATGGTGAAAGGCTTTTCTAACCGTGCTCGGAA AGCCCGACTTGCTGAGCCCCAGCTGCGGGAAGAAAATGACCTGGGCCTGTTTGGCTACTGGCAGACAGAGGAGTATCAGCCCCCAGTGGC CGTGGACGGGAAGGTGCCCCGGAACGAGTTTGGGAATGTGTACCTCTTCCTGCCCAGCATGATGCCTATTGGCTGTGTCCAGCTGAACCT GCCCAATCTACACCGCGTGGCCCGCAAGCTGGACATCGACTGTGTCCAGGCCATCACTGGCTTTGATTTCCATGGCGGCTACTCCCATCC CGTGACTGATGGATACATCGTCTGCGAGGAATTCAAAGACGTGCTCCTGACTGCCTGGGAAAATGAGCAGGCAGTCATTGAAAGGAAGGA GAAGGAGAAAAAGGAGAAGCGGGCTCTAGGGAACTGGAAGTTGCTGGCCAAAGGTCTGCTCATCAGGGAGAGGCTGAAGCGTCGCTACGG GCCCAAGAGTGAGGCAGCAGCTCCCCACACAGATGCAGGAGGTGGACTCTCTTCTGATGAAGAGGAGGGGACCAGCTCTCAAGCAGAAGC GGCCAGGATACTGGCTGCCTCCTGGCCTCAAAACCGAGAAGATGAAGAAAAGCAGAAGCTGAAGGGTGGGCCCAAGAAGACCAAAAGGGA AAAGAAAGCAGCAGCTTCCCACCTGTTCCCATTTGAGCAGCTGTGAGCTGAGCGCCCACTAGAGGGGCACCCACCAGTTGCTGCTGCCCC ACTACAGGCCCCACACCTGCCCTGGGCATGCCCAGCCCCTGGTGGTGGGGGCTTCTCTGCTGAGAAGGCAAACTGAGGCAGCATGCACGG AGGCGGGGTCAGGGGAGACGAGGCCAAGCTGAGGAGGTGCTGCAGGTCCCGTCTGGCTCCAGCCCTTGTCAGATTCACCCAGGGTGAAGC CTTCAAAGCTTTTTGCTACCAAAGCCCACTCACCCTTTGAGCTACAGAACACTTTGCTAGGAGATACTCTTCTGCCTCCTAGACCTGTTC TTTCCATCTTTAGAAACATCAGTTTTTGTATGGAAGCCACCGGGAGATTTCTGGATGGTGGTGCATCCGTGAATGCGCTGATCGTTTCTT CCAGTTAGAGTCTTCATCTGTCCGACAAGTTCACTCGCCTCGGTTGCGGACCTAGGACCATTTCTCTGCAGGCCACTTACCTTCCCCTGA GTCAGGCTTACTAATGCTGCCCTCACTGCCTCTTTGCAGTAGGGGAGAGAGCAGAGAAGTACAGGTCATCTGCTGGGATCTAGTTTTCCA AGTAACATTTTGTGGTGACAGAAGCCTAAAAAAAGCTAAAATCAGGAAAGAAAAGGAAAAATACGAATTGAAAATTAAGGAAATGTTAGT AAAATAGATGAGTGTTAAACTAGATTGTATTCATTACTAGATAAAATGTATAAAGCTCTCTGTACTAAGGAGAAATGACTTTTATAACAT >90466_90466_1_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000295754_XPC_chr3_14214562_ENST00000285021_length(amino acids)=937AA_BP=31 MGRGLLRGLWPLHIVLWTRIASTIPPHVQKSDAFEDEKPPKKSLLSKVSQGKRKRGCSHPGGSADGPAKKKVAKVTVKSENLKVIKDEAL SDGDDLRDFPSDLKKAHHLKRGATMNEDSNEEEEESENDWEEVEELSEPVLGDVRESTAFSRSLLPVKPVEIEIETPEQAKTRERSEKIK LEFETYLRRAMKRFNKGVHEDTHKVHLLCLLANGFYRNNICSQPDLHAIGLSIIPARFTRVLPRDVDTYYLSNLVKWFIGTFTVNAELSA SEQDNLQTTLERRFAIYSARDDEELVHIFLLILRALQLLTRLVLSLQPIPLKSATAKGKKPSKERLTADPGGSSETSSQVLENHTKPKTS KGTKQEETFAKGTCRPSAKGKRNKGGRKKRSKPSSSEEDEGPGDKQEKATQRRPHGRERRVASRVSYKEESGSDEAGSGSDFELSSGEAS DPSDEDSEPGPPKQRKAPAPQRTKAGSKSASRTHRGSHRKDPSLPAASSSSSSSKRGKKMCSDGEKAEKRSIAGIDQWLEVFCEQEEKWV CVDCVHGVVGQPLTCYKYATKPMTYVVGIDSDGWVRDVTQRYDPVWMTVTRKCRVDAEWWAETLRPYQSPFMDREKKEDLEFQAKHMDQP LPTAIGLYKNHPLYALKRHLLKYEAIYPETAAILGYCRGEAVYSRDCVHTLHSRDTWLKKARVVRLGEVPYKMVKGFSNRARKARLAEPQ LREENDLGLFGYWQTEEYQPPVAVDGKVPRNEFGNVYLFLPSMMPIGCVQLNLPNLHRVARKLDIDCVQAITGFDFHGGYSHPVTDGYIV CEEFKDVLLTAWENEQAVIERKEKEKKEKRALGNWKLLAKGLLIRERLKRRYGPKSEAAAPHTDAGGGLSSDEEEGTSSQAEAARILAAS -------------------------------------------------------------- >90466_90466_2_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000295754_XPC_chr3_14214562_ENST00000449060_length(transcript)=3085nt_BP=476nt GGAGAGGGAGAAGGCTCTCGGGCGGAGAGAGGTCCTGCCCAGCTGTTGGCGAGGAGTTTCCTGTTTCCCCCGCAGCGCTGAGTTGAAGTT GAGTGAGTCACTCGCGCGCACGGAGCGACGACACCCCCGCGCGTGCACCCGCTCGGGACAGGAGCCGGACTCCTGTGCAGCTTCCCTCGG CCGCCGGGGGCCTCCCCGCGCCTCGCCGGCCTCCAGGCCCCCTCCTGGCTGGCGAGCGGGCGCCACATCTGGCCCGCACATCTGCGCTGC CGGCCCGGCGCGGGGTCCGGAGAGGGCGCGGCGCGGAGGCGCAGCCAGGGGTCCGGGAAGGCGCCGTCCGCTGCGCTGGGGGCTCGGTCT ATGACGAGCAGCGGGGTCTGCCATGGGTCGGGGGCTGCTCAGGGGCCTGTGGCCGCTGCACATCGTCCTGTGGACGCGTATCGCCAGCAC GATCCCACCGCACGTTCAGAAGTCGGATGCCTTTGAAGATGAGAAACCCCCAAAGAAGAGCCTTCTCTCCAAAGTTTCACAAGGAAAGAG GAAAAGAGGCTGCAGTCATCCTGGGGGTTCAGCAGATGGTCCAGCAAAAAAGAAAGTGGCCAAGGTGACTGTTAAATCTGAAAACCTCAA GGTTATAAAGGATGAAGCCCTCAGCGATGGGGATGACCTCAGGGACTTTCCAAGTGACCTCAAGAAGGCACACCATCTGAAGAGAGGGGC TACCATGAATGAAGACAGCAATGAAGAAGAGGAAGAAAGTGAAAATGATTGGGAAGAGGCGAAGACAAGAGAAAGAAGTGAAAAGATAAA ACTGGAGTTTGAGACATATCTTCGGAGGGCGATGAAACGTTTCAATAAAGGGGTCCATGAGGACACACACAAGGTTCACCTTCTCTGCCT GCTAGCAAATGGCTTCTATCGAAATAACATCTGCAGCCAGCCAGATCTGCATGCTATTGGCCTGTCCATCATCCCAGCCCGCTTTACCAG AGTGCTGCCTCGAGATGTGGACACCTACTACCTCTCAAACCTGGTGAAGTGGTTCATTGGAACATTTACAGTTAATGCAGAACTTTCAGC CAGTGAACAAGATAACCTGCAGACTACATTGGAAAGGAGATTTGCTATTTACTCTGCTCGAGATGATGAGGAATTGGTCCATATATTCTT ACTGATTCTCCGGGCTCTGCAGCTCTTGACCCGGCTGGTATTGTCTCTACAGCCAATTCCTCTGAAGTCAGCAACAGCAAAGGGAAAGAA ACCTTCCAAGGAAAGATTGACTGCGGATCCAGGAGGCTCCTCAGAAACTTCCAGCCAAGTTCTAGAAAACCACACCAAACCAAAGACCAG CAAAGGAACCAAACAAGAGGAAACCTTTGCTAAGGGCACCTGCAGGCCAAGTGCCAAAGGGAAGAGGAACAAGGGAGGCAGAAAGAAACG GAGCAAGCCCTCCTCCAGCGAGGAAGATGAGGGCCCAGGAGACAAGCAGGAGAAGGCAACCCAGCGACGTCCGCATGGCCGGGAGCGGCG GGTGGCCTCCAGGGTGTCTTATAAAGAGGAGAGTGGGAGTGATGAGGCTGGCAGCGGCTCTGATTTTGAGCTCTCCAGTGGAGAAGCCTC TGATCCCTCTGATGAGGATTCCGAACCTGGCCCTCCAAAGCAGAGGAAAGCCCCCGCTCCTCAGAGGACAAAGGCTGGGTCCAAGAGTGC CTCCAGGACCCATCGTGGGAGCCATCGTAAGGACCCAAGCTTGCCAGCGGCATCCTCAAGCTCTTCAAGCAGTAAAAGAGGCAAGAAAAT GTGCAGCGATGGTGAGAAGGCAGAAAAAAGAAGCATAGCTGGTATAGACCAGTGGCTAGAGGTGTTCTGTGAGCAGGAGGAAAAGTGGGT ATGTGTAGACTGTGTGCACGGTGTGGTGGGCCAGCCTCTGACCTGTTACAAGTACGCCACCAAGCCCATGACCTATGTGGTGGGCATTGA CAGTGACGGCTGGGTCCGAGATGTCACACAGAGGTACGACCCAGTCTGGATGACAGTGACCCGCAAGTGCCGGGTTGATGCTGAGTGGTG GGCCGAGACCTTGAGACCATACCAGAGCCCATTTATGGACAGGGAGAAGAAAGAAGACTTGGAGTTTCAGGCTAAACACATGGACCAGCC TTTGCCCACTGCCATTGGCTTATATAAGAACCACCCTCTGTATGCCCTGAAGCGGCATCTCCTGAAATATGAGGCCATCTATCCCGAGAC AGCTGCCATCCTTGGGTATTGTCGTGGAGAAGCGGTCTACTCCAGGGATTGTGTGCACACTCTGCATTCCAGGGACACGTGGCTGAAGAA AGCAAGAGTGGTGAGGCTTGGAGAAGTACCCTACAAGATGGTGAAAGGCTTTTCTAACCGTGCTCGGAAAGCCCGACTTGCTGAGCCCCA GCTGCGGGAAGAAAATGACCTGGGCCTGTTTGGCTACTGGCAGACAGAGGAGTATCAGCCCCCAGTGGCCGTGGACGGGAAGGTGCCCCG GAACGAGTTTGGGAATGTGTACCTCTTCCTGCCCAGCATGATGCCTATTGGCTGTGTCCAGCTGAACCTGCCCAATCTACACCGCGTGGC CCGCAAGCTGGACATCGACTGTGTCCAGGCCATCACTGGCTTTGATTTCCATGGCGGCTACTCCCATCCCGTGACTGATGGATACATCGT CTGCGAGGAATTCAAAGACGTGCTCCTGACTGCCTGGGAAAATGAGCAGGCAGTCATTGAAAGGAAGGAGAAGGAGAAAAAGGAGAAGCG GGCTCTAGGGAACTGGAAGTTGCTGGCCAAAGGTCTGCTCATCAGGGAGAGGCTGAAGCGTCGCTACGGGCCCAAGAGTGAGGCAGCAGC TCCCCACACAGATGCAGGAGGTGGACTCTCTTCTGATGAAGAGGAGGGGACCAGCTCTCAAGCAGAAGCGGCCAGGATACTGGCTGCCTC CTGGCCTCAAAACCGAGAAGATGAAGAAAAGCAGAAGCTGAAGGGTGGGCCCAAGAAGACCAAAAGGGAAAAGAAAGCAGCAGCTTCCCA >90466_90466_2_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000295754_XPC_chr3_14214562_ENST00000449060_length(amino acids)=901AA_BP=31 MGRGLLRGLWPLHIVLWTRIASTIPPHVQKSDAFEDEKPPKKSLLSKVSQGKRKRGCSHPGGSADGPAKKKVAKVTVKSENLKVIKDEAL SDGDDLRDFPSDLKKAHHLKRGATMNEDSNEEEEESENDWEEAKTRERSEKIKLEFETYLRRAMKRFNKGVHEDTHKVHLLCLLANGFYR NNICSQPDLHAIGLSIIPARFTRVLPRDVDTYYLSNLVKWFIGTFTVNAELSASEQDNLQTTLERRFAIYSARDDEELVHIFLLILRALQ LLTRLVLSLQPIPLKSATAKGKKPSKERLTADPGGSSETSSQVLENHTKPKTSKGTKQEETFAKGTCRPSAKGKRNKGGRKKRSKPSSSE EDEGPGDKQEKATQRRPHGRERRVASRVSYKEESGSDEAGSGSDFELSSGEASDPSDEDSEPGPPKQRKAPAPQRTKAGSKSASRTHRGS HRKDPSLPAASSSSSSSKRGKKMCSDGEKAEKRSIAGIDQWLEVFCEQEEKWVCVDCVHGVVGQPLTCYKYATKPMTYVVGIDSDGWVRD VTQRYDPVWMTVTRKCRVDAEWWAETLRPYQSPFMDREKKEDLEFQAKHMDQPLPTAIGLYKNHPLYALKRHLLKYEAIYPETAAILGYC RGEAVYSRDCVHTLHSRDTWLKKARVVRLGEVPYKMVKGFSNRARKARLAEPQLREENDLGLFGYWQTEEYQPPVAVDGKVPRNEFGNVY LFLPSMMPIGCVQLNLPNLHRVARKLDIDCVQAITGFDFHGGYSHPVTDGYIVCEEFKDVLLTAWENEQAVIERKEKEKKEKRALGNWKL LAKGLLIRERLKRRYGPKSEAAAPHTDAGGGLSSDEEEGTSSQAEAARILAASWPQNREDEEKQKLKGGPKKTKREKKAAASHLFPFEQL -------------------------------------------------------------- >90466_90466_3_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000359013_XPC_chr3_14214562_ENST00000285021_length(transcript)=3891nt_BP=377nt ACTCGCGCGCACGGAGCGACGACACCCCCGCGCGTGCACCCGCTCGGGACAGGAGCCGGACTCCTGTGCAGCTTCCCTCGGCCGCCGGGG GCCTCCCCGCGCCTCGCCGGCCTCCAGGCCCCCTCCTGGCTGGCGAGCGGGCGCCACATCTGGCCCGCACATCTGCGCTGCCGGCCCGGC GCGGGGTCCGGAGAGGGCGCGGCGCGGAGGCGCAGCCAGGGGTCCGGGAAGGCGCCGTCCGCTGCGCTGGGGGCTCGGTCTATGACGAGC AGCGGGGTCTGCCATGGGTCGGGGGCTGCTCAGGGGCCTGTGGCCGCTGCACATCGTCCTGTGGACGCGTATCGCCAGCACGATCCCACC GCACGTTCAGAAGTCGGATGCCTTTGAAGATGAGAAACCCCCAAAGAAGAGCCTTCTCTCCAAAGTTTCACAAGGAAAGAGGAAAAGAGG CTGCAGTCATCCTGGGGGTTCAGCAGATGGTCCAGCAAAAAAGAAAGTGGCCAAGGTGACTGTTAAATCTGAAAACCTCAAGGTTATAAA GGATGAAGCCCTCAGCGATGGGGATGACCTCAGGGACTTTCCAAGTGACCTCAAGAAGGCACACCATCTGAAGAGAGGGGCTACCATGAA TGAAGACAGCAATGAAGAAGAGGAAGAAAGTGAAAATGATTGGGAAGAGGTTGAAGAACTTAGTGAGCCTGTGCTGGGTGACGTGAGAGA AAGTACAGCCTTCTCTCGATCTCTTCTGCCTGTGAAGCCAGTGGAGATAGAGATTGAAACGCCAGAGCAGGCGAAGACAAGAGAAAGAAG TGAAAAGATAAAACTGGAGTTTGAGACATATCTTCGGAGGGCGATGAAACGTTTCAATAAAGGGGTCCATGAGGACACACACAAGGTTCA CCTTCTCTGCCTGCTAGCAAATGGCTTCTATCGAAATAACATCTGCAGCCAGCCAGATCTGCATGCTATTGGCCTGTCCATCATCCCAGC CCGCTTTACCAGAGTGCTGCCTCGAGATGTGGACACCTACTACCTCTCAAACCTGGTGAAGTGGTTCATTGGAACATTTACAGTTAATGC AGAACTTTCAGCCAGTGAACAAGATAACCTGCAGACTACATTGGAAAGGAGATTTGCTATTTACTCTGCTCGAGATGATGAGGAATTGGT CCATATATTCTTACTGATTCTCCGGGCTCTGCAGCTCTTGACCCGGCTGGTATTGTCTCTACAGCCAATTCCTCTGAAGTCAGCAACAGC AAAGGGAAAGAAACCTTCCAAGGAAAGATTGACTGCGGATCCAGGAGGCTCCTCAGAAACTTCCAGCCAAGTTCTAGAAAACCACACCAA ACCAAAGACCAGCAAAGGAACCAAACAAGAGGAAACCTTTGCTAAGGGCACCTGCAGGCCAAGTGCCAAAGGGAAGAGGAACAAGGGAGG CAGAAAGAAACGGAGCAAGCCCTCCTCCAGCGAGGAAGATGAGGGCCCAGGAGACAAGCAGGAGAAGGCAACCCAGCGACGTCCGCATGG CCGGGAGCGGCGGGTGGCCTCCAGGGTGTCTTATAAAGAGGAGAGTGGGAGTGATGAGGCTGGCAGCGGCTCTGATTTTGAGCTCTCCAG TGGAGAAGCCTCTGATCCCTCTGATGAGGATTCCGAACCTGGCCCTCCAAAGCAGAGGAAAGCCCCCGCTCCTCAGAGGACAAAGGCTGG GTCCAAGAGTGCCTCCAGGACCCATCGTGGGAGCCATCGTAAGGACCCAAGCTTGCCAGCGGCATCCTCAAGCTCTTCAAGCAGTAAAAG AGGCAAGAAAATGTGCAGCGATGGTGAGAAGGCAGAAAAAAGAAGCATAGCTGGTATAGACCAGTGGCTAGAGGTGTTCTGTGAGCAGGA GGAAAAGTGGGTATGTGTAGACTGTGTGCACGGTGTGGTGGGCCAGCCTCTGACCTGTTACAAGTACGCCACCAAGCCCATGACCTATGT GGTGGGCATTGACAGTGACGGCTGGGTCCGAGATGTCACACAGAGGTACGACCCAGTCTGGATGACAGTGACCCGCAAGTGCCGGGTTGA TGCTGAGTGGTGGGCCGAGACCTTGAGACCATACCAGAGCCCATTTATGGACAGGGAGAAGAAAGAAGACTTGGAGTTTCAGGCTAAACA CATGGACCAGCCTTTGCCCACTGCCATTGGCTTATATAAGAACCACCCTCTGTATGCCCTGAAGCGGCATCTCCTGAAATATGAGGCCAT CTATCCCGAGACAGCTGCCATCCTTGGGTATTGTCGTGGAGAAGCGGTCTACTCCAGGGATTGTGTGCACACTCTGCATTCCAGGGACAC GTGGCTGAAGAAAGCAAGAGTGGTGAGGCTTGGAGAAGTACCCTACAAGATGGTGAAAGGCTTTTCTAACCGTGCTCGGAAAGCCCGACT TGCTGAGCCCCAGCTGCGGGAAGAAAATGACCTGGGCCTGTTTGGCTACTGGCAGACAGAGGAGTATCAGCCCCCAGTGGCCGTGGACGG GAAGGTGCCCCGGAACGAGTTTGGGAATGTGTACCTCTTCCTGCCCAGCATGATGCCTATTGGCTGTGTCCAGCTGAACCTGCCCAATCT ACACCGCGTGGCCCGCAAGCTGGACATCGACTGTGTCCAGGCCATCACTGGCTTTGATTTCCATGGCGGCTACTCCCATCCCGTGACTGA TGGATACATCGTCTGCGAGGAATTCAAAGACGTGCTCCTGACTGCCTGGGAAAATGAGCAGGCAGTCATTGAAAGGAAGGAGAAGGAGAA AAAGGAGAAGCGGGCTCTAGGGAACTGGAAGTTGCTGGCCAAAGGTCTGCTCATCAGGGAGAGGCTGAAGCGTCGCTACGGGCCCAAGAG TGAGGCAGCAGCTCCCCACACAGATGCAGGAGGTGGACTCTCTTCTGATGAAGAGGAGGGGACCAGCTCTCAAGCAGAAGCGGCCAGGAT ACTGGCTGCCTCCTGGCCTCAAAACCGAGAAGATGAAGAAAAGCAGAAGCTGAAGGGTGGGCCCAAGAAGACCAAAAGGGAAAAGAAAGC AGCAGCTTCCCACCTGTTCCCATTTGAGCAGCTGTGAGCTGAGCGCCCACTAGAGGGGCACCCACCAGTTGCTGCTGCCCCACTACAGGC CCCACACCTGCCCTGGGCATGCCCAGCCCCTGGTGGTGGGGGCTTCTCTGCTGAGAAGGCAAACTGAGGCAGCATGCACGGAGGCGGGGT CAGGGGAGACGAGGCCAAGCTGAGGAGGTGCTGCAGGTCCCGTCTGGCTCCAGCCCTTGTCAGATTCACCCAGGGTGAAGCCTTCAAAGC TTTTTGCTACCAAAGCCCACTCACCCTTTGAGCTACAGAACACTTTGCTAGGAGATACTCTTCTGCCTCCTAGACCTGTTCTTTCCATCT TTAGAAACATCAGTTTTTGTATGGAAGCCACCGGGAGATTTCTGGATGGTGGTGCATCCGTGAATGCGCTGATCGTTTCTTCCAGTTAGA GTCTTCATCTGTCCGACAAGTTCACTCGCCTCGGTTGCGGACCTAGGACCATTTCTCTGCAGGCCACTTACCTTCCCCTGAGTCAGGCTT ACTAATGCTGCCCTCACTGCCTCTTTGCAGTAGGGGAGAGAGCAGAGAAGTACAGGTCATCTGCTGGGATCTAGTTTTCCAAGTAACATT TTGTGGTGACAGAAGCCTAAAAAAAGCTAAAATCAGGAAAGAAAAGGAAAAATACGAATTGAAAATTAAGGAAATGTTAGTAAAATAGAT GAGTGTTAAACTAGATTGTATTCATTACTAGATAAAATGTATAAAGCTCTCTGTACTAAGGAGAAATGACTTTTATAACATTTTGAGAAA >90466_90466_3_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000359013_XPC_chr3_14214562_ENST00000285021_length(amino acids)=937AA_BP=31 MGRGLLRGLWPLHIVLWTRIASTIPPHVQKSDAFEDEKPPKKSLLSKVSQGKRKRGCSHPGGSADGPAKKKVAKVTVKSENLKVIKDEAL SDGDDLRDFPSDLKKAHHLKRGATMNEDSNEEEEESENDWEEVEELSEPVLGDVRESTAFSRSLLPVKPVEIEIETPEQAKTRERSEKIK LEFETYLRRAMKRFNKGVHEDTHKVHLLCLLANGFYRNNICSQPDLHAIGLSIIPARFTRVLPRDVDTYYLSNLVKWFIGTFTVNAELSA SEQDNLQTTLERRFAIYSARDDEELVHIFLLILRALQLLTRLVLSLQPIPLKSATAKGKKPSKERLTADPGGSSETSSQVLENHTKPKTS KGTKQEETFAKGTCRPSAKGKRNKGGRKKRSKPSSSEEDEGPGDKQEKATQRRPHGRERRVASRVSYKEESGSDEAGSGSDFELSSGEAS DPSDEDSEPGPPKQRKAPAPQRTKAGSKSASRTHRGSHRKDPSLPAASSSSSSSKRGKKMCSDGEKAEKRSIAGIDQWLEVFCEQEEKWV CVDCVHGVVGQPLTCYKYATKPMTYVVGIDSDGWVRDVTQRYDPVWMTVTRKCRVDAEWWAETLRPYQSPFMDREKKEDLEFQAKHMDQP LPTAIGLYKNHPLYALKRHLLKYEAIYPETAAILGYCRGEAVYSRDCVHTLHSRDTWLKKARVVRLGEVPYKMVKGFSNRARKARLAEPQ LREENDLGLFGYWQTEEYQPPVAVDGKVPRNEFGNVYLFLPSMMPIGCVQLNLPNLHRVARKLDIDCVQAITGFDFHGGYSHPVTDGYIV CEEFKDVLLTAWENEQAVIERKEKEKKEKRALGNWKLLAKGLLIRERLKRRYGPKSEAAAPHTDAGGGLSSDEEEGTSSQAEAARILAAS -------------------------------------------------------------- >90466_90466_4_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000359013_XPC_chr3_14214562_ENST00000449060_length(transcript)=2986nt_BP=377nt ACTCGCGCGCACGGAGCGACGACACCCCCGCGCGTGCACCCGCTCGGGACAGGAGCCGGACTCCTGTGCAGCTTCCCTCGGCCGCCGGGG GCCTCCCCGCGCCTCGCCGGCCTCCAGGCCCCCTCCTGGCTGGCGAGCGGGCGCCACATCTGGCCCGCACATCTGCGCTGCCGGCCCGGC GCGGGGTCCGGAGAGGGCGCGGCGCGGAGGCGCAGCCAGGGGTCCGGGAAGGCGCCGTCCGCTGCGCTGGGGGCTCGGTCTATGACGAGC AGCGGGGTCTGCCATGGGTCGGGGGCTGCTCAGGGGCCTGTGGCCGCTGCACATCGTCCTGTGGACGCGTATCGCCAGCACGATCCCACC GCACGTTCAGAAGTCGGATGCCTTTGAAGATGAGAAACCCCCAAAGAAGAGCCTTCTCTCCAAAGTTTCACAAGGAAAGAGGAAAAGAGG CTGCAGTCATCCTGGGGGTTCAGCAGATGGTCCAGCAAAAAAGAAAGTGGCCAAGGTGACTGTTAAATCTGAAAACCTCAAGGTTATAAA GGATGAAGCCCTCAGCGATGGGGATGACCTCAGGGACTTTCCAAGTGACCTCAAGAAGGCACACCATCTGAAGAGAGGGGCTACCATGAA TGAAGACAGCAATGAAGAAGAGGAAGAAAGTGAAAATGATTGGGAAGAGGCGAAGACAAGAGAAAGAAGTGAAAAGATAAAACTGGAGTT TGAGACATATCTTCGGAGGGCGATGAAACGTTTCAATAAAGGGGTCCATGAGGACACACACAAGGTTCACCTTCTCTGCCTGCTAGCAAA TGGCTTCTATCGAAATAACATCTGCAGCCAGCCAGATCTGCATGCTATTGGCCTGTCCATCATCCCAGCCCGCTTTACCAGAGTGCTGCC TCGAGATGTGGACACCTACTACCTCTCAAACCTGGTGAAGTGGTTCATTGGAACATTTACAGTTAATGCAGAACTTTCAGCCAGTGAACA AGATAACCTGCAGACTACATTGGAAAGGAGATTTGCTATTTACTCTGCTCGAGATGATGAGGAATTGGTCCATATATTCTTACTGATTCT CCGGGCTCTGCAGCTCTTGACCCGGCTGGTATTGTCTCTACAGCCAATTCCTCTGAAGTCAGCAACAGCAAAGGGAAAGAAACCTTCCAA GGAAAGATTGACTGCGGATCCAGGAGGCTCCTCAGAAACTTCCAGCCAAGTTCTAGAAAACCACACCAAACCAAAGACCAGCAAAGGAAC CAAACAAGAGGAAACCTTTGCTAAGGGCACCTGCAGGCCAAGTGCCAAAGGGAAGAGGAACAAGGGAGGCAGAAAGAAACGGAGCAAGCC CTCCTCCAGCGAGGAAGATGAGGGCCCAGGAGACAAGCAGGAGAAGGCAACCCAGCGACGTCCGCATGGCCGGGAGCGGCGGGTGGCCTC CAGGGTGTCTTATAAAGAGGAGAGTGGGAGTGATGAGGCTGGCAGCGGCTCTGATTTTGAGCTCTCCAGTGGAGAAGCCTCTGATCCCTC TGATGAGGATTCCGAACCTGGCCCTCCAAAGCAGAGGAAAGCCCCCGCTCCTCAGAGGACAAAGGCTGGGTCCAAGAGTGCCTCCAGGAC CCATCGTGGGAGCCATCGTAAGGACCCAAGCTTGCCAGCGGCATCCTCAAGCTCTTCAAGCAGTAAAAGAGGCAAGAAAATGTGCAGCGA TGGTGAGAAGGCAGAAAAAAGAAGCATAGCTGGTATAGACCAGTGGCTAGAGGTGTTCTGTGAGCAGGAGGAAAAGTGGGTATGTGTAGA CTGTGTGCACGGTGTGGTGGGCCAGCCTCTGACCTGTTACAAGTACGCCACCAAGCCCATGACCTATGTGGTGGGCATTGACAGTGACGG CTGGGTCCGAGATGTCACACAGAGGTACGACCCAGTCTGGATGACAGTGACCCGCAAGTGCCGGGTTGATGCTGAGTGGTGGGCCGAGAC CTTGAGACCATACCAGAGCCCATTTATGGACAGGGAGAAGAAAGAAGACTTGGAGTTTCAGGCTAAACACATGGACCAGCCTTTGCCCAC TGCCATTGGCTTATATAAGAACCACCCTCTGTATGCCCTGAAGCGGCATCTCCTGAAATATGAGGCCATCTATCCCGAGACAGCTGCCAT CCTTGGGTATTGTCGTGGAGAAGCGGTCTACTCCAGGGATTGTGTGCACACTCTGCATTCCAGGGACACGTGGCTGAAGAAAGCAAGAGT GGTGAGGCTTGGAGAAGTACCCTACAAGATGGTGAAAGGCTTTTCTAACCGTGCTCGGAAAGCCCGACTTGCTGAGCCCCAGCTGCGGGA AGAAAATGACCTGGGCCTGTTTGGCTACTGGCAGACAGAGGAGTATCAGCCCCCAGTGGCCGTGGACGGGAAGGTGCCCCGGAACGAGTT TGGGAATGTGTACCTCTTCCTGCCCAGCATGATGCCTATTGGCTGTGTCCAGCTGAACCTGCCCAATCTACACCGCGTGGCCCGCAAGCT GGACATCGACTGTGTCCAGGCCATCACTGGCTTTGATTTCCATGGCGGCTACTCCCATCCCGTGACTGATGGATACATCGTCTGCGAGGA ATTCAAAGACGTGCTCCTGACTGCCTGGGAAAATGAGCAGGCAGTCATTGAAAGGAAGGAGAAGGAGAAAAAGGAGAAGCGGGCTCTAGG GAACTGGAAGTTGCTGGCCAAAGGTCTGCTCATCAGGGAGAGGCTGAAGCGTCGCTACGGGCCCAAGAGTGAGGCAGCAGCTCCCCACAC AGATGCAGGAGGTGGACTCTCTTCTGATGAAGAGGAGGGGACCAGCTCTCAAGCAGAAGCGGCCAGGATACTGGCTGCCTCCTGGCCTCA AAACCGAGAAGATGAAGAAAAGCAGAAGCTGAAGGGTGGGCCCAAGAAGACCAAAAGGGAAAAGAAAGCAGCAGCTTCCCACCTGTTCCC >90466_90466_4_TGFBR2-XPC_TGFBR2_chr3_30648469_ENST00000359013_XPC_chr3_14214562_ENST00000449060_length(amino acids)=901AA_BP=31 MGRGLLRGLWPLHIVLWTRIASTIPPHVQKSDAFEDEKPPKKSLLSKVSQGKRKRGCSHPGGSADGPAKKKVAKVTVKSENLKVIKDEAL SDGDDLRDFPSDLKKAHHLKRGATMNEDSNEEEEESENDWEEAKTRERSEKIKLEFETYLRRAMKRFNKGVHEDTHKVHLLCLLANGFYR NNICSQPDLHAIGLSIIPARFTRVLPRDVDTYYLSNLVKWFIGTFTVNAELSASEQDNLQTTLERRFAIYSARDDEELVHIFLLILRALQ LLTRLVLSLQPIPLKSATAKGKKPSKERLTADPGGSSETSSQVLENHTKPKTSKGTKQEETFAKGTCRPSAKGKRNKGGRKKRSKPSSSE EDEGPGDKQEKATQRRPHGRERRVASRVSYKEESGSDEAGSGSDFELSSGEASDPSDEDSEPGPPKQRKAPAPQRTKAGSKSASRTHRGS HRKDPSLPAASSSSSSSKRGKKMCSDGEKAEKRSIAGIDQWLEVFCEQEEKWVCVDCVHGVVGQPLTCYKYATKPMTYVVGIDSDGWVRD VTQRYDPVWMTVTRKCRVDAEWWAETLRPYQSPFMDREKKEDLEFQAKHMDQPLPTAIGLYKNHPLYALKRHLLKYEAIYPETAAILGYC RGEAVYSRDCVHTLHSRDTWLKKARVVRLGEVPYKMVKGFSNRARKARLAEPQLREENDLGLFGYWQTEEYQPPVAVDGKVPRNEFGNVY LFLPSMMPIGCVQLNLPNLHRVARKLDIDCVQAITGFDFHGGYSHPVTDGYIVCEEFKDVLLTAWENEQAVIERKEKEKKEKRALGNWKL LAKGLLIRERLKRRYGPKSEAAAPHTDAGGGLSSDEEEGTSSQAEAARILAASWPQNREDEEKQKLKGGPKKTKREKKAAASHLFPFEQL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TGFBR2-XPC |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 847_866 | 34.333333333333336 | 941.0 | CETN2 | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 847_866 | 34.333333333333336 | 904.0 | CETN2 | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 816_940 | 34.333333333333336 | 941.0 | ERCC2 and GTF2H1 | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 816_940 | 34.333333333333336 | 904.0 | ERCC2 and GTF2H1 | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000285021 | 0 | 16 | 496_734 | 34.333333333333336 | 941.0 | RAD23B | |
| Tgene | XPC | chr3:30648469 | chr3:14214562 | ENST00000449060 | 0 | 16 | 496_734 | 34.333333333333336 | 904.0 | RAD23B |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TGFBR2-XPC |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TGFBR2-XPC |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
