
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:THAP6-SETD2 (FusionGDB2 ID:90607) |
Fusion Gene Summary for THAP6-SETD2 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: THAP6-SETD2 | Fusion gene ID: 90607 | Hgene | Tgene | Gene symbol | THAP6 | SETD2 | Gene ID | 152815 | 29072 |
| Gene name | THAP domain containing 6 | SET domain containing 2, histone lysine methyltransferase | |
| Synonyms | - | HBP231|HIF-1|HIP-1|HSPC069|HYPB|KMT3A|LLS|SET2|p231HBP | |
| Cytomap | 4q21.1 | 3p21.31 | |
| Type of gene | protein-coding | protein-coding | |
| Description | THAP domain-containing protein 6 | histone-lysine N-methyltransferase SETD2SET domain containing 2huntingtin interacting protein 1huntingtin yeast partner Bhuntingtin-interacting protein Blysine N-methyltransferase 3Aprotein-lysine N-methyltransferase SETD2 | |
| Modification date | 20200313 | 20200315 | |
| UniProtAcc | . | Q9BYW2 | |
| Ensembl transtripts involved in fusion gene | ENST00000311638, ENST00000380837, ENST00000502620, ENST00000504190, ENST00000507556, ENST00000507557, ENST00000507885, ENST00000508105, ENST00000514480, | ENST00000492397, ENST00000409792, | |
| Fusion gene scores | * DoF score | 5 X 5 X 3=75 | 14 X 12 X 8=1344 |
| # samples | 5 | 17 | |
| ** MAII score | log2(5/75*10)=-0.584962500721156 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(17/1344*10)=-2.98292648664106 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: THAP6 [Title/Abstract] AND SETD2 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | THAP6(76442189)-SETD2(47061330), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | SETD2 | GO:0010569 | regulation of double-strand break repair via homologous recombination | 24843002 |
| Tgene | SETD2 | GO:0018023 | peptidyl-lysine trimethylation | 27518565 |
| Tgene | SETD2 | GO:0018026 | peptidyl-lysine monomethylation | 28753426 |
| Tgene | SETD2 | GO:0032465 | regulation of cytokinesis | 27518565 |
| Tgene | SETD2 | GO:0032727 | positive regulation of interferon-alpha production | 28753426 |
| Tgene | SETD2 | GO:0034340 | response to type I interferon | 28753426 |
| Tgene | SETD2 | GO:0051607 | defense response to virus | 28753426 |
| Tgene | SETD2 | GO:0097198 | histone H3-K36 trimethylation | 23043551|24843002|26002201|27474439|28753426 |
| Tgene | SETD2 | GO:0097676 | histone H3-K36 dimethylation | 26002201 |
| Tgene | SETD2 | GO:1902850 | microtubule cytoskeleton organization involved in mitosis | 27518565 |
| Tgene | SETD2 | GO:1905634 | regulation of protein localization to chromatin | 24843002 |
 Fusion gene breakpoints across THAP6 (5'-gene) Fusion gene breakpoints across THAP6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
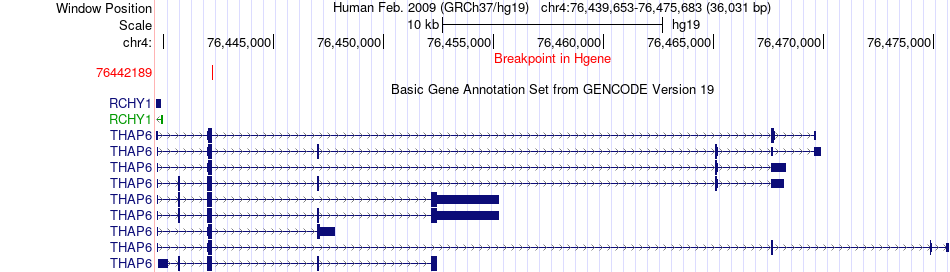 |
 Fusion gene breakpoints across SETD2 (3'-gene) Fusion gene breakpoints across SETD2 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
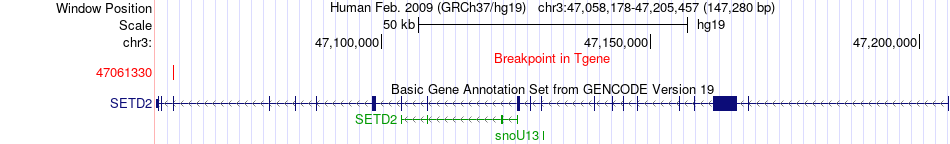 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-HU-A4GT-01A | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
Top |
Fusion Gene ORF analysis for THAP6-SETD2 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000311638 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000380837 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000502620 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000504190 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000507556 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000507557 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000507885 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000508105 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| 5CDS-intron | ENST00000514480 | ENST00000492397 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000311638 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000380837 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000502620 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000504190 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000507556 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000507557 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000507885 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000508105 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
| In-frame | ENST00000514480 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000507557 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1034 | 285 | 120 | 629 | 169 |
| ENST00000508105 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1005 | 256 | 91 | 600 | 169 |
| ENST00000311638 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1105 | 356 | 68 | 700 | 210 |
| ENST00000380837 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1105 | 356 | 68 | 700 | 210 |
| ENST00000507556 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1105 | 356 | 68 | 700 | 210 |
| ENST00000504190 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1005 | 256 | 91 | 600 | 169 |
| ENST00000507885 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1005 | 256 | 91 | 600 | 169 |
| ENST00000502620 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 999 | 250 | 85 | 594 | 169 |
| ENST00000514480 | THAP6 | chr4 | 76442189 | + | ENST00000409792 | SETD2 | chr3 | 47061330 | - | 1529 | 780 | 492 | 1124 | 210 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000507557 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.009256204 | 0.9907438 |
| ENST00000508105 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.008933778 | 0.99106616 |
| ENST00000311638 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.003006435 | 0.99699354 |
| ENST00000380837 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.003006435 | 0.99699354 |
| ENST00000507556 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.003006435 | 0.99699354 |
| ENST00000504190 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.008933778 | 0.99106616 |
| ENST00000507885 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.008933778 | 0.99106616 |
| ENST00000502620 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.010391295 | 0.9896087 |
| ENST00000514480 | ENST00000409792 | THAP6 | chr4 | 76442189 | + | SETD2 | chr3 | 47061330 | - | 0.003718934 | 0.9962811 |
Top |
Fusion Genomic Features for THAP6-SETD2 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
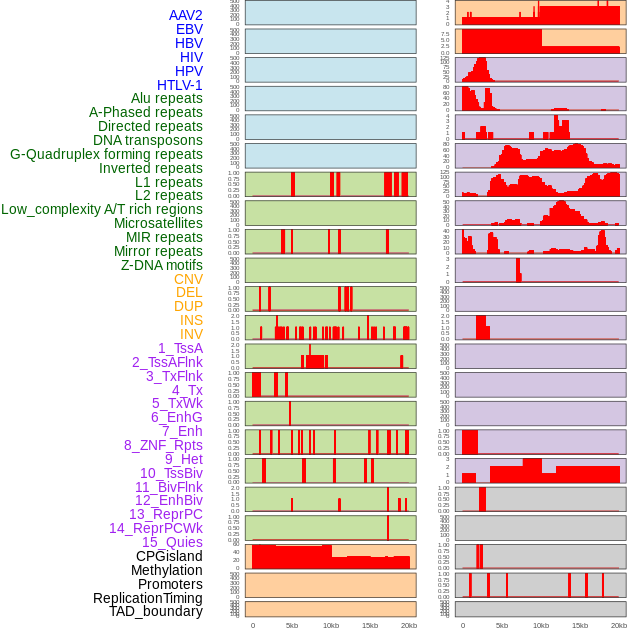 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for THAP6-SETD2 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr4:76442189/chr3:47061330) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | SETD2 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Histone methyltransferase that specifically trimethylates 'Lys-36' of histone H3 (H3K36me3) using dimethylated 'Lys-36' (H3K36me2) as substrate (PubMed:16118227, PubMed:19141475, PubMed:21526191, PubMed:21792193, PubMed:23043551, PubMed:27474439). It is capable of trimethylating unmethylated H3K36 (H3K36me0) in vitro (PubMed:19332550). Represents the main enzyme generating H3K36me3, a specific tag for epigenetic transcriptional activation (By similarity). Plays a role in chromatin structure modulation during elongation by coordinating recruitment of the FACT complex and by interacting with hyperphosphorylated POLR2A (PubMed:23325844). Acts as a key regulator of DNA mismatch repair in G1 and early S phase by generating H3K36me3, a mark required to recruit MSH6 subunit of the MutS alpha complex: early recruitment of the MutS alpha complex to chromatin to be replicated allows a quick identification of mismatch DNA to initiate the mismatch repair reaction (PubMed:23622243). Required for DNA double-strand break repair in response to DNA damage: acts by mediating formation of H3K36me3, promoting recruitment of RAD51 and DNA repair via homologous recombination (HR) (PubMed:24843002). Acts as a tumor suppressor (PubMed:24509477). H3K36me3 also plays an essential role in the maintenance of a heterochromatic state, by recruiting DNA methyltransferase DNMT3A (PubMed:27317772). H3K36me3 is also enhanced in intron-containing genes, suggesting that SETD2 recruitment is enhanced by splicing and that splicing is coupled to recruitment of elongating RNA polymerase (PubMed:21792193). Required during angiogenesis (By similarity). Required for endoderm development by promoting embryonic stem cell differentiation toward endoderm: acts by mediating formation of H3K36me3 in distal promoter regions of FGFR3, leading to regulate transcription initiation of FGFR3 (By similarity). In addition to histones, also mediates methylation of other proteins, such as tubulins and STAT1 (PubMed:27518565, PubMed:28753426). Trimethylates 'Lys-40' of alpha-tubulins such as TUBA1B (alpha-TubK40me3); alpha-TubK40me3 is required for normal mitosis and cytokinesis and may be a specific tag in cytoskeletal remodeling (PubMed:27518565). Involved in interferon-alpha-induced antiviral defense by mediating both monomethylation of STAT1 at 'Lys-525' and catalyzing H3K36me3 on promoters of some interferon-stimulated genes (ISGs) to activate gene transcription (PubMed:28753426). {ECO:0000250|UniProtKB:E9Q5F9, ECO:0000269|PubMed:16118227, ECO:0000269|PubMed:19141475, ECO:0000269|PubMed:21526191, ECO:0000269|PubMed:21792193, ECO:0000269|PubMed:23043551, ECO:0000269|PubMed:23325844, ECO:0000269|PubMed:23622243, ECO:0000269|PubMed:24509477, ECO:0000269|PubMed:24843002, ECO:0000269|PubMed:27317772, ECO:0000269|PubMed:27474439, ECO:0000269|PubMed:27518565, ECO:0000269|PubMed:28753426}.; FUNCTION: (Microbial infection) Recruited to the promoters of adenovirus 12 E1A gene in case of infection, possibly leading to regulate its expression. {ECO:0000269|PubMed:11461154}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000311638 | + | 3 | 5 | 1_89 | 96 | 223.0 | Zinc finger | THAP-type |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000380837 | + | 3 | 4 | 1_89 | 96 | 181.0 | Zinc finger | THAP-type |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000514480 | + | 3 | 5 | 1_89 | 96 | 223.0 | Zinc finger | THAP-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000311638 | + | 3 | 5 | 149_194 | 96 | 223.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000380837 | + | 3 | 4 | 149_194 | 96 | 181.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000514480 | + | 3 | 5 | 149_194 | 96 | 223.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000311638 | + | 3 | 5 | 139_142 | 96 | 223.0 | Motif | HCFC1-binding motif (HBM) |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000380837 | + | 3 | 4 | 139_142 | 96 | 181.0 | Motif | HCFC1-binding motif (HBM) |
| Hgene | THAP6 | chr4:76442189 | chr3:47061330 | ENST00000514480 | + | 3 | 5 | 139_142 | 96 | 223.0 | Motif | HCFC1-binding motif (HBM) |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 2117_2146 | 2450 | 2565.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 166_247 | 2450 | 2565.0 | Compositional bias | Note=Pro-rich | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 2149_2232 | 2450 | 2565.0 | Compositional bias | Note=Pro-rich | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 2266_2365 | 2450 | 2565.0 | Compositional bias | Note=Gln-rich | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 385_456 | 2450 | 2565.0 | Compositional bias | Note=Arg-rich | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1494_1548 | 2450 | 2565.0 | Domain | AWS | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1550_1667 | 2450 | 2565.0 | Domain | SET | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1674_1690 | 2450 | 2565.0 | Domain | Post-SET | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 2389_2422 | 2450 | 2565.0 | Domain | WW | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1560_1562 | 2450 | 2565.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1603_1605 | 2450 | 2565.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1628_1629 | 2450 | 2565.0 | Region | S-adenosyl-L-methionine binding | |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 2137_2366 | 2450 | 2565.0 | Region | Low charge region |
Top |
Fusion Gene Sequence for THAP6-SETD2 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >90607_90607_1_THAP6-SETD2_THAP6_chr4_76442189_ENST00000311638_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1105nt_BP=356nt AGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGTTTTGTTATGAGTTGCTAAAATGGTGAAATGCTGCTCCGCCA TTGGATGTGCTTCTCGCTGCTTGCCAAATTCGAAGTTAAAAGGACTGACATTTCACGTATTCCCCACAGATGAAAACATCAAAAGGAAAT GGGTATTAGCAATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGA AGACAGATTTTGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCT CGAAAAAGCCCAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCA TCGTCCAGTGCCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGC TGACTCACGGTGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGG AGTACATTAAGAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGA GGATGGGTGGTCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGAT ACATCACCCTGGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTG TTGGCATGTGGTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCC TTGGCGGTGCTGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTG >90607_90607_1_THAP6-SETD2_THAP6_chr4_76442189_ENST00000311638_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=210AA_BP=96 MVKCCSAIGCASRCLPNSKLKGLTFHVFPTDENIKRKWVLAMKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFD SPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQCLNPYRKPDCKVGRITTTEDFKHLARKLTHGVMNKELKYCKNPEDLECNE -------------------------------------------------------------- >90607_90607_2_THAP6-SETD2_THAP6_chr4_76442189_ENST00000380837_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1105nt_BP=356nt AGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGTTTTGTTATGAGTTGCTAAAATGGTGAAATGCTGCTCCGCCA TTGGATGTGCTTCTCGCTGCTTGCCAAATTCGAAGTTAAAAGGACTGACATTTCACGTATTCCCCACAGATGAAAACATCAAAAGGAAAT GGGTATTAGCAATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGA AGACAGATTTTGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCT CGAAAAAGCCCAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCA TCGTCCAGTGCCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGC TGACTCACGGTGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGG AGTACATTAAGAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGA GGATGGGTGGTCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGAT ACATCACCCTGGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTG TTGGCATGTGGTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCC TTGGCGGTGCTGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTG >90607_90607_2_THAP6-SETD2_THAP6_chr4_76442189_ENST00000380837_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=210AA_BP=96 MVKCCSAIGCASRCLPNSKLKGLTFHVFPTDENIKRKWVLAMKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFD SPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQCLNPYRKPDCKVGRITTTEDFKHLARKLTHGVMNKELKYCKNPEDLECNE -------------------------------------------------------------- >90607_90607_3_THAP6-SETD2_THAP6_chr4_76442189_ENST00000502620_SETD2_chr3_47061330_ENST00000409792_length(transcript)=999nt_BP=250nt AGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGATTCCCCACAGATGAAAACATCAAAAGGAAATGGGTATTAGCAATGAA AAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGAAGACAGATTTTGACAG AAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCTCGAAAAAGCCCAAGAC AGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCATCGTCCAGTGCCTGAA CCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGCTGACTCACGGTGTTAT GAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGGAGTACATTAAGAAGTA CATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGAGGATGGGTGGTCAGGT AAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGATACATCACCCTGGGGAA TTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTGTTGGCATGTGGTTAGA AGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCCTTGGCGGTGCTGTCAG CGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTGAATATATTGAAAATAA >90607_90607_3_THAP6-SETD2_THAP6_chr4_76442189_ENST00000502620_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=169AA_BP=55 MKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFDSPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQC -------------------------------------------------------------- >90607_90607_4_THAP6-SETD2_THAP6_chr4_76442189_ENST00000504190_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1005nt_BP=256nt AGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGATTCCCCACAGATGAAAACATCAAAAGGAAATGGGTATTAGC AATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGAAGACAGATTT TGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCTCGAAAAAGCC CAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCATCGTCCAGTG CCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGCTGACTCACGG TGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGGAGTACATTAA GAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGAGGATGGGTGG TCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGATACATCACCCT GGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTGTTGGCATGTG GTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCCTTGGCGGTGC TGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTGAATATATTGA >90607_90607_4_THAP6-SETD2_THAP6_chr4_76442189_ENST00000504190_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=169AA_BP=55 MKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFDSPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQC -------------------------------------------------------------- >90607_90607_5_THAP6-SETD2_THAP6_chr4_76442189_ENST00000507556_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1105nt_BP=356nt AGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGTTTTGTTATGAGTTGCTAAAATGGTGAAATGCTGCTCCGCCA TTGGATGTGCTTCTCGCTGCTTGCCAAATTCGAAGTTAAAAGGACTGACATTTCACGTATTCCCCACAGATGAAAACATCAAAAGGAAAT GGGTATTAGCAATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGA AGACAGATTTTGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCT CGAAAAAGCCCAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCA TCGTCCAGTGCCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGC TGACTCACGGTGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGG AGTACATTAAGAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGA GGATGGGTGGTCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGAT ACATCACCCTGGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTG TTGGCATGTGGTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCC TTGGCGGTGCTGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTG >90607_90607_5_THAP6-SETD2_THAP6_chr4_76442189_ENST00000507556_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=210AA_BP=96 MVKCCSAIGCASRCLPNSKLKGLTFHVFPTDENIKRKWVLAMKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFD SPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQCLNPYRKPDCKVGRITTTEDFKHLARKLTHGVMNKELKYCKNPEDLECNE -------------------------------------------------------------- >90607_90607_6_THAP6-SETD2_THAP6_chr4_76442189_ENST00000507557_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1034nt_BP=285nt TCCCGGGGCTACGAGGCGGAAGCGAAGGCAGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGATTCCCCACAGAT GAAAACATCAAAAGGAAATGGGTATTAGCAATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTG TGTTCGAGGCACTTTAAGAAGACAGATTTTGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCT CCATATCACCTACAGGCCTCGAAAAAGCCCAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGA AAAGAGATGTCCCAGTTCATCGTCCAGTGCCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTT AAACATCTGGCTCGCAAGCTGACTCACGGTGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAAT GTGAAACACAAAACCAAGGAGTACATTAAGAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGA CTGTTGGGCCAGGGTGGGAGGATGGGTGGTCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCA GCACTGTGCTACTGATGATACATCACCCTGGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTG ATTACCTAGTTGTGAGCTGTTGGCATGTGGTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTC TTAAACCTGGGTCTTCTCCTTGGCGGTGCTGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATT >90607_90607_6_THAP6-SETD2_THAP6_chr4_76442189_ENST00000507557_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=169AA_BP=55 MKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFDSPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQC -------------------------------------------------------------- >90607_90607_7_THAP6-SETD2_THAP6_chr4_76442189_ENST00000507885_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1005nt_BP=256nt AGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGATTCCCCACAGATGAAAACATCAAAAGGAAATGGGTATTAGC AATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGAAGACAGATTT TGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCTCGAAAAAGCC CAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCATCGTCCAGTG CCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGCTGACTCACGG TGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGGAGTACATTAA GAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGAGGATGGGTGG TCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGATACATCACCCT GGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTGTTGGCATGTG GTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCCTTGGCGGTGC TGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTGAATATATTGA >90607_90607_7_THAP6-SETD2_THAP6_chr4_76442189_ENST00000507885_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=169AA_BP=55 MKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFDSPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQC -------------------------------------------------------------- >90607_90607_8_THAP6-SETD2_THAP6_chr4_76442189_ENST00000508105_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1005nt_BP=256nt AGACGCAGTCTCCGTCGTTGACGTTAGTCGCAGTCTTCGCTGCTAACGATTCCCCACAGATGAAAACATCAAAAGGAAATGGGTATTAGC AATGAAAAGACTTGATGTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGAAGACAGATTT TGACAGAAGTGCTCCAAATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCTCGAAAAAGCC CAAGACAGCAGAAGCAGACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCATCGTCCAGTG CCTGAACCCTTACCGGAAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGCTGACTCACGG TGTTATGAATAAGGAGCTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGGAGTACATTAA GAAGTACATGCAGAAGTTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGAGGATGGGTGG TCAGGTAAGACAGACTCTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGATACATCACCCT GGGGAATTCAACCCTGCAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTGTTGGCATGTG GTTAGAAGCCATCAGAGGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCCTTGGCGGTGC TGTCAGCGCACAGACCCATGCGCATCCCCACCCACAACCCTTTACCCTGATGATCTGTATTATATTTTAATGTATATGTGAATATATTGA >90607_90607_8_THAP6-SETD2_THAP6_chr4_76442189_ENST00000508105_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=169AA_BP=55 MKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFDSPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQC -------------------------------------------------------------- >90607_90607_9_THAP6-SETD2_THAP6_chr4_76442189_ENST00000514480_SETD2_chr3_47061330_ENST00000409792_length(transcript)=1529nt_BP=780nt GGTAATAACTCTTAGGTTGCCTGTTTTCCCCCAATCGCTACCCGCCCAAATCTCAGGGCGTAGTTCGGCGCGAGGTTGGCGGCAGCGGCG CGGAGACGCTGGTTGGCCCCAGAGGAAAGGACGCTCACTGGGCGCATCGTTACCTGTGACGGGTCCTCCCTCTCTCCCTCCGGTGGCGGG GAGCCTGGAGAAAGGGGCGGAGCTCGGCCCGGGCCCTGGGCTCAGCGGGTCCCGCGCGCTGCTCTTGGACGCCAGGGTGCTCGCTGGCAC CTCGAAGTGCTTCTTGCTTCCTCCCTTTCCCGTTTACTTGCTGTTCTTTCAAAATATGTACTGACATTCACTATTGCCAGTCACCAGCTA AACCGTGAGGACAAAAAGTAAGGAAAAATAGACACACTCCACCATGACCTCATGGAACTAACAAGACTGGTGGAGGAGGAAGACATTAAT AAGAGCAATCACTGCAATAAATTTTTGTTATGAGTTGCTAAAATGGTGAAATGCTGCTCCGCCATTGGATGTGCTTCTCGCTGCTTGCCA AATTCGAAGTTAAAAGGACTGACATTTCACGTATTCCCCACAGATGAAAACATCAAAAGGAAATGGGTATTAGCAATGAAAAGACTTGAT GTGAATGCAGCCGGCATTTGGGAGCCTAAAAAAGGAGATGTGTTGTGTTCGAGGCACTTTAAGAAGACAGATTTTGACAGAAGTGCTCCA AATATTAAACTGAAACCTGGAGTCATACCTTCTATCTTTGATTCTCCATATCACCTACAGGCCTCGAAAAAGCCCAAGACAGCAGAAGCA GACACCTCCAGTGAACTAGCAAAGAAAAGCAAAGAAGTATTCAGAAAAGAGATGTCCCAGTTCATCGTCCAGTGCCTGAACCCTTACCGG AAACCTGACTGCAAAGTGGGAAGAATTACCACAACTGAAGACTTTAAACATCTGGCTCGCAAGCTGACTCACGGTGTTATGAATAAGGAG CTGAAGTACTGTAAGAATCCTGAGGACCTGGAGTGCAATGAGAATGTGAAACACAAAACCAAGGAGTACATTAAGAAGTACATGCAGAAG TTTGGGGCTGTTTACAAACCCAAAGAGGACACTGAATTAGAGTGACTGTTGGGCCAGGGTGGGAGGATGGGTGGTCAGGTAAGACAGACT CTAGGGAGAGGAAATCCTGTGGGCCTTTCTGTCCCACCCCTGTCAGCACTGTGCTACTGATGATACATCACCCTGGGGAATTCAACCCTG CAGATGTCAACTGAAGGCCACAAAAATGAACTCCATCTACAAGTGATTACCTAGTTGTGAGCTGTTGGCATGTGGTTAGAAGCCATCAGA GGTGCAAGGGCTTAGAAAAGACCCTGGCCAGACCTGACTCCACTCTTAAACCTGGGTCTTCTCCTTGGCGGTGCTGTCAGCGCACAGACC >90607_90607_9_THAP6-SETD2_THAP6_chr4_76442189_ENST00000514480_SETD2_chr3_47061330_ENST00000409792_length(amino acids)=210AA_BP=96 MVKCCSAIGCASRCLPNSKLKGLTFHVFPTDENIKRKWVLAMKRLDVNAAGIWEPKKGDVLCSRHFKKTDFDRSAPNIKLKPGVIPSIFD SPYHLQASKKPKTAEADTSSELAKKSKEVFRKEMSQFIVQCLNPYRKPDCKVGRITTTEDFKHLARKLTHGVMNKELKYCKNPEDLECNE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for THAP6-SETD2 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 2457_2564 | 2450.0 | 2565.0 | POLR2A |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | SETD2 | chr4:76442189 | chr3:47061330 | ENST00000409792 | 17 | 21 | 1418_1714 | 2450.0 | 2565.0 | TUBA1A |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for THAP6-SETD2 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for THAP6-SETD2 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
