
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:THOP1-NOTCH3 (FusionGDB2 ID:90689) |
Fusion Gene Summary for THOP1-NOTCH3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: THOP1-NOTCH3 | Fusion gene ID: 90689 | Hgene | Tgene | Gene symbol | THOP1 | NOTCH3 | Gene ID | 7064 | 4854 |
| Gene name | thimet oligopeptidase 1 | notch receptor 3 | |
| Synonyms | EP24.15|MEPD_HUMAN|MP78|TOP | CADASIL|CADASIL1|CASIL|IMF2|LMNS | |
| Cytomap | 19p13.3 | 19p13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | thimet oligopeptidaseendopeptidase 24.15 | neurogenic locus notch homolog protein 3Notch homolog 3notch 3 | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | Q9UM47 | |
| Ensembl transtripts involved in fusion gene | ENST00000307741, ENST00000395212, ENST00000586677, ENST00000591149, | ENST00000263388, | |
| Fusion gene scores | * DoF score | 4 X 4 X 3=48 | 4 X 5 X 5=100 |
| # samples | 4 | 6 | |
| ** MAII score | log2(4/48*10)=-0.263034405833794 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(6/100*10)=-0.736965594166206 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: THOP1 [Title/Abstract] AND NOTCH3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | THOP1(2785676)-NOTCH3(15308389), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across THOP1 (5'-gene) Fusion gene breakpoints across THOP1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
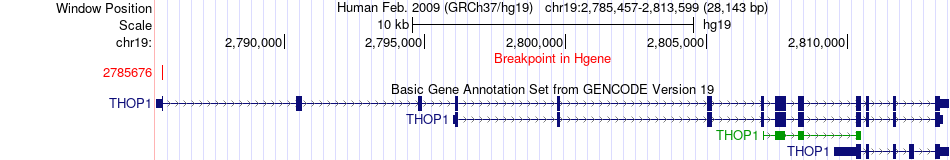 |
 Fusion gene breakpoints across NOTCH3 (3'-gene) Fusion gene breakpoints across NOTCH3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
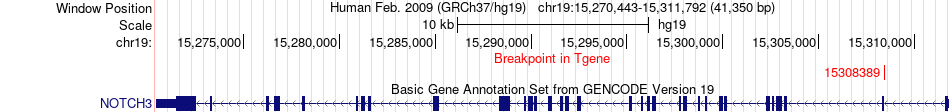 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-FX-A3NK-01A | THOP1 | chr19 | 2785676 | - | NOTCH3 | chr19 | 15308389 | - |
| ChimerDB4 | SARC | TCGA-FX-A3NK-01A | THOP1 | chr19 | 2785676 | + | NOTCH3 | chr19 | 15308389 | - |
Top |
Fusion Gene ORF analysis for THOP1-NOTCH3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000307741 | ENST00000263388 | THOP1 | chr19 | 2785676 | + | NOTCH3 | chr19 | 15308389 | - |
| intron-3CDS | ENST00000395212 | ENST00000263388 | THOP1 | chr19 | 2785676 | + | NOTCH3 | chr19 | 15308389 | - |
| intron-3CDS | ENST00000586677 | ENST00000263388 | THOP1 | chr19 | 2785676 | + | NOTCH3 | chr19 | 15308389 | - |
| intron-3CDS | ENST00000591149 | ENST00000263388 | THOP1 | chr19 | 2785676 | + | NOTCH3 | chr19 | 15308389 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000307741 | THOP1 | chr19 | 2785676 | + | ENST00000263388 | NOTCH3 | chr19 | 15308389 | - | 8096 | 219 | 203 | 7066 | 2287 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000307741 | ENST00000263388 | THOP1 | chr19 | 2785676 | + | NOTCH3 | chr19 | 15308389 | - | 0.002880413 | 0.99711967 |
Top |
Fusion Genomic Features for THOP1-NOTCH3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
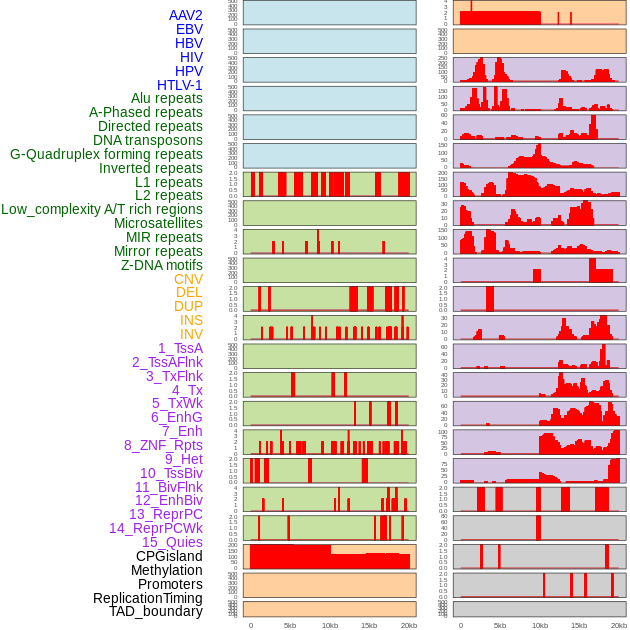 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for THOP1-NOTCH3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:2785676/chr19:15308389) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | NOTCH3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Functions as a receptor for membrane-bound ligands Jagged1, Jagged2 and Delta1 to regulate cell-fate determination (PubMed:15350543). Upon ligand activation through the released notch intracellular domain (NICD) it forms a transcriptional activator complex with RBPJ/RBPSUH and activates genes of the enhancer of split locus. Affects the implementation of differentiation, proliferation and apoptotic programs (By similarity). {ECO:0000250|UniProtKB:Q9R172, ECO:0000269|PubMed:15350543}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1000_1034 | 39 | 2322.0 | Domain | EGF-like 26 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1036_1082 | 39 | 2322.0 | Domain | EGF-like 27 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1084_1120 | 39 | 2322.0 | Domain | EGF-like 28 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1122_1158 | 39 | 2322.0 | Domain | EGF-like 29%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1160_1203 | 39 | 2322.0 | Domain | EGF-like 30%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 119_156 | 39 | 2322.0 | Domain | EGF-like 3 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1205_1244 | 39 | 2322.0 | Domain | EGF-like 31 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1246_1287 | 39 | 2322.0 | Domain | EGF-like 32 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1289_1325 | 39 | 2322.0 | Domain | EGF-like 33 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1335_1373 | 39 | 2322.0 | Domain | EGF-like 34 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 158_195 | 39 | 2322.0 | Domain | EGF-like 4%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 197_234 | 39 | 2322.0 | Domain | EGF-like 5 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 236_272 | 39 | 2322.0 | Domain | EGF-like 6%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 274_312 | 39 | 2322.0 | Domain | EGF-like 7 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 314_350 | 39 | 2322.0 | Domain | EGF-like 8%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 351_389 | 39 | 2322.0 | Domain | EGF-like 9 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 391_429 | 39 | 2322.0 | Domain | EGF-like 10%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 40_77 | 39 | 2322.0 | Domain | EGF-like 1 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 431_467 | 39 | 2322.0 | Domain | EGF-like 11%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 469_505 | 39 | 2322.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 507_543 | 39 | 2322.0 | Domain | EGF-like 13%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 545_580 | 39 | 2322.0 | Domain | EGF-like 14%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 582_618 | 39 | 2322.0 | Domain | EGF-like 15%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 620_655 | 39 | 2322.0 | Domain | EGF-like 16%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 657_693 | 39 | 2322.0 | Domain | EGF-like 17%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 695_730 | 39 | 2322.0 | Domain | EGF-like 18 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 734_770 | 39 | 2322.0 | Domain | EGF-like 19 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 771_808 | 39 | 2322.0 | Domain | EGF-like 20 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 78_118 | 39 | 2322.0 | Domain | EGF-like 2 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 810_847 | 39 | 2322.0 | Domain | EGF-like 21%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 849_885 | 39 | 2322.0 | Domain | EGF-like 22%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 887_922 | 39 | 2322.0 | Domain | EGF-like 23%3B calcium-binding | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 924_960 | 39 | 2322.0 | Domain | EGF-like 24 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 962_998 | 39 | 2322.0 | Domain | EGF-like 25 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1387_1427 | 39 | 2322.0 | Repeat | Note=LNR 1 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1428_1458 | 39 | 2322.0 | Repeat | Note=LNR 2 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1467_1505 | 39 | 2322.0 | Repeat | Note=LNR 3 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1838_1867 | 39 | 2322.0 | Repeat | Note=ANK 1 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1871_1901 | 39 | 2322.0 | Repeat | Note=ANK 2 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1905_1934 | 39 | 2322.0 | Repeat | Note=ANK 3 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1938_1967 | 39 | 2322.0 | Repeat | Note=ANK 4 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1971_2000 | 39 | 2322.0 | Repeat | Note=ANK 5 | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1665_2321 | 39 | 2322.0 | Topological domain | Cytoplasmic | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 40_1643 | 39 | 2322.0 | Topological domain | Extracellular | |
| Tgene | NOTCH3 | chr19:2785676 | chr19:15308389 | ENST00000263388 | 0 | 33 | 1644_1664 | 39 | 2322.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for THOP1-NOTCH3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >90689_90689_1_THOP1-NOTCH3_THOP1_chr19_2785676_ENST00000307741_NOTCH3_chr19_15308389_ENST00000263388_length(transcript)=8096nt_BP=219nt CAGGGGCGCGTGCGTCCTGCCCTCCCTGTGCGCGCGCCGCCCCAGCATGCCCCGGGAGCGCGGGCGGCGGGCCCCTTGGTCCTCAGGCGG CCGTGGCGGCGGTGGCGGCGGTTGGGCCGAGGCAGGCGGCCTCAGTGGCCGAGGTGGCTGGACGCGTAGCAGGTGGAAGGAGGGAGGGAG CCGCAGGCGCAGACCCACCCGCCATGAAGCCCCCCGCAGCCCCCCCTTGCCTGGACGGAAGCCCGTGTGCAAATGGAGGTCGTTGCACCC AGCTGCCCTCCCGGGAGGCTGCCTGCCTGTGCCCGCCTGGCTGGGTGGGTGAGCGGTGTCAGCTGGAGGACCCCTGTCACTCAGGCCCCT GTGCTGGCCGTGGTGTCTGCCAGAGTTCAGTGGTGGCTGGCACCGCCCGATTCTCATGCCGGTGCCCCCGTGGCTTCCGAGGCCCTGACT GCTCCCTGCCAGATCCCTGCCTCAGCAGCCCTTGTGCCCACGGTGCCCGCTGCTCAGTGGGGCCCGATGGACGCTTCCTCTGCTCCTGCC CACCTGGCTACCAGGGCCGCAGCTGCCGAAGCGACGTGGATGAGTGCCGGGTGGGTGAGCCCTGCCGCCATGGTGGCACCTGCCTCAACA CACCTGGCTCCTTCCGCTGCCAGTGTCCAGCTGGCTACACAGGGCCACTATGTGAGAACCCCGCGGTGCCCTGTGCACCCTCACCATGCC GTAACGGGGGCACCTGCAGGCAGAGTGGCGACCTCACTTACGACTGTGCCTGTCTTCCTGGGTTTGAGGGTCAGAATTGTGAAGTGAACG TGGACGACTGTCCAGGACACCGATGTCTCAATGGGGGGACATGCGTGGATGGCGTCAACACCTATAACTGCCAGTGCCCTCCTGAGTGGA CAGGCCAGTTCTGCACGGAGGACGTGGATGAGTGTCAGCTGCAGCCCAACGCCTGCCACAATGGGGGTACCTGCTTCAACACGCTGGGTG GCCACAGCTGCGTGTGTGTCAATGGCTGGACAGGCGAGAGCTGCAGTCAGAATATCGATGACTGTGCCACAGCCGTGTGCTTCCATGGGG CCACCTGCCATGACCGCGTGGCTTCTTTCTACTGTGCCTGCCCCATGGGCAAGACTGGCCTCCTGTGTCACCTGGATGACGCCTGTGTCA GCAACCCCTGCCACGAGGATGCTATCTGTGACACAAATCCGGTGAACGGCCGGGCCATTTGCACCTGTCCTCCCGGCTTCACGGGTGGGG CATGTGACCAGGATGTGGACGAGTGCTCTATCGGCGCCAACCCCTGCGAGCACTTGGGCAGGTGCGTGAACACGCAGGGCTCCTTCCTGT GCCAGTGCGGTCGTGGCTACACTGGACCTCGCTGTGAGACCGATGTCAACGAGTGTCTGTCGGGGCCCTGCCGAAACCAGGCCACGTGCC TCGACCGCATAGGCCAGTTCACCTGTATCTGTATGGCAGGCTTCACAGGAACCTATTGCGAGGTGGACATTGACGAGTGTCAGAGTAGCC CCTGTGTCAACGGTGGGGTCTGCAAGGACCGAGTCAATGGCTTCAGCTGCACCTGCCCCTCGGGCTTCAGCGGCTCCACGTGTCAGCTGG ACGTGGACGAATGCGCCAGCACGCCCTGCAGGAATGGCGCCAAATGCGTGGACCAGCCCGATGGCTACGAGTGCCGCTGTGCCGAGGGCT TTGAGGGCACGCTGTGTGATCGCAACGTGGACGACTGCTCCCCTGACCCATGCCACCATGGTCGCTGCGTGGATGGCATCGCCAGCTTCT CATGTGCCTGTGCTCCTGGCTACACGGGCACACGCTGCGAGAGCCAGGTGGACGAATGCCGCAGCCAGCCCTGCCGCCATGGCGGCAAAT GCCTAGACCTGGTGGACAAGTACCTCTGCCGCTGCCCTTCTGGGACCACAGGTGTGAACTGCGAAGTGAACATTGACGACTGTGCCAGCA ACCCCTGCACCTTTGGAGTCTGCCGTGATGGCATCAACCGCTACGACTGTGTCTGCCAACCTGGCTTCACAGGGCCCCTTTGTAACGTGG AGATCAATGAGTGTGCTTCCAGCCCATGCGGCGAGGGAGGTTCCTGTGTGGATGGGGAAAATGGCTTCCGCTGCCTCTGCCCGCCTGGCT CCTTGCCCCCACTCTGCCTCCCCCCGAGCCATCCCTGTGCCCATGAGCCCTGCAGTCACGGCATCTGCTATGATGCACCTGGCGGGTTCC GCTGTGTGTGTGAGCCTGGCTGGAGTGGCCCCCGCTGCAGCCAGAGCCTGGCCCGAGACGCCTGTGAGTCCCAGCCGTGCAGGGCCGGTG GGACATGCAGCAGCGATGGAATGGGTTTCCACTGCACCTGCCCGCCTGGTGTCCAGGGACGTCAGTGTGAACTCCTCTCCCCCTGCACCC CGAACCCCTGTGAGCATGGGGGCCGCTGCGAGTCTGCCCCTGGCCAGCTGCCTGTCTGCTCCTGCCCCCAGGGCTGGCAAGGCCCACGAT GCCAGCAGGATGTGGACGAGTGTGCTGGCCCCGCACCCTGTGGCCCTCATGGTATCTGCACCAACCTGGCAGGGAGTTTCAGCTGCACCT GCCATGGAGGGTACACTGGCCCTTCCTGCGATCAGGACATCAATGACTGTGACCCCAACCCATGCCTGAACGGTGGCTCGTGCCAAGACG GCGTGGGCTCCTTTTCCTGCTCCTGCCTCCCTGGTTTCGCCGGCCCACGATGCGCCCGCGATGTGGATGAGTGCCTGAGCAACCCCTGCG GCCCGGGCACCTGTACCGACCACGTGGCCTCCTTCACCTGCACCTGCCCGCCAGGCTACGGAGGCTTCCACTGCGAACAGGACCTGCCCG ACTGCAGCCCCAGCTCCTGCTTCAATGGCGGGACCTGTGTGGACGGCGTGAACTCGTTCAGCTGCCTGTGCCGTCCCGGCTACACAGGAG CCCACTGCCAACATGAGGCAGACCCCTGCCTCTCGCGGCCCTGCCTACACGGGGGCGTCTGCAGCGCCGCCCACCCTGGCTTCCGCTGCA CCTGCCTCGAGAGCTTCACGGGCCCGCAGTGCCAGACGCTGGTGGATTGGTGCAGCCGCCAGCCTTGTCAAAACGGGGGTCGCTGCGTCC AGACTGGGGCCTATTGCCTTTGTCCCCCTGGATGGAGCGGACGCCTCTGTGACATCCGAAGCTTGCCCTGCAGGGAGGCCGCAGCCCAGA TCGGGGTGCGGCTGGAGCAGCTGTGTCAGGCGGGTGGGCAGTGTGTGGATGAAGACAGCTCCCACTACTGCGTGTGCCCAGAGGGCCGTA CTGGTAGCCACTGTGAGCAGGAGGTGGACCCCTGCTTGGCCCAGCCCTGCCAGCATGGGGGGACCTGCCGTGGCTATATGGGGGGCTACA TGTGTGAGTGTCTTCCTGGCTACAATGGTGATAACTGTGAGGACGACGTGGACGAGTGTGCCTCCCAGCCCTGCCAGCACGGGGGTTCAT GCATTGACCTCGTGGCCCGCTATCTCTGCTCCTGTCCCCCAGGAACGCTGGGGGTGCTCTGCGAGATTAATGAGGATGACTGCGGCCCAG GCCCACCGCTGGACTCAGGGCCCCGGTGCCTACACAATGGCACCTGCGTGGACCTGGTGGGTGGTTTCCGCTGCACCTGTCCCCCAGGAT ACACTGGTTTGCGCTGCGAGGCAGACATCAATGAGTGTCGCTCAGGTGCCTGCCACGCGGCACACACCCGGGACTGCCTGCAGGACCCAG GCGGAGGTTTCCGTTGCCTTTGTCATGCTGGCTTCTCAGGTCCTCGCTGTCAGACTGTCCTGTCTCCCTGCGAGTCCCAGCCATGCCAGC ATGGAGGCCAGTGCCGTCCTAGCCCGGGTCCTGGGGGTGGGCTGACCTTCACCTGTCACTGTGCCCAGCCGTTCTGGGGTCCGCGTTGCG AGCGGGTGGCGCGCTCCTGCCGGGAGCTGCAGTGCCCGGTGGGCGTCCCATGCCAGCAGACGCCCCGCGGGCCGCGCTGCGCCTGCCCCC CAGGGTTGTCGGGACCCTCCTGCCGCAGCTTCCCGGGGTCGCCGCCGGGGGCCAGCAACGCCAGCTGCGCGGCCGCCCCCTGTCTCCACG GGGGCTCCTGCCGCCCCGCGCCGCTCGCGCCCTTCTTCCGCTGCGCTTGCGCGCAGGGCTGGACCGGGCCGCGCTGCGAGGCGCCCGCCG CGGCACCCGAGGTCTCGGAGGAGCCGCGGTGCCCGCGCGCCGCCTGCCAGGCCAAGCGCGGGGACCAGCGCTGCGACCGCGAGTGCAACA GCCCAGGCTGCGGCTGGGACGGCGGCGACTGCTCGCTGAGCGTGGGCGACCCCTGGCGGCAATGCGAGGCGCTGCAGTGCTGGCGCCTCT TCAACAACAGCCGCTGCGACCCCGCCTGCAGCTCGCCCGCCTGCCTCTACGACAACTTCGACTGCCACGCCGGTGGCCGCGAGCGCACTT GCAACCCGGTGTACGAGAAGTACTGCGCCGACCACTTTGCCGACGGCCGCTGCGACCAGGGCTGCAACACGGAGGAGTGCGGCTGGGATG GGCTGGATTGTGCCAGCGAGGTGCCGGCCCTGCTGGCCCGCGGCGTGCTGGTGCTCACAGTGCTGCTGCCGCCAGAGGAGCTACTGCGTT CCAGCGCCGACTTTCTGCAGCGGCTCAGCGCCATCCTGCGCACCTCGCTGCGCTTCCGCCTGGACGCGCACGGCCAGGCCATGGTCTTCC CTTACCACCGGCCTAGTCCTGGCTCCGAACCCCGGGCCCGTCGGGAGCTGGCCCCCGAGGTGATCGGCTCGGTAGTAATGCTGGAGATTG ACAACCGGCTCTGCCTGCAGTCGCCTGAGAATGATCACTGCTTCCCCGATGCCCAGAGCGCCGCTGACTACCTGGGAGCGTTGTCAGCGG TGGAGCGCCTGGACTTCCCGTACCCACTGCGGGACGTGCGGGGGGAGCCGCTGGAGCCTCCAGAACCCAGCGTCCCGCTGCTGCCACTGC TAGTGGCGGGCGCTGTCTTGCTGCTGGTCATTCTCGTCCTGGGTGTCATGGTGGCCCGGCGCAAGCGCGAGCACAGCACCCTCTGGTTCC CTGAGGGCTTCTCACTGCACAAGGACGTGGCCTCTGGTCACAAGGGCCGGCGGGAACCCGTGGGCCAGGACGCGCTGGGCATGAAGAACA TGGCCAAGGGTGAGAGCCTGATGGGGGAGGTGGCCACAGACTGGATGGACACAGAGTGCCCAGAGGCCAAGCGGCTAAAGGTAGAGGAGC CAGGCATGGGGGCTGAGGAGGCTGTGGATTGCCGTCAGTGGACTCAACACCATCTGGTTGCTGCTGACATCCGCGTGGCACCAGCCATGG CACTGACACCACCACAGGGCGACGCAGATGCTGATGGCATGGATGTCAATGTGCGTGGCCCAGATGGCTTCACCCCGCTAATGCTGGCTT CCTTCTGTGGGGGGGCTCTGGAGCCAATGCCAACTGAAGAGGATGAGGCAGATGACACATCAGCTAGCATCATCTCCGACCTGATCTGCC AGGGGGCTCAGCTTGGGGCACGGACTGACCGTACTGGCGAGACTGCTTTGCACCTGGCTGCCCGTTATGCCCGTGCTGATGCAGCCAAGC GGCTGCTGGATGCTGGGGCAGACACCAATGCCCAGGACCACTCAGGCCGCACTCCCCTGCACACAGCTGTCACAGCCGATGCCCAGGGTG TCTTCCAGATTCTCATCCGAAACCGCTCTACAGACTTGGATGCCCGCATGGCAGATGGCTCAACGGCACTGATCCTGGCGGCCCGCCTGG CAGTAGAGGGCATGGTGGAAGAGCTCATCGCCAGCCATGCTGATGTCAATGCTGTGGATGAGCTTGGGAAATCAGCCTTACACTGGGCTG CGGCTGTGAACAACGTGGAAGCCACTTTGGCCCTGCTCAAAAATGGAGCCAATAAGGACATGCAGGATAGCAAGGAGGAGACCCCCCTAT TCCTGGCCGCCCGCGAGGGCAGCTATGAGGCTGCCAAGCTGCTGTTGGACCACTTTGCCAACCGTGAGATCACCGACCACCTGGACAGGC TGCCGCGGGACGTAGCCCAGGAGAGACTGCACCAGGACATCGTGCGCTTGCTGGATCAACCCAGTGGGCCCCGCAGCCCCCCCGGTCCCC ACGGCCTGGGGCCTCTGCTCTGTCCTCCAGGGGCCTTCCTCCCTGGCCTCAAAGCGGCACAGTCGGGGTCCAAGAAGAGCAGGAGGCCCC CCGGGAAGGCGGGGCTGGGGCCGCAGGGGCCCCGGGGGCGGGGCAAGAAGCTGACGCTGGCCTGCCCGGGCCCCCTGGCTGACAGCTCGG TCACGCTGTCGCCCGTGGACTCGCTGGACTCCCCGCGGCCTTTCGGTGGGCCCCCTGCTTCCCCTGGTGGCTTCCCCCTTGAGGGGCCCT ATGCAGCTGCCACTGCCACTGCAGTGTCTCTGGCACAGCTTGGTGGCCCAGGCCGGGCGGGTCTAGGGCGCCAGCCCCCTGGAGGATGTG TACTCAGCCTGGGCCTGCTGAACCCTGTGGCTGTGCCCCTCGATTGGGCCCGGCTGCCCCCACCTGCCCCTCCAGGCCCCTCGTTCCTGC TGCCACTGGCGCCGGGACCCCAGCTGCTCAACCCAGGGACCCCCGTCTCCCCGCAGGAGCGGCCCCCGCCTTACCTGGCAGTCCCAGGAC ATGGCGAGGAGTACCCGGCGGCTGGGGCACACAGCAGCCCCCCAAAGGCCCGCTTCCTGCGGGTTCCCAGTGAGCACCCTTACCTGACCC CATCCCCCGAATCCCCTGAGCACTGGGCCAGCCCCTCACCTCCCTCCCTCTCAGACTGGTCCGAATCCACGCCTAGCCCAGCCACTGCCA CTGGGGCCATGGCCACCACCACTGGGGCACTGCCTGCCCAGCCACTTCCCTTGTCTGTTCCCAGCTCCCTTGCTCAGGCCCAGACCCAGC TGGGGCCCCAGCCGGAAGTTACCCCCAAGAGGCAAGTGTTGGCCTGAGACGCTCGTCAGTTCTTAGATCTTGGGGGCCTAAAGAGACCCC CGTCCTGCCTCCTTTCTTTCTCTGTCTCTTCCTTCCTTTTAGTCTTTTTCATCCTCTTCTCTTTCCACCAACCCTCCTGCATCCTTGCCT TGCAGCGTGACCGAGATAGGTCATCAGCCCAGGGCTTCAGTCTTCCTTTATTTATAATGGGTGGGGGCTACCACCCACCCTCTCAGTCTT GTGAAGAGTCTGGGACCTCCTTCTTCCCCACTTCTCTCTTCCCTCATTCCTTTCTCTCTCCTTCTGGCCTCTCATTTCCTTACACTCTGA CATGAATGAATTATTATTATTTTTATTTTTCTTTTTTTTTTTACATTTTGTATAGAAACAAATTCATTTAAACAAACTTATTATTATTAT TTTTTACAAAATATATATATGGAGATGCTCCCTCCCCCTGTGAACCCCCCAGTGCCCCCGTGGGGCTGAGTCTGTGGGCCCATTCGGCCA AGCTGGATTCTGTGTACCTAGTACACAGGCATGACTGGGATCCCGTGTACCGAGTACACGACCCAGGTATGTACCAAGTAGGCACCCTTG GGCGCACCCACTGGGGCCAGGGGTCGGGGGAGTGTTGGGAGCCTCCTCCCCACCCCACCTCCCTCACTTCACTGCATTCCAGATGGGACA TGTTCCATAGCCTTGCTGGGGAAGGGCCCACTGCCAACTCCCTCTGCCCCAGCCCCACCCTTGGCCATCTCCCTTTGGGAACTAGGGGGC TGCTGGTGGGAAATGGGAGCCAGGGCAGATGTATGCATTCCTTTGTGTCCCTGTAAATGTGGGACTACAAGAAGAGGAGCTGCCTGAGTG GTACTTTCTCTTCCTGGTAATCCTCTGGCCCAGCCTCATGGCAGAATAGAGGTATTTTTAGGCTATTTTTGTAATATGGCTTCTGGTCAA >90689_90689_1_THOP1-NOTCH3_THOP1_chr19_2785676_ENST00000307741_NOTCH3_chr19_15308389_ENST00000263388_length(amino acids)=2287AA_BP=5 MKPPAAPPCLDGSPCANGGRCTQLPSREAACLCPPGWVGERCQLEDPCHSGPCAGRGVCQSSVVAGTARFSCRCPRGFRGPDCSLPDPCL SSPCAHGARCSVGPDGRFLCSCPPGYQGRSCRSDVDECRVGEPCRHGGTCLNTPGSFRCQCPAGYTGPLCENPAVPCAPSPCRNGGTCRQ SGDLTYDCACLPGFEGQNCEVNVDDCPGHRCLNGGTCVDGVNTYNCQCPPEWTGQFCTEDVDECQLQPNACHNGGTCFNTLGGHSCVCVN GWTGESCSQNIDDCATAVCFHGATCHDRVASFYCACPMGKTGLLCHLDDACVSNPCHEDAICDTNPVNGRAICTCPPGFTGGACDQDVDE CSIGANPCEHLGRCVNTQGSFLCQCGRGYTGPRCETDVNECLSGPCRNQATCLDRIGQFTCICMAGFTGTYCEVDIDECQSSPCVNGGVC KDRVNGFSCTCPSGFSGSTCQLDVDECASTPCRNGAKCVDQPDGYECRCAEGFEGTLCDRNVDDCSPDPCHHGRCVDGIASFSCACAPGY TGTRCESQVDECRSQPCRHGGKCLDLVDKYLCRCPSGTTGVNCEVNIDDCASNPCTFGVCRDGINRYDCVCQPGFTGPLCNVEINECASS PCGEGGSCVDGENGFRCLCPPGSLPPLCLPPSHPCAHEPCSHGICYDAPGGFRCVCEPGWSGPRCSQSLARDACESQPCRAGGTCSSDGM GFHCTCPPGVQGRQCELLSPCTPNPCEHGGRCESAPGQLPVCSCPQGWQGPRCQQDVDECAGPAPCGPHGICTNLAGSFSCTCHGGYTGP SCDQDINDCDPNPCLNGGSCQDGVGSFSCSCLPGFAGPRCARDVDECLSNPCGPGTCTDHVASFTCTCPPGYGGFHCEQDLPDCSPSSCF NGGTCVDGVNSFSCLCRPGYTGAHCQHEADPCLSRPCLHGGVCSAAHPGFRCTCLESFTGPQCQTLVDWCSRQPCQNGGRCVQTGAYCLC PPGWSGRLCDIRSLPCREAAAQIGVRLEQLCQAGGQCVDEDSSHYCVCPEGRTGSHCEQEVDPCLAQPCQHGGTCRGYMGGYMCECLPGY NGDNCEDDVDECASQPCQHGGSCIDLVARYLCSCPPGTLGVLCEINEDDCGPGPPLDSGPRCLHNGTCVDLVGGFRCTCPPGYTGLRCEA DINECRSGACHAAHTRDCLQDPGGGFRCLCHAGFSGPRCQTVLSPCESQPCQHGGQCRPSPGPGGGLTFTCHCAQPFWGPRCERVARSCR ELQCPVGVPCQQTPRGPRCACPPGLSGPSCRSFPGSPPGASNASCAAAPCLHGGSCRPAPLAPFFRCACAQGWTGPRCEAPAAAPEVSEE PRCPRAACQAKRGDQRCDRECNSPGCGWDGGDCSLSVGDPWRQCEALQCWRLFNNSRCDPACSSPACLYDNFDCHAGGRERTCNPVYEKY CADHFADGRCDQGCNTEECGWDGLDCASEVPALLARGVLVLTVLLPPEELLRSSADFLQRLSAILRTSLRFRLDAHGQAMVFPYHRPSPG SEPRARRELAPEVIGSVVMLEIDNRLCLQSPENDHCFPDAQSAADYLGALSAVERLDFPYPLRDVRGEPLEPPEPSVPLLPLLVAGAVLL LVILVLGVMVARRKREHSTLWFPEGFSLHKDVASGHKGRREPVGQDALGMKNMAKGESLMGEVATDWMDTECPEAKRLKVEEPGMGAEEA VDCRQWTQHHLVAADIRVAPAMALTPPQGDADADGMDVNVRGPDGFTPLMLASFCGGALEPMPTEEDEADDTSASIISDLICQGAQLGAR TDRTGETALHLAARYARADAAKRLLDAGADTNAQDHSGRTPLHTAVTADAQGVFQILIRNRSTDLDARMADGSTALILAARLAVEGMVEE LIASHADVNAVDELGKSALHWAAAVNNVEATLALLKNGANKDMQDSKEETPLFLAAREGSYEAAKLLLDHFANREITDHLDRLPRDVAQE RLHQDIVRLLDQPSGPRSPPGPHGLGPLLCPPGAFLPGLKAAQSGSKKSRRPPGKAGLGPQGPRGRGKKLTLACPGPLADSSVTLSPVDS LDSPRPFGGPPASPGGFPLEGPYAAATATAVSLAQLGGPGRAGLGRQPPGGCVLSLGLLNPVAVPLDWARLPPPAPPGPSFLLPLAPGPQ LLNPGTPVSPQERPPPYLAVPGHGEEYPAAGAHSSPPKARFLRVPSEHPYLTPSPESPEHWASPSPPSLSDWSESTPSPATATGAMATTT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for THOP1-NOTCH3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for THOP1-NOTCH3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for THOP1-NOTCH3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
