
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TMBIM6-ARPC3 (FusionGDB2 ID:91308) |
Fusion Gene Summary for TMBIM6-ARPC3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TMBIM6-ARPC3 | Fusion gene ID: 91308 | Hgene | Tgene | Gene symbol | TMBIM6 | ARPC3 | Gene ID | 7009 | 10094 |
| Gene name | transmembrane BAX inhibitor motif containing 6 | actin related protein 2/3 complex subunit 3 | |
| Synonyms | BAXI1|BI-1|TEGT | ARC21|p21-Arc | |
| Cytomap | 12q13.12 | 12q24.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | bax inhibitor 1testis enhanced gene transcripttestis-enhanced gene transcript proteintransmembrane BAX inhibitor motif-containing protein 6 | actin-related protein 2/3 complex subunit 3ARP2/3 protein complex subunit p21actin related protein 2/3 complex, subunit 3, 21kDaarp2/3 complex 21 kDa subunit | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O15145 | |
| Ensembl transtripts involved in fusion gene | ENST00000267115, ENST00000395006, ENST00000423828, ENST00000547798, ENST00000549385, ENST00000552699, ENST00000548589, | ENST00000471641, ENST00000228825, | |
| Fusion gene scores | * DoF score | 18 X 20 X 7=2520 | 6 X 4 X 4=96 |
| # samples | 21 | 7 | |
| ** MAII score | log2(21/2520*10)=-3.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(7/96*10)=-0.45567948377619 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TMBIM6 [Title/Abstract] AND ARPC3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TMBIM6(50149538)-ARPC3(110878193), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TMBIM6 | GO:0010523 | negative regulation of calcium ion transport into cytosol | 26582200 |
| Hgene | TMBIM6 | GO:0032091 | negative regulation of protein binding | 19328063 |
| Hgene | TMBIM6 | GO:0033119 | negative regulation of RNA splicing | 19328063 |
| Hgene | TMBIM6 | GO:0034620 | cellular response to unfolded protein | 19328063 |
| Hgene | TMBIM6 | GO:0060702 | negative regulation of endoribonuclease activity | 19328063 |
| Hgene | TMBIM6 | GO:1902065 | response to L-glutamate | 21075086 |
| Hgene | TMBIM6 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway | 21075086 |
| Hgene | TMBIM6 | GO:1903298 | negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway | 21075086 |
| Hgene | TMBIM6 | GO:2001234 | negative regulation of apoptotic signaling pathway | 21075086 |
| Tgene | ARPC3 | GO:0034314 | Arp2/3 complex-mediated actin nucleation | 11741539 |
 Fusion gene breakpoints across TMBIM6 (5'-gene) Fusion gene breakpoints across TMBIM6 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
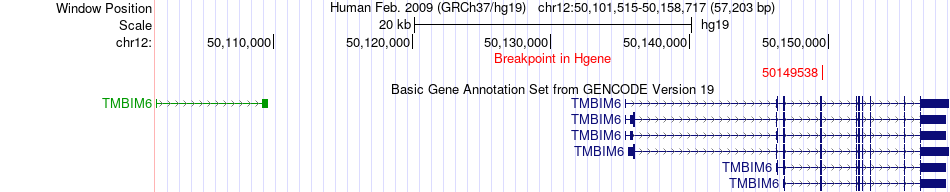 |
 Fusion gene breakpoints across ARPC3 (3'-gene) Fusion gene breakpoints across ARPC3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
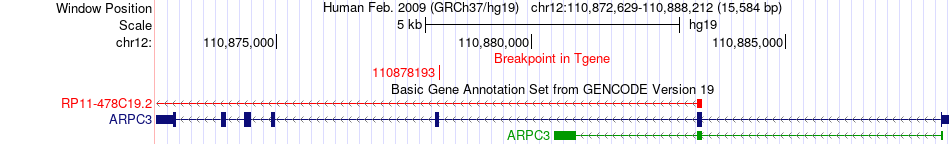 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | Non-Cancer | 61N | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
Top |
Fusion Gene ORF analysis for TMBIM6-ARPC3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000267115 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000395006 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000423828 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000547798 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000549385 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| 5CDS-intron | ENST00000552699 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000267115 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000395006 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000423828 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000547798 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000549385 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| In-frame | ENST00000552699 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| intron-3CDS | ENST00000548589 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
| intron-intron | ENST00000548589 | ENST00000471641 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000552699 | TMBIM6 | chr12 | 50149538 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1456 | 695 | 235 | 1125 | 296 |
| ENST00000267115 | TMBIM6 | chr12 | 50149538 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1132 | 371 | 34 | 801 | 255 |
| ENST00000549385 | TMBIM6 | chr12 | 50149538 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1274 | 513 | 227 | 943 | 238 |
| ENST00000423828 | TMBIM6 | chr12 | 50149538 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1549 | 788 | 328 | 1218 | 296 |
| ENST00000395006 | TMBIM6 | chr12 | 50149538 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1085 | 324 | 2 | 754 | 250 |
| ENST00000547798 | TMBIM6 | chr12 | 50149538 | + | ENST00000228825 | ARPC3 | chr12 | 110878193 | - | 1000 | 239 | 31 | 669 | 212 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000552699 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - | 0.001484149 | 0.9985159 |
| ENST00000267115 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - | 0.002930576 | 0.9970694 |
| ENST00000549385 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - | 0.00415884 | 0.9958411 |
| ENST00000423828 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - | 0.00159997 | 0.99840003 |
| ENST00000395006 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - | 0.003138189 | 0.9968618 |
| ENST00000547798 | ENST00000228825 | TMBIM6 | chr12 | 50149538 | + | ARPC3 | chr12 | 110878193 | - | 0.00303699 | 0.99696296 |
Top |
Fusion Genomic Features for TMBIM6-ARPC3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
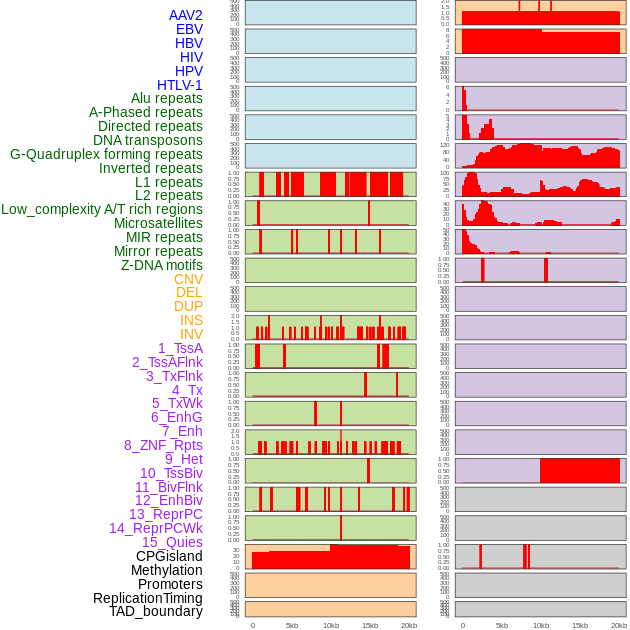 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TMBIM6-ARPC3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr12:50149538/chr12:110878193) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | ARPC3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Component of the Arp2/3 complex, a multiprotein complex that mediates actin polymerization upon stimulation by nucleation-promoting factor (NPF) (PubMed:9230079). The Arp2/3 complex mediates the formation of branched actin networks in the cytoplasm, providing the force for cell motility (PubMed:9230079). In addition to its role in the cytoplasmic cytoskeleton, the Arp2/3 complex also promotes actin polymerization in the nucleus, thereby regulating gene transcription and repair of damaged DNA (PubMed:29925947). The Arp2/3 complex promotes homologous recombination (HR) repair in response to DNA damage by promoting nuclear actin polymerization, leading to drive motility of double-strand breaks (DSBs) (PubMed:29925947). {ECO:0000269|PubMed:29925947, ECO:0000269|PubMed:9230079}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 1_29 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 51_52 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 74_86 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 1_29 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 51_52 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 74_86 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 1_29 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 51_52 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 74_86 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 30_50 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 53_73 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 30_50 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 53_73 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 30_50 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 53_73 | 95 | 238.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 207_227 | 95 | 238.0 | Intramembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 207_227 | 95 | 238.0 | Intramembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 207_227 | 95 | 238.0 | Intramembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 108_112 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 134_139 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 161_166 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 188_206 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 228_237 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 108_112 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 134_139 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 161_166 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 188_206 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 228_237 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 108_112 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 134_139 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 161_166 | 95 | 238.0 | Topological domain | Lumenal |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 188_206 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 228_237 | 95 | 238.0 | Topological domain | Cytoplasmic |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 113_133 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 140_160 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 167_187 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000267115 | + | 4 | 10 | 87_107 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 113_133 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 140_160 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 167_187 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000395006 | + | 3 | 9 | 87_107 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 113_133 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 140_160 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 167_187 | 95 | 238.0 | Transmembrane | Helical |
| Hgene | TMBIM6 | chr12:50149538 | chr12:110878193 | ENST00000549385 | + | 5 | 11 | 87_107 | 95 | 238.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for TMBIM6-ARPC3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >91308_91308_1_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000267115_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1132nt_BP=371nt AGAGCACATCCGGTGTTAGAAGCGCTGGTAGGCCTTGGAGAGGCGGGTTAGGAAGAGTGGAGACTGCTGCACGGACTCTGGAACCATGAA CATATTTGATCGAAAGATCAACTTTGATGCGCTTTTAAAATTTTCTCATATAACCCCGTCAACGCAGCAGCACCTGAAGAAGGTCTATGC AAGTTTTGCCCTTTGTATGTTTGTGGCGGCTGCAGGGGCCTATGTCCATATGGTCACTCATTTCATTCAGGCTGGCCTGCTGTCTGCCTT GGGCTCCCTGATATTGATGATTTGGCTGATGGCAACACCTCATAGCCATGAAACTGAACAGAAAAGACTGGGACTTCTTGCTGGATTTGC ATTCCTTACAGCAAAAGATACAGATATTGTGGATGAAGCCATCTATTACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAA TGAAGCTGATAGGACCTTGATATATATAACTCTCTACATTTCTGAATGTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGA GAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCATTCCTGGAGAGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAA CAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAACAGCTAAGGCAAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCA GAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGAAGAGACAGTTCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCC CGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTTCCAGCAAGATGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGA AGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGATCAAGAATTTGGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAAT TTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAGTAATAATAAAGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGT >91308_91308_1_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000267115_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=255AA_BP=112 MERRVRKSGDCCTDSGTMNIFDRKINFDALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWLMA TPHSHETEQKRLGLLAGFAFLTAKDTDIVDEAIYYFKANVFFKNYEIKNEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNF -------------------------------------------------------------- >91308_91308_2_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000395006_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1085nt_BP=324nt TTCTGCAGAGTGGAGACTGCTGCACGGACTCTGGAACCATGAACATATTTGATCGAAAGATCAACTTTGATGCGCTTTTAAAATTTTCTC ATATAACCCCGTCAACGCAGCAGCACCTGAAGAAGGTCTATGCAAGTTTTGCCCTTTGTATGTTTGTGGCGGCTGCAGGGGCCTATGTCC ATATGGTCACTCATTTCATTCAGGCTGGCCTGCTGTCTGCCTTGGGCTCCCTGATATTGATGATTTGGCTGATGGCAACACCTCATAGCC ATGAAACTGAACAGAAAAGACTGGGACTTCTTGCTGGATTTGCATTCCTTACAGCAAAAGATACAGATATTGTGGATGAAGCCATCTATT ACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTACATTTCTGAAT GTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCATTCCTGGAG AGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAACAGCTAAGGC AAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGAAGAGACAGT TCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTTCCAGCAAGA TGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGATCAAGAATTT GGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAGTAATAATAA AGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAATTCACATTCCA >91308_91308_2_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000395006_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=250AA_BP=107 LQSGDCCTDSGTMNIFDRKINFDALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWLMATPHSH ETEQKRLGLLAGFAFLTAKDTDIVDEAIYYFKANVFFKNYEIKNEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGE -------------------------------------------------------------- >91308_91308_3_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000423828_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1549nt_BP=788nt ACAGTGCTGAAGCGGGGGTGGGGCGGAGGCGAGTCTGCGGGGGTTTTGGGGGGTGTCGAGGCCTCTATTCTGCCCCAGAGCGCTGGCGAA GGCCCCTTCTCAGCCCGCCTTTTCCTTTCTCCCGCTCCTTCTCCTCTACTAAGTGTAGACGCAGGGCCCCTTGGCCTAGCTTCGATCGTT CGAATTCAGAGCACGTCCTTCCGAGGTGAAGGAACGCGAAACTCCACCCATCCGATTGCTGTTCGGCTGCGGGCGGGTCCTTTGGTCGGG CTGACCCTGGGTGAGCGGCCCGGAGCCAAGACTCGAGGTAGGGCCTGGCGGGCGGGTGATGTCACACTCCTCTGTGACACGCGAGGCTCC TCAGTTACTTAGCCAACGGCAGAGGCGGGAAGTGAGAGGAGTCTGGGGCTGGGGCTGCCTTCCAGGCCCACGGGGCGGCCCCGCTCTTTT CGGATTGGTTACCTTTGGGCAGAGTGGAGACTGCTGCACGGACTCTGGAACCATGAACATATTTGATCGAAAGATCAACTTTGATGCGCT TTTAAAATTTTCTCATATAACCCCGTCAACGCAGCAGCACCTGAAGAAGGTCTATGCAAGTTTTGCCCTTTGTATGTTTGTGGCGGCTGC AGGGGCCTATGTCCATATGGTCACTCATTTCATTCAGGCTGGCCTGCTGTCTGCCTTGGGCTCCCTGATATTGATGATTTGGCTGATGGC AACACCTCATAGCCATGAAACTGAACAGAAAAGACTGGGACTTCTTGCTGGATTTGCATTCCTTACAGCAAAAGATACAGATATTGTGGA TGAAGCCATCTATTACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCT CTACATTTCTGAATGTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTT TCCCATTCCTGGAGAGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTT ACAACAGCTAAGGCAAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTT TGTGAAGAGACAGTTCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGC ATTTTCCAGCAAGATGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTC TTGATCAAGAATTTGGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGA ATAAGTAATAATAAAGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATA >91308_91308_3_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000423828_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=296AA_BP=153 MSHSSVTREAPQLLSQRQRREVRGVWGWGCLPGPRGGPALFGLVTFGQSGDCCTDSGTMNIFDRKINFDALLKFSHITPSTQQHLKKVYA SFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWLMATPHSHETEQKRLGLLAGFAFLTAKDTDIVDEAIYYFKANVFFKNYEIKN EADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKPANKQEDEVMRAYLQQLRQETGLRLCEKVFDPQ -------------------------------------------------------------- >91308_91308_4_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000547798_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1000nt_BP=239nt TTTTCTTAGAACCCCGTCAACGCAGCAGCACCTGAAGAAGGTCTATGCAAGTTTTGCCCTTTGTATGTTTGTGGCGGCTGCAGGGGCCTA TGTCCATATGGTCACTCATTTCATTCAGGCTGGCCTGCTGTCTGCCTTGGGCTCCCTGATATTGATGATTTGGCTGATGGCAACACCTCA TAGCCATGAAACTGAACAGAAAAGACTGGGACTTCTTGCTGGATTTGCATTCCTTACAGCAAAAGATACAGATATTGTGGATGAAGCCAT CTATTACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTACATTTC TGAATGTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCATTCC TGGAGAGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAACAGCT AAGGCAAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGAAGAG ACAGTTCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTTCCAG CAAGATGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGATCAAG AATTTGGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAGTAAT AATAAAGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAATTCACA >91308_91308_4_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000547798_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=212AA_BP=69 MKKVYASFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWLMATPHSHETEQKRLGLLAGFAFLTAKDTDIVDEAIYYFKANVFFK NYEIKNEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKPANKQEDEVMRAYLQQLRQETGLRLCE -------------------------------------------------------------- >91308_91308_5_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000549385_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1274nt_BP=513nt AGAAGCGCTGGTAGGCCTTGGAGAGGCGGGTTAGGAAGACGCAGGGCCCCTTGGCCTAGCTTCGATCGTTCGAATTCAGAGCACGTCCTT CCGAGGTGAAGGAACGCGAAACTCCACCCATCCGATTGCTGTTCGGCTGCGGGCGGGTCCTTTGGTCGGGCTGACCCTGGGTGAGCGGCC CGGAGCCAAGACTCGAGAGTGGAGACTGCTGCACGGACTCTGGAACCATGAACATATTTGATCGAAAGATCAACTTTGATGCGCTTTTAA AATTTTCTCATATAACCCCGTCAACGCAGCAGCACCTGAAGAAGGTCTATGCAAGTTTTGCCCTTTGTATGTTTGTGGCGGCTGCAGGGG CCTATGTCCATATGGTCACTCATTTCATTCAGGCTGGCCTGCTGTCTGCCTTGGGCTCCCTGATATTGATGATTTGGCTGATGGCAACAC CTCATAGCCATGAAACTGAACAGAAAAGACTGGGACTTCTTGCTGGATTTGCATTCCTTACAGCAAAAGATACAGATATTGTGGATGAAG CCATCTATTACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTACA TTTCTGAATGTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCCCA TTCCTGGAGAGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACAAC AGCTAAGGCAAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGTGA AGAGACAGTTCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATTTT CCAGCAAGATGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTGAT CAAGAATTTGGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATAAG TAATAATAAAGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAATT >91308_91308_5_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000549385_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=238AA_BP=95 MNIFDRKINFDALLKFSHITPSTQQHLKKVYASFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWLMATPHSHETEQKRLGLLAG FAFLTAKDTDIVDEAIYYFKANVFFKNYEIKNEADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKP -------------------------------------------------------------- >91308_91308_6_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000552699_ARPC3_chr12_110878193_ENST00000228825_length(transcript)=1456nt_BP=695nt AGAGCACATCCGGTGTTAGAAGCGCTGGTAGGCCTTGGAGAGGCGGGTTAGGAAGACGCAGGGCCCCTTGGCCTAGCTTCGATCGTTCGA ATTCAGAGCACGTCCTTCCGAGGTGAAGGAACGCGAAACTCCACCCATCCGATTGCTGTTCGGCTGCGGGCGGGTCCTTTGGTCGGGCTG ACCCTGGGTGAGCGGCCCGGAGCCAAGACTCGAGGTAGGGCCTGGCGGGCGGGTGATGTCACACTCCTCTGTGACACGCGAGGCTCCTCA GTTACTTAGCCAACGGCAGAGGCGGGAAGTGAGAGGAGTCTGGGGCTGGGGCTGCCTTCCAGGCCCACGGGGCGGCCCCGCTCTTTTCGG ATTGGTTACCTTTGGGCAGAGTGGAGACTGCTGCACGGACTCTGGAACCATGAACATATTTGATCGAAAGATCAACTTTGATGCGCTTTT AAAATTTTCTCATATAACCCCGTCAACGCAGCAGCACCTGAAGAAGGTCTATGCAAGTTTTGCCCTTTGTATGTTTGTGGCGGCTGCAGG GGCCTATGTCCATATGGTCACTCATTTCATTCAGGCTGGCCTGCTGTCTGCCTTGGGCTCCCTGATATTGATGATTTGGCTGATGGCAAC ACCTCATAGCCATGAAACTGAACAGAAAAGACTGGGACTTCTTGCTGGATTTGCATTCCTTACAGCAAAAGATACAGATATTGTGGATGA AGCCATCTATTACTTCAAGGCCAATGTCTTCTTCAAAAACTATGAAATTAAGAATGAAGCTGATAGGACCTTGATATATATAACTCTCTA CATTTCTGAATGTCTGAAGAAACTGCAAAAGTGCAATTCCAAAAGCCAAGGTGAGAAAGAAATGTATACGCTGGGAATCACTAATTTTCC CATTCCTGGAGAGCCTGGTTTTCCACTTAACGCAATTTATGCCAAACCTGCAAACAAACAGGAAGATGAAGTGATGAGAGCCTATTTACA ACAGCTAAGGCAAGAGACTGGACTGAGACTTTGTGAGAAAGTTTTCGACCCTCAGAATGATAAACCCAGCAAGTGGTGGACTTGCTTTGT GAAGAGACAGTTCATGAACAAGAGTCTTTCAGGACCTGGACAGTGAAGGGAGCCCGGGCAGCCACCGTCTCCAGAGCCCTGGGCAGCATT TTCCAGCAAGATGTACACAATCTTTTGCCTTTATTTCGTAAAGTTTTATACAGAAGAGAGAAGAGCATGTCTTTACTTGAAAAACTCTTG ATCAAGAATTTGGGTGGGAGAAAAGAAAGTGGGTTATCAAGGGTGATTTGAAATTTTCTGCAGCATTAAAGCTGGCGCTTAATAAGAATA AGTAATAATAAAGAAATTTCTAACATTCCATGTCAGAAAAACTTTATTGCCAGTCTTTGACATTCACTTACAATTTGTTTTGAGATAAAA >91308_91308_6_TMBIM6-ARPC3_TMBIM6_chr12_50149538_ENST00000552699_ARPC3_chr12_110878193_ENST00000228825_length(amino acids)=296AA_BP=153 MSHSSVTREAPQLLSQRQRREVRGVWGWGCLPGPRGGPALFGLVTFGQSGDCCTDSGTMNIFDRKINFDALLKFSHITPSTQQHLKKVYA SFALCMFVAAAGAYVHMVTHFIQAGLLSALGSLILMIWLMATPHSHETEQKRLGLLAGFAFLTAKDTDIVDEAIYYFKANVFFKNYEIKN EADRTLIYITLYISECLKKLQKCNSKSQGEKEMYTLGITNFPIPGEPGFPLNAIYAKPANKQEDEVMRAYLQQLRQETGLRLCEKVFDPQ -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TMBIM6-ARPC3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TMBIM6-ARPC3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TMBIM6-ARPC3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
