
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TMEM9-MYO5B (FusionGDB2 ID:92304) |
Fusion Gene Summary for TMEM9-MYO5B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TMEM9-MYO5B | Fusion gene ID: 92304 | Hgene | Tgene | Gene symbol | TMEM9 | MYO5B | Gene ID | 252839 | 4645 |
| Gene name | transmembrane protein 9 | myosin VB | |
| Synonyms | DERM4|TMEM9A | - | |
| Cytomap | 1q32.1 | 18q21.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transmembrane protein 9dermal papilla-derived protein 4 | unconventional myosin-VbMYO5B variant proteinmyosin-Vb | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9ULV0 | |
| Ensembl transtripts involved in fusion gene | ENST00000367330, ENST00000367332, ENST00000367333, ENST00000367334, ENST00000485839, ENST00000472411, | ENST00000587895, ENST00000592688, ENST00000285039, ENST00000324581, | |
| Fusion gene scores | * DoF score | 5 X 6 X 2=60 | 11 X 10 X 7=770 |
| # samples | 7 | 14 | |
| ** MAII score | log2(7/60*10)=0.222392421336448 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(14/770*10)=-2.4594316186373 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TMEM9 [Title/Abstract] AND MYO5B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TMEM9(201112948)-MYO5B(47421544), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across TMEM9 (5'-gene) Fusion gene breakpoints across TMEM9 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
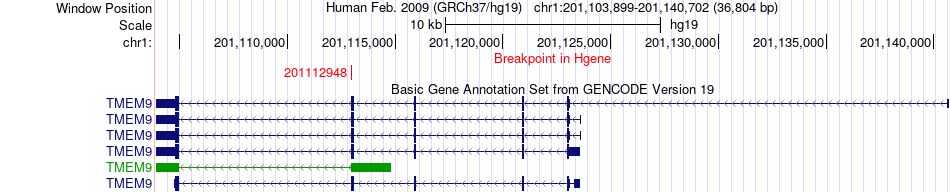 |
 Fusion gene breakpoints across MYO5B (3'-gene) Fusion gene breakpoints across MYO5B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
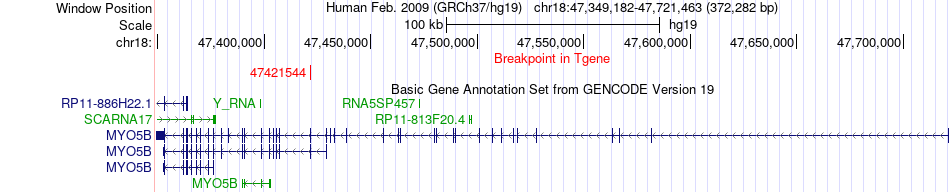 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | LUAD | TCGA-91-6848-01A | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
Top |
Fusion Gene ORF analysis for TMEM9-MYO5B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000367330 | ENST00000587895 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367330 | ENST00000592688 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367332 | ENST00000587895 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367332 | ENST00000592688 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367333 | ENST00000587895 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367333 | ENST00000592688 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367334 | ENST00000587895 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000367334 | ENST00000592688 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000485839 | ENST00000587895 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5CDS-intron | ENST00000485839 | ENST00000592688 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5UTR-3CDS | ENST00000472411 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5UTR-3CDS | ENST00000472411 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5UTR-intron | ENST00000472411 | ENST00000587895 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| 5UTR-intron | ENST00000472411 | ENST00000592688 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000367332 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000367332 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000367333 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000367333 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000367334 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000367334 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000485839 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| Frame-shift | ENST00000485839 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| In-frame | ENST00000367330 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
| In-frame | ENST00000367330 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000367330 | TMEM9 | chr1 | 201112948 | - | ENST00000285039 | MYO5B | chr18 | 47421544 | - | 7310 | 916 | 427 | 3651 | 1074 |
| ENST00000367330 | TMEM9 | chr1 | 201112948 | - | ENST00000324581 | MYO5B | chr18 | 47421544 | - | 3785 | 916 | 427 | 3573 | 1048 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000367330 | ENST00000285039 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - | 0.000593758 | 0.9994062 |
| ENST00000367330 | ENST00000324581 | TMEM9 | chr1 | 201112948 | - | MYO5B | chr18 | 47421544 | - | 0.007092727 | 0.9929073 |
Top |
Fusion Genomic Features for TMEM9-MYO5B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
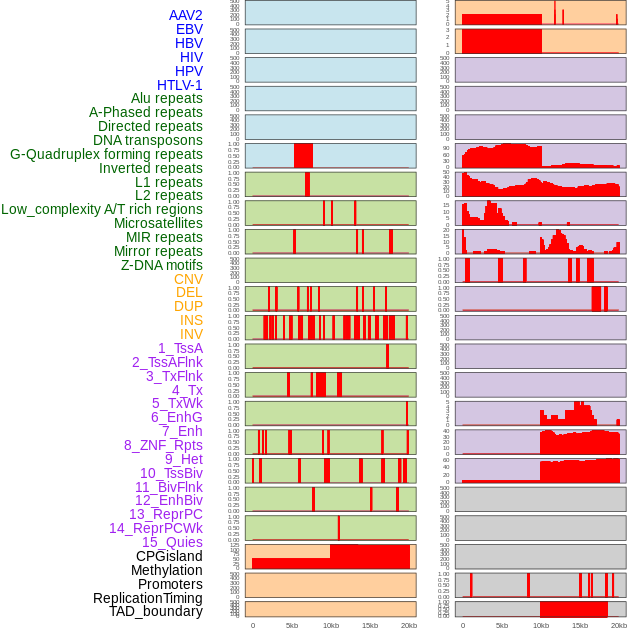 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TMEM9-MYO5B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr1:201112948/chr18:47421544) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MYO5B |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: May be involved in vesicular trafficking via its association with the CART complex. The CART complex is necessary for efficient transferrin receptor recycling but not for EGFR degradation. Required in a complex with RAB11A and RAB11FIP2 for the transport of NPC1L1 to the plasma membrane. Together with RAB11A participates in CFTR trafficking to the plasma membrane and TF (transferrin) recycling in nonpolarized cells. Together with RAB11A and RAB8A participates in epithelial cell polarization. Together with RAB25 regulates transcytosis. {ECO:0000269|PubMed:21206382, ECO:0000269|PubMed:21282656}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TMEM9 | chr1:201112948 | chr18:47421544 | ENST00000367330 | - | 4 | 5 | 21_89 | 133 | 184.0 | Topological domain | Extracellular |
| Hgene | TMEM9 | chr1:201112948 | chr18:47421544 | ENST00000367334 | - | 5 | 6 | 21_89 | 133 | 184.0 | Topological domain | Extracellular |
| Hgene | TMEM9 | chr1:201112948 | chr18:47421544 | ENST00000367330 | - | 4 | 5 | 90_110 | 133 | 184.0 | Transmembrane | Helical |
| Hgene | TMEM9 | chr1:201112948 | chr18:47421544 | ENST00000367334 | - | 5 | 6 | 90_110 | 133 | 184.0 | Transmembrane | Helical |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 1341_1471 | 937 | 1849.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 1526_1803 | 937 | 1849.0 | Domain | Dilute |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TMEM9 | chr1:201112948 | chr18:47421544 | ENST00000367330 | - | 4 | 5 | 111_183 | 133 | 184.0 | Topological domain | Cytoplasmic |
| Hgene | TMEM9 | chr1:201112948 | chr18:47421544 | ENST00000367334 | - | 5 | 6 | 111_183 | 133 | 184.0 | Topological domain | Cytoplasmic |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 899_1266 | 937 | 1849.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 801_916 | 937 | 1849.0 | Compositional bias | Note=Arg-rich | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 69_761 | 937 | 1849.0 | Domain | Myosin motor | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 769_798 | 937 | 1849.0 | Domain | IQ 1 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 792_821 | 937 | 1849.0 | Domain | IQ 2 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 817_848 | 937 | 1849.0 | Domain | IQ 3 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 840_869 | 937 | 1849.0 | Domain | IQ 4 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 865_896 | 937 | 1849.0 | Domain | IQ 5 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 888_917 | 937 | 1849.0 | Domain | IQ 6 | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 8_60 | 937 | 1849.0 | Domain | Myosin N-terminal SH3-like | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 163_170 | 937 | 1849.0 | Nucleotide binding | ATP | |
| Tgene | MYO5B | chr1:201112948 | chr18:47421544 | ENST00000285039 | 20 | 40 | 640_662 | 937 | 1849.0 | Region | Actin-binding |
Top |
Fusion Gene Sequence for TMEM9-MYO5B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >92304_92304_1_TMEM9-MYO5B_TMEM9_chr1_201112948_ENST00000367330_MYO5B_chr18_47421544_ENST00000285039_length(transcript)=7310nt_BP=916nt GTGGACGCGGCATTGGGGGAGGGGACGGGGGTCGCGGGGCTATTGGGGGTCCCAGTCCGGCTTAAGACCTCTGGGCGTGGTGGGCACCGG CCTTCGCCGTCGGAGCAGGGGTCCCAGACTCGCACCCAAAAGTTCCCGTCGGGCCCGAGCGGGGAGGAGCTGCGGGGCGGGCAGAGGCGG AGATGGCGGAGGGCGGTGGGACGTGATGCGCGGGTCAGAGCCGGGCCTTGAGAAGGAACTGGAGGCCCCTGGCAGCGGTGTCCCCTCGAG GACCCCTCTGCCGGGCTCACCAGGTGGCTGGCGGAGAGCCGGAGAGTTGGGCGGGAGGGAAGCAGGGTTTGGCGACCAGGTTTAGCAGGC CTTGGAATCCTGGAGCCCAGCCCCCAACCCCATTCCCACTTGGCCGGATGGTTAAGGGCCTAGCGTGCTGGCTGCGGTGTGGCCACCTTC CCCTTCTCCGACTCCGTATCTCTTTTTCCAGGTGTCCGGCTTTGCTGGCCCAGCAAGCCTGATAAGCATGAAGCTCTTATCTTTGGTGGC TGTGGTCGGGTGTTTGCTGGTGCCCCCAGCTGAAGCCAACAAGAGTTCTGAAGATATCCGGTGCAAATGCATCTGTCCACCTTATAGAAA CATCAGTGGGCACATTTACAACCAGAATGTATCCCAGAAGGACTGCAACTGCCTGCACGTGGTGGAGCCCATGCCAGTGCCTGGCCATGA CGTGGAGGCCTACTGCCTGCTGTGCGAGTGCAGGTACGAGGAGCGCAGCACCACCACCATCAAGGTCATCATTGTCATCTACCTGTCCGT GGTGGGTGCCCTGTTGCTCTACATGGCCTTCCTGATGCTGGTGGACCCTCTGATCCGAAAGCCGGATGCATATACTGAGCAACTGCACAA TGAGGAGGAGAATGAGAACAAAGAGTTCAAGACACTTTCAGAGCAGTTGTCCGTGACCACCTCAACATACACCATGGAGGTAGAGCGGCT GAAGAAGGAGCTGGTGCACTACCAGCAGAGCCCAGGTGAGGACACCAGCCTCAGGCTGCAGGAGGAGGTGGAGAGCCTGCGCACAGAGCT GCAGAGGGCCCACTCGGAGCGCAAGATCTTGGAGGACGCCCACAGCAGGGAGAAAGATGAGCTGAGGAAGCGAGTTGCAGACCTGGAGCA AGAAAATGCTCTCTTGAAAGATGAGAAAGAACAGCTCAACAACCAAATCCTGTGCCAGTCTAAAGATGAATTTGCCCAGAACTCTGTGAA GGAAAATCTCATGAAGAAAGAACTGGAGGAGGAGCGATCCCGGTACCAGAACCTTGTGAAGGAATATTCACAGTTGGAGCAGAGATACGA CAACCTTCGGGATGAAATGACCATCATAAAGCAAACTCCAGGTCATAGGCGGAACCCATCAAACCAAAGTAGCTTAGAATCTGACTCCAA TTACCCCTCCATCTCCACATCTGAGATCGGAGACACTGAGGATGCCCTCCAGCAGGTGGAGGAAATTGGCCTGGAGAAGGCAGCCATGGA CATGACGGTCTTCCTGAAGCTGCAGAAGAGAGTACGGGAGCTGGAGCAGGAGAGGAAAAAGCTGCAAGTGCAGCTGGAGAAGAGAGAACA GCAGGACAGCAAGAAAGTCCAGGCGGAACCACCACAGACTGACATAGATTTGGACCCGAATGCAGATCTGGCCTACAATAGTCTGAAGAG GCAAGAGCTGGAGTCAGAGAACAAAAAGCTGAAGAATGACCTGAATGAGCTGAGGAAAGCCGTGGCCGACCAAGCCACGCAGAATAACTC CAGCCACGGCTCCCCAGATAGCTACAGCCTCCTGCTGAACCAGCTCAAGCTGGCCCACGAGGAGCTCGAGGTGCGCAAGGAGGAGGTGCT CATCCTCAGGACCCAGATCGTGAGCGCCGACCAGCGGCGACTCGCCGGCAGGAACGCGGAGCCGAACATTAATGCCAGATCAAGTTGGCC TAACAGTGAAAAGCATGTTGACCAGGAGGATGCCATTGAGGCCTATCACGGGGTCTGCCAGACAAACAGCAAGACTGAGGATTGGGGATA TTTAAATGAAGATGGAGAACTCGGCTTGGCCTACCAAGGCCTAAAGCAAGTTGCCAGGCTGCTGGAGGCTCAGCTGCAGGCCCAGAGCCT GGAGCATGAGGAGGAGGTGGAGCATCTCAAGGCTCAGCTCGAGGCCCTGAAGGAGGAGATGGACAAACAGCAGCAGACCTTCTGCCAGAC GCTACTGCTCTCCCCAGAGGCCCAGGTGGAATTCGGCGTTCAGCAGGAAATATCCCGGCTGACCAACGAGAATCTGGACCTTAAAGAACT GGTAGAAAAGCTGGAAAAGAATGAGAGGAAGCTCAAAAAGCAACTGAAGATTTACATGAAGAAAGCCCAGGACCTAGAAGCTGCCCAGGC ATTGGCCCAGAGTGAGAGGAAGCGCCATGAGCTCAACAGGCAGGTCACGGTCCAGCGGAAAGAGAAGGATTTCCAGGGCATGCTGGAGTA CCACAAAGAGGACGAGGCCCTCCTCATCCGGAACCTGGTGACAGACTTGAAGCCCCAGATGCTGTCGGGCACAGTGCCCTGTCTCCCCGC CTACATCCTCTACATGTGCATCCGGCACGCGGACTACACCAACGACGATCTCAAGGTGCACTCCCTGCTGACCTCCACCATCAACGGCAT TAAGAAAGTCCTGAAAAAGCACAATGATGACTTTGAGATGACGTCATTCTGGTTATCCAACACCTGCCGCCTTCTTCACTGTCTGAAGCA GTACAGCGGGGATGAGGGCTTCATGACTCAGAACACTGCAAAGCAGAATGAACACTGTCTTAAGAATTTTGACCTCACCGAATACCGTCA GGTGCTGAGTGACCTTTCCATTCAGATCTACCAGCAGCTCATTAAAATTGCCGAGGGCGTGTTACAGCCGATGATAGTTTCTGCCATGTT GGAAAATGAGAGCATTCAGGGTCTATCTGGTGTGAAGCCCACCGGCTACCGGAAGCGCTCCTCCAGCATGGCAGATGGGGATAACTCATA CTGCCTGGAAGCTATCATCCGCCAGATGAATGCCTTTCATACAGTCATGTGTGACCAGGGCTTGGACCCTGAGATCATCCTGCAGGTATT CAAACAGCTCTTCTACATGATCAACGCAGTGACTCTTAACAACCTGCTCTTGCGGAAGGACGTCTGCTCTTGGAGCACAGGCATGCAACT CAGGTACAATATAAGTCAGCTTGAGGAGTGGCTTCGGGGAAGAAACCTTCACCAGAGTGGAGCAGTTCAGACCATGGAACCTCTGATCCA AGCAGCCCAGCTCCTGCAATTAAAGAAGAAAACCCAGGAGGACGCAGAGGCTATCTGCTCCCTGTGTACCTCCCTCAGCACCCAGCAGAT TGTCAAAATTTTAAACCTTTATACTCCCCTGAATGAATTTGAAGAACGGGTAACAGTGGCCTTTATACGAACAATCCAGGCACAACTACA AGAGCGGAATGACCCTCAGCAACTGCTATTAGATGCCAAGCACATGTTTCCTGTTTTGTTTCCATTTAATCCATCTTCTCTAACCATGGA CTCAATCCACATCCCAGCGTGTCTCAATCTGGAATTCCTCAATGAAGTCTGAAGATGCATGTTTCCAGCATTAGTTTGATTCCCAATGTG AGCAAGAAGGAAGTATATACAGTAAAGTAAATTCAAGGATCTGTTAAATCTGGTAAAAGTAGATCAAATCAGAGATTGACAGCCTGTGGA GGGTGCTGAACTATACAGAATTAGACACAACTATGTCATTATTTTTTGTACCTACTGCTCAGAATAAAAACACTTGAAATATGGAAGATT TTAAGTTTGATTTCAGTCCAACACATATACATAATTTATAGACACCAAGCAGTCCCCATAGACATATAAAAGGTGTCAATTCTATAAAAC GAAGCTGCCTAGTTTTGATCTTTGCATAGAACTAGAGAATGTCCAAATTAAAATACCAAATATATATAAGTCACATAAATTGCCTTCAAA GGGCTTTAACAAATAATGGTACTAATAACCATGATAATGGCATATACTGACATTTCCCAAAGTTTGCAAACCATAGGTGTGGTTGAGTTT GTGGTGAGATGTTTTAAGAACAAAAATATGGGGATGAGACTTCTGAGAAATATTCCCAAAATATTTTTTAATGGCTGATTATACACAGAC AGTGGTGTAACTGACCTCCAGACCAGACATTTTGAGTACTGGTTTCTGAAGCAAAATTAGAAGTGCCAGTCCTCAGTGTGCTCAAACGCT TTTGTGTTATCTTGATTTAATGGAAGAGATTATTAAAATGCTGCTATCCCAAACTCCAAGTGAGAAAGATGGAAAAATATTTTGTTTCTG ATGCTAGTCCATACACTTTCCAAGTCCCACAAAACTTTCACAAAAATGTATATAAGCTAAATATTAGAAACGGATAACAAACTTGTTTTA TTTATAGATGTAAAAACCAAACAAGTCAATATGAAAGCTTTTAATCTCTTAATACCATTAAGCTTCCAGTAAGAGCATCACATAATGCTC TACTGTTCCAGAAACCAAATAGTAAAACAAACTAAAGTTCGCACATCAGATCATCTGAAAAACCTTCAAAAATAATCAGTTCAGGGATAT TATACAAAAGTTTGGGTTTTTTTTTTTTAAGAGAATAAAATGGCTTAGGTCAACTTTCCCTTTTCAGGTTATTTTCAACGTTTTTCAAAT TTAGCACACAAAAAATTGTAAATATCTCTCCCACAAAATAAGGATTTTAAAAAAGTAATTCAGTAATATAACAGGCTTAGATGTTTGCTG CTCTTAGAATTTTTTTTAACTTGTTTTTGGTTTCTTCAAAAGCAAGCATTCAATTGGAAACCCATATTCTTTCCACACTTTTTTTTACTG TCTTTTCTGTATTTCTTGATAGCAGTATGCTGTTCCCATAAGAAAAAAATGGTATTTGCAAATCATGGAAGAACAGCCTCTGTATTACAT TGAGAAAATAAGATTTATCCATGAATTGGAAGTAGAACAGCCTGCCTTCACCCTCTTTTACTCAACCACCCAACTTAAAAGGCTCTTGGA AACACAGCACACTCCACGCTACCTTCTGCACTGTGCCCTTAGAGCACAGCTTCCTCAGTTGTTCTCTGCATCTCCTGGGGCTTAGGCCAG TCTTAGCCTGGGTTAAGGCTGCTGACATTGTGTTCCAATCAGTTGTCATGGGCATTATCCCCTCTACATCCACATTAAACATGCCGGCTT CTCTTGGGCATCGGCAGAGCTGTGCCTTTTTCTTTCAGTTACAGTTACATAATCACTGACGTCCATGACACTTACCCATGGATCCATGTG CTGACTTCATTTAGAAGGCCAATCTAAAACAACTGGGTTTGTGGCTACCTCTTTAAAGTTGTTTGTGAAGGATAATTTGTTTTTTAATGC ACTTTAGTTTGAAAGTGAGTCTCTTATGTAAGGACCATCCTTAAAAGACCAAAAATGCCTTGTTAGAGTGTTAAGGAGTTTTGACATGCA GTGGTTCCACAAACACAGTGGCTTACTATCCTTATACACTGTCTTATACCATCATTCTCTCCATCTCTCTTGGTCACTACTCTCTGCTGT CACTGGTTAATCACTAGGTGCCAAGAGCTTACTGAATAAAAGCTTGGCAATTAGAATAAATGGGGAGGGAAGGACCTTATGAATAGTCCA TTTAGCCTAAGAAATGGCAGATTTAGTTCTTCTCTTCCAAAAGATAAAGGTATATCCTGGAATTGTACTTAAAACTTACAGATGACTAAC AAATATATACTTTATATGTAGTTAATATTTAGATCTGTCTTATTTAATACTTGGAGGCTAGAAGAAGCATCTTTAGGGGAACTATATAAT CTTTTGTTAGCATTTTCTCTGCATTTTAAAAAATCATTTCAATTCAAACATTTATCAGTGTCATGAAATCAGTAATGACTCTTTAACAAT TCAGGTTTGAACTCTGCATTAGATGTCTCTTTAATTTTTTAATATTTAAAATTTAGTTGACATTTTTTTCACCAGGTGCCTTTAGCGGTT ACTAAGATAACTGACATCAGTTGTTTCTCTGAAATAAGTGTTGCTGTGGGAATAATTTTAATGTTCAAGGTGATATCATGGGGGAGTTTT GTCTTTTAAAACATTAGAAGCATTTTAAATATTAAGAATCAAATATTTATAGATCAAAACTTGTGTTTTAAGTATTATACGGGACCTGTT TACTTATAGTAAATGTGAATGTACACATGAGTTGTTGCTGAAGCTGACAAGCATATTACATACATGCATTTTCCCTGTGCCCTCATAGTT GCAGTTAGAGTTCCAGTACCTGTAGGCTCACCTGGGAGGCAGATTAGACCCAAAGGTAGATGTTTTTCCCCTTTCCATGAAGCATGTCAG TGGGAGTTGCTTCCTTTGATTTCCCTAGTACTAAATTTTAAGGCTTTTGTAAAAACAAAACAAAACTAGGAGCTTGGAACAGTTAAAAAT CAACACTGCTACCATCAATTCATCAAATATTTACTTAGAGCTTTCATACATTAAGATTCCAGTAACCAATAAATTAGAATTCATTTCTTC TGCATAAAGTAAATTTTCATACACTTGACCTACTAAGACAGCAAGGGTGTCCTAAATTGAGGCATTTGTATAATGCCTGCATAACTAAAT GGTCACTAAAATGGGACAGCATGGGGCAAGACCTTGTAGTTCTTCACAGAATATTTGTGGTCAGTTTCTCCAATTAATTTGCTGCATGAG CCAAATAACCATAATTCACTTTTTATACCCACTGGTGCCATAATTAGAGAATTAGAGGGTGTAGACAGAGGTTAATGCCAATGAGAAACA CAGGACAGGGTTTTTTTTATTATAAAGGTCATTAGATACAAAAGATTGTTTTTCAAAAAATTTCTAATTCTAACAAAGGGGATCAATCAG AAATGAAACTAAGCTACTTTCTAAAGTGACACTGTATCAGAATAATCCAGATTTGAATATAACATTTTGCCACCAACTGACATTTAGATG AAGGACTGCCTCTCTGAAAGAGTTCAGATCATATTCAGGGGTGAATCCAACACCATGGAAGAAAGACTACTGATGAAAATATTTTCCCAC TTTGCACAAATCTGTAAACTACACCTTTGTTTATAGAAAAATGCTTGTAATAGTCACTGTAATATTTAGCTGTGGATAAAAATTTGTGGA >92304_92304_1_TMEM9-MYO5B_TMEM9_chr1_201112948_ENST00000367330_MYO5B_chr18_47421544_ENST00000285039_length(amino acids)=1074AA_BP=163 MAAVWPPSPSPTPYLFFQVSGFAGPASLISMKLLSLVAVVGCLLVPPAEANKSSEDIRCKCICPPYRNISGHIYNQNVSQKDCNCLHVVE PMPVPGHDVEAYCLLCECRYEERSTTTIKVIIVIYLSVVGALLLYMAFLMLVDPLIRKPDAYTEQLHNEEENENKEFKTLSEQLSVTTST YTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKD EFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEI GLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVA DQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTN SKTEDWGYLNEDGELGLAYQGLKQVARLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTN ENLDLKELVEKLEKNERKLKKQLKIYMKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLS GTVPCLPAYILYMCIRHADYTNDDLKVHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKN FDLTEYRQVLSDLSIQIYQQLIKIAEGVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLD PEIILQVFKQLFYMINAVTLNNLLLRKDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLC -------------------------------------------------------------- >92304_92304_2_TMEM9-MYO5B_TMEM9_chr1_201112948_ENST00000367330_MYO5B_chr18_47421544_ENST00000324581_length(transcript)=3785nt_BP=916nt GTGGACGCGGCATTGGGGGAGGGGACGGGGGTCGCGGGGCTATTGGGGGTCCCAGTCCGGCTTAAGACCTCTGGGCGTGGTGGGCACCGG CCTTCGCCGTCGGAGCAGGGGTCCCAGACTCGCACCCAAAAGTTCCCGTCGGGCCCGAGCGGGGAGGAGCTGCGGGGCGGGCAGAGGCGG AGATGGCGGAGGGCGGTGGGACGTGATGCGCGGGTCAGAGCCGGGCCTTGAGAAGGAACTGGAGGCCCCTGGCAGCGGTGTCCCCTCGAG GACCCCTCTGCCGGGCTCACCAGGTGGCTGGCGGAGAGCCGGAGAGTTGGGCGGGAGGGAAGCAGGGTTTGGCGACCAGGTTTAGCAGGC CTTGGAATCCTGGAGCCCAGCCCCCAACCCCATTCCCACTTGGCCGGATGGTTAAGGGCCTAGCGTGCTGGCTGCGGTGTGGCCACCTTC CCCTTCTCCGACTCCGTATCTCTTTTTCCAGGTGTCCGGCTTTGCTGGCCCAGCAAGCCTGATAAGCATGAAGCTCTTATCTTTGGTGGC TGTGGTCGGGTGTTTGCTGGTGCCCCCAGCTGAAGCCAACAAGAGTTCTGAAGATATCCGGTGCAAATGCATCTGTCCACCTTATAGAAA CATCAGTGGGCACATTTACAACCAGAATGTATCCCAGAAGGACTGCAACTGCCTGCACGTGGTGGAGCCCATGCCAGTGCCTGGCCATGA CGTGGAGGCCTACTGCCTGCTGTGCGAGTGCAGGTACGAGGAGCGCAGCACCACCACCATCAAGGTCATCATTGTCATCTACCTGTCCGT GGTGGGTGCCCTGTTGCTCTACATGGCCTTCCTGATGCTGGTGGACCCTCTGATCCGAAAGCCGGATGCATATACTGAGCAACTGCACAA TGAGGAGGAGAATGAGAACAAAGAGTTCAAGACACTTTCAGAGCAGTTGTCCGTGACCACCTCAACATACACCATGGAGGTAGAGCGGCT GAAGAAGGAGCTGGTGCACTACCAGCAGAGCCCAGGTGAGGACACCAGCCTCAGGCTGCAGGAGGAGGTGGAGAGCCTGCGCACAGAGCT GCAGAGGGCCCACTCGGAGCGCAAGATCTTGGAGGACGCCCACAGCAGGGAGAAAGATGAGCTGAGGAAGCGAGTTGCAGACCTGGAGCA AGAAAATGCTCTCTTGAAAGATGAGAAAGAACAGCTCAACAACCAAATCCTGTGCCAGTCTAAAGATGAATTTGCCCAGAACTCTGTGAA GGAAAATCTCATGAAGAAAGAACTGGAGGAGGAGCGATCCCGGTACCAGAACCTTGTGAAGGAATATTCACAGTTGGAGCAGAGATACGA CAACCTTCGGGATGAAATGACCATCATAAAGCAAACTCCAGGTCATAGGCGGAACCCATCAAACCAAAGTAGCTTAGAATCTGACTCCAA TTACCCCTCCATCTCCACATCTGAGATCGGAGACACTGAGGATGCCCTCCAGCAGGTGGAGGAAATTGGCCTGGAGAAGGCAGCCATGGA CATGACGGTCTTCCTGAAGCTGCAGAAGAGAGTACGGGAGCTGGAGCAGGAGAGGAAAAAGCTGCAAGTGCAGCTGGAGAAGAGAGAACA GCAGGACAGCAAGAAAGTCCAGGCGGAACCACCACAGACTGACATAGATTTGGACCCGAATGCAGATCTGGCCTACAATAGTCTGAAGAG GCAAGAGCTGGAGTCAGAGAACAAAAAGCTGAAGAATGACCTGAATGAGCTGAGGAAAGCCGTGGCCGACCAAGCCACGCAGAATAACTC CAGCCACGGCTCCCCAGATAGCTACAGCCTCCTGCTGAACCAGCTCAAGCTGGCCCACGAGGAGCTCGAGGTGCGCAAGGAGGAGGTGCT CATCCTCAGGACCCAGATCGTGAGCGCCGACCAGCGGCGACTCGCCGGCAGGAACGCGGAGCCGAACATTAATGCCAGATCAAGTTGGCC TAACAGTGAAAAGCATGTTGACCAGGAGGATGCCATTGAGGCCTATCACGGGGTCTGCCAGACAAACAGGCTGCTGGAGGCTCAGCTGCA GGCCCAGAGCCTGGAGCATGAGGAGGAGGTGGAGCATCTCAAGGCTCAGCTCGAGGCCCTGAAGGAGGAGATGGACAAACAGCAGCAGAC CTTCTGCCAGACGCTACTGCTCTCCCCAGAGGCCCAGGTGGAATTCGGCGTTCAGCAGGAAATATCCCGGCTGACCAACGAGAATCTGGA CCTTAAAGAACTGGTAGAAAAGCTGGAAAAGAATGAGAGGAAGCTCAAAAAGCAACTGAAGATTTACATGAAGAAAGCCCAGGACCTAGA AGCTGCCCAGGCATTGGCCCAGAGTGAGAGGAAGCGCCATGAGCTCAACAGGCAGGTCACGGTCCAGCGGAAAGAGAAGGATTTCCAGGG CATGCTGGAGTACCACAAAGAGGACGAGGCCCTCCTCATCCGGAACCTGGTGACAGACTTGAAGCCCCAGATGCTGTCGGGCACAGTGCC CTGTCTCCCCGCCTACATCCTCTACATGTGCATCCGGCACGCGGACTACACCAACGACGATCTCAAGGTGCACTCCCTGCTGACCTCCAC CATCAACGGCATTAAGAAAGTCCTGAAAAAGCACAATGATGACTTTGAGATGACGTCATTCTGGTTATCCAACACCTGCCGCCTTCTTCA CTGTCTGAAGCAGTACAGCGGGGATGAGGGCTTCATGACTCAGAACACTGCAAAGCAGAATGAACACTGTCTTAAGAATTTTGACCTCAC CGAATACCGTCAGGTGCTGAGTGACCTTTCCATTCAGATCTACCAGCAGCTCATTAAAATTGCCGAGGGCGTGTTACAGCCGATGATAGT TTCTGCCATGTTGGAAAATGAGAGCATTCAGGGTCTATCTGGTGTGAAGCCCACCGGCTACCGGAAGCGCTCCTCCAGCATGGCAGATGG GGATAACTCATACTGCCTGGAAGCTATCATCCGCCAGATGAATGCCTTTCATACAGTCATGTGTGACCAGGGCTTGGACCCTGAGATCAT CCTGCAGGTATTCAAACAGCTCTTCTACATGATCAACGCAGTGACTCTTAACAACCTGCTCTTGCGGAAGGACGTCTGCTCTTGGAGCAC AGGCATGCAACTCAGGTACAATATAAGTCAGCTTGAGGAGTGGCTTCGGGGAAGAAACCTTCACCAGAGTGGAGCAGTTCAGACCATGGA ACCTCTGATCCAAGCAGCCCAGCTCCTGCAATTAAAGAAGAAAACCCAGGAGGACGCAGAGGCTATCTGCTCCCTGTGTACCTCCCTCAG CACCCAGCAGATTGTCAAAATTTTAAACCTTTATACTCCCCTGAATGAATTTGAAGAACGGGTAACAGTGGCCTTTATACGAACAATCCA GGCACAACTACAAGAGCGGAATGACCCTCAGCAACTGCTATTAGATGCCAAGCACATGTTTCCTGTTTTGTTTCCATTTAATCCATCTTC TCTAACCATGGACTCAATCCACATCCCAGCGTGTCTCAATCTGGAATTCCTCAATGAAGTCTGAAGATGCATGTTTCCAGCATTAGTTTG ATTCCCAATGTGAGCAAGAAGGAAGTATATACAGTAAAGTAAATTCAAGGATCTGTTAAATCTGGTAAAAGTAGATCAAATCAGAGATTG ACAGCCTGTGGAGGGTGCTGAACTATACAGAATTAGACACAACTATGTCATTATTTTTTGTACCTACTGCTCAGAATAAAAACACTTGAA >92304_92304_2_TMEM9-MYO5B_TMEM9_chr1_201112948_ENST00000367330_MYO5B_chr18_47421544_ENST00000324581_length(amino acids)=1048AA_BP=163 MAAVWPPSPSPTPYLFFQVSGFAGPASLISMKLLSLVAVVGCLLVPPAEANKSSEDIRCKCICPPYRNISGHIYNQNVSQKDCNCLHVVE PMPVPGHDVEAYCLLCECRYEERSTTTIKVIIVIYLSVVGALLLYMAFLMLVDPLIRKPDAYTEQLHNEEENENKEFKTLSEQLSVTTST YTMEVERLKKELVHYQQSPGEDTSLRLQEEVESLRTELQRAHSERKILEDAHSREKDELRKRVADLEQENALLKDEKEQLNNQILCQSKD EFAQNSVKENLMKKELEEERSRYQNLVKEYSQLEQRYDNLRDEMTIIKQTPGHRRNPSNQSSLESDSNYPSISTSEIGDTEDALQQVEEI GLEKAAMDMTVFLKLQKRVRELEQERKKLQVQLEKREQQDSKKVQAEPPQTDIDLDPNADLAYNSLKRQELESENKKLKNDLNELRKAVA DQATQNNSSHGSPDSYSLLLNQLKLAHEELEVRKEEVLILRTQIVSADQRRLAGRNAEPNINARSSWPNSEKHVDQEDAIEAYHGVCQTN RLLEAQLQAQSLEHEEEVEHLKAQLEALKEEMDKQQQTFCQTLLLSPEAQVEFGVQQEISRLTNENLDLKELVEKLEKNERKLKKQLKIY MKKAQDLEAAQALAQSERKRHELNRQVTVQRKEKDFQGMLEYHKEDEALLIRNLVTDLKPQMLSGTVPCLPAYILYMCIRHADYTNDDLK VHSLLTSTINGIKKVLKKHNDDFEMTSFWLSNTCRLLHCLKQYSGDEGFMTQNTAKQNEHCLKNFDLTEYRQVLSDLSIQIYQQLIKIAE GVLQPMIVSAMLENESIQGLSGVKPTGYRKRSSSMADGDNSYCLEAIIRQMNAFHTVMCDQGLDPEIILQVFKQLFYMINAVTLNNLLLR KDVCSWSTGMQLRYNISQLEEWLRGRNLHQSGAVQTMEPLIQAAQLLQLKKKTQEDAEAICSLCTSLSTQQIVKILNLYTPLNEFEERVT -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TMEM9-MYO5B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TMEM9-MYO5B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TMEM9-MYO5B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
