
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TMPRSS2-MORC3 (FusionGDB2 ID:92395) |
Fusion Gene Summary for TMPRSS2-MORC3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TMPRSS2-MORC3 | Fusion gene ID: 92395 | Hgene | Tgene | Gene symbol | TMPRSS2 | MORC3 | Gene ID | 7113 | 23515 |
| Gene name | transmembrane serine protease 2 | MORC family CW-type zinc finger 3 | |
| Synonyms | PP9284|PRSS10 | NXP2|ZCW5|ZCWCC3 | |
| Cytomap | 21q22.3 | 21q22.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | transmembrane protease serine 2epitheliasinserine protease 10transmembrane protease, serine 2 | MORC family CW-type zinc finger protein 3nuclear matrix protein 2nuclear matrix protein NXP2zinc finger CW-type coiled-coil domain protein 3zinc finger, CW type with coiled-coil domain 3 | |
| Modification date | 20200313 | 20200320 | |
| UniProtAcc | . | Q14149 | |
| Ensembl transtripts involved in fusion gene | ENST00000332149, ENST00000398585, ENST00000458356, ENST00000497881, | ENST00000487909, ENST00000400485, | |
| Fusion gene scores | * DoF score | 40 X 73 X 10=29200 | 8 X 11 X 4=352 |
| # samples | 130 | 11 | |
| ** MAII score | log2(130/29200*10)=-4.48938484073892 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/352*10)=-1.67807190511264 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TMPRSS2 [Title/Abstract] AND MORC3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TMPRSS2(42870046)-MORC3(37705944), # samples:2 TMPRSS2(42880007)-MORC3(37705943), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | TMPRSS2-MORC3 seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. TMPRSS2-MORC3 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. TMPRSS2-MORC3 seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TMPRSS2 | GO:0006508 | proteolysis | 21068237|24227843 |
| Hgene | TMPRSS2 | GO:0046598 | positive regulation of viral entry into host cell | 21068237|24227843 |
| Tgene | MORC3 | GO:0006468 | protein phosphorylation | 17332504 |
| Tgene | MORC3 | GO:0007569 | cell aging | 17332504 |
| Tgene | MORC3 | GO:0018105 | peptidyl-serine phosphorylation | 17332504 |
| Tgene | MORC3 | GO:0048147 | negative regulation of fibroblast proliferation | 17332504 |
| Tgene | MORC3 | GO:0050821 | protein stabilization | 17332504 |
| Tgene | MORC3 | GO:0051457 | maintenance of protein location in nucleus | 17332504 |
 Fusion gene breakpoints across TMPRSS2 (5'-gene) Fusion gene breakpoints across TMPRSS2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
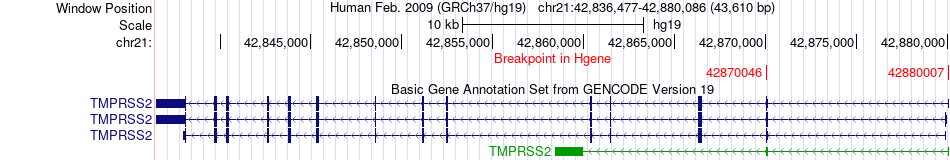 |
 Fusion gene breakpoints across MORC3 (3'-gene) Fusion gene breakpoints across MORC3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
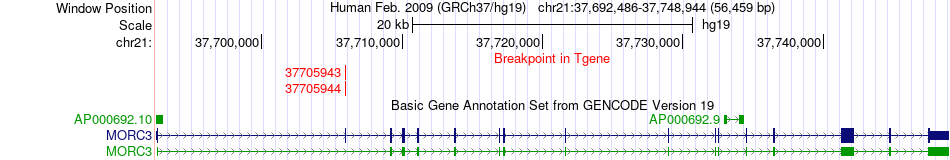 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | PRAD | TCGA-CH-5762-01A | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| ChimerDB4 | PRAD | TCGA-CH-5762 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
Top |
Fusion Gene ORF analysis for TMPRSS2-MORC3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000332149 | ENST00000487909 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| 5CDS-intron | ENST00000398585 | ENST00000487909 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| 5CDS-intron | ENST00000458356 | ENST00000487909 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| 5UTR-3CDS | ENST00000332149 | ENST00000400485 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| 5UTR-3CDS | ENST00000497881 | ENST00000400485 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| 5UTR-3CDS | ENST00000497881 | ENST00000400485 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| 5UTR-intron | ENST00000332149 | ENST00000487909 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| 5UTR-intron | ENST00000497881 | ENST00000487909 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| 5UTR-intron | ENST00000497881 | ENST00000487909 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| Frame-shift | ENST00000458356 | ENST00000400485 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| In-frame | ENST00000332149 | ENST00000400485 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| In-frame | ENST00000398585 | ENST00000400485 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + |
| intron-3CDS | ENST00000398585 | ENST00000400485 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| intron-3CDS | ENST00000458356 | ENST00000400485 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| intron-intron | ENST00000398585 | ENST00000487909 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
| intron-intron | ENST00000458356 | ENST00000487909 | TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000332149 | TMPRSS2 | chr21 | 42870046 | - | ENST00000400485 | MORC3 | chr21 | 37705944 | + | 4281 | 150 | 135 | 2930 | 931 |
| ENST00000398585 | TMPRSS2 | chr21 | 42870046 | - | ENST00000400485 | MORC3 | chr21 | 37705944 | + | 4318 | 187 | 40 | 2967 | 975 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000332149 | ENST00000400485 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + | 0.000197243 | 0.99980277 |
| ENST00000398585 | ENST00000400485 | TMPRSS2 | chr21 | 42870046 | - | MORC3 | chr21 | 37705944 | + | 0.000146029 | 0.99985397 |
Top |
Fusion Genomic Features for TMPRSS2-MORC3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + | 0.000104626 | 0.99989533 |
| TMPRSS2 | chr21 | 42870045 | - | MORC3 | chr21 | 37705943 | + | 3.15E-06 | 0.9999969 |
| TMPRSS2 | chr21 | 42880007 | - | MORC3 | chr21 | 37705943 | + | 0.000104626 | 0.99989533 |
| TMPRSS2 | chr21 | 42870045 | - | MORC3 | chr21 | 37705943 | + | 3.15E-06 | 0.9999969 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
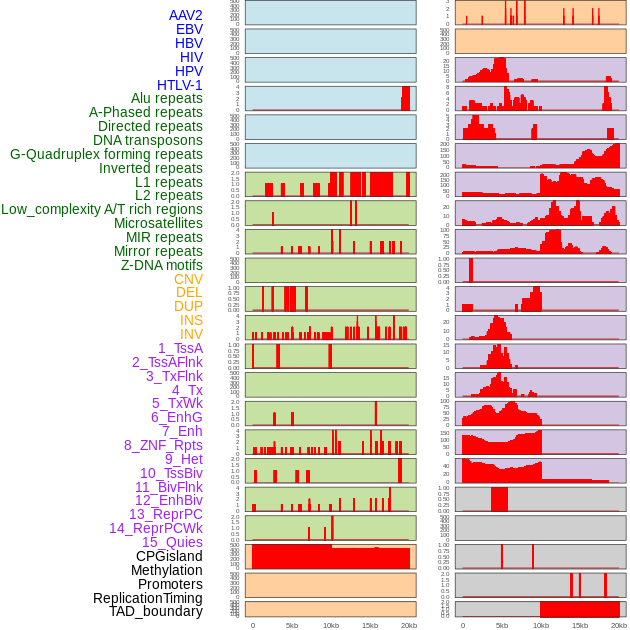 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
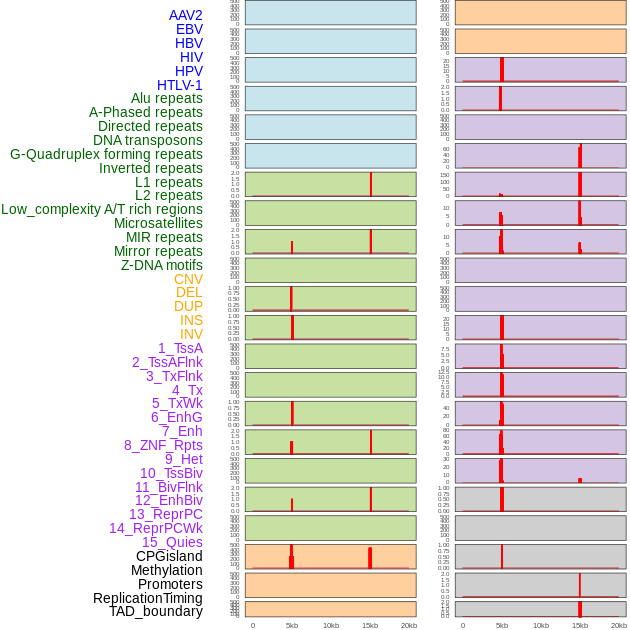 |
Top |
Fusion Protein Features for TMPRSS2-MORC3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr21:42870046/chr21:37705944) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | MORC3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Nuclear factor which forms MORC3-NBs (nuclear bodies) via an ATP-dependent mechanism (PubMed:20501696). Sumoylated MORC3-NBs can also associate with PML-NBs (PubMed:20501696). Recruits TP53 and SP100 to PML-NBs, thus regulating TP53 activity (PubMed:17332504). Binds RNA in vitro (PubMed:11927593). May be required for influenza A transcription during viral infection (PubMed:26202233). Histone methylation reader which binds to non-methylated (H3K4me0), monomethylated (H3K4me1), dimethylated (H3K4me2) and trimethylated (H3K4me3) 'Lys-4' on histone H3 (PubMed:26933034). The order of binding preference is H3K4me3 > H3K4me2 > H3K4me1 > H3K4me0 (PubMed:26933034). {ECO:0000269|PubMed:11927593, ECO:0000269|PubMed:17332504, ECO:0000269|PubMed:20501696, ECO:0000269|PubMed:26202233, ECO:0000269|PubMed:26933034}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MORC3 | chr21:42870046 | chr21:37705944 | ENST00000400485 | 0 | 17 | 686_877 | 13 | 940.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MORC3 | chr21:42870046 | chr21:37705944 | ENST00000400485 | 0 | 17 | 326_353 | 13 | 940.0 | Region | Nuclear matrix binding | |
| Tgene | MORC3 | chr21:42870046 | chr21:37705944 | ENST00000400485 | 0 | 17 | 500_591 | 13 | 940.0 | Region | RNA binding | |
| Tgene | MORC3 | chr21:42870046 | chr21:37705944 | ENST00000400485 | 0 | 17 | 404_454 | 13 | 940.0 | Zinc finger | CW-type |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000332149 | - | 2 | 14 | 112_149 | 5 | 493.0 | Domain | LDL-receptor class A |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000332149 | - | 2 | 14 | 150_242 | 5 | 493.0 | Domain | SRCR |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000332149 | - | 2 | 14 | 256_489 | 5 | 493.0 | Domain | Peptidase S1 |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000398585 | - | 2 | 14 | 112_149 | 42 | 530.0 | Domain | LDL-receptor class A |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000398585 | - | 2 | 14 | 150_242 | 42 | 530.0 | Domain | SRCR |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000398585 | - | 2 | 14 | 256_489 | 42 | 530.0 | Domain | Peptidase S1 |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000458356 | - | 2 | 14 | 112_149 | 5 | 493.0 | Domain | LDL-receptor class A |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000458356 | - | 2 | 14 | 150_242 | 5 | 493.0 | Domain | SRCR |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000458356 | - | 2 | 14 | 256_489 | 5 | 493.0 | Domain | Peptidase S1 |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000332149 | - | 2 | 14 | 106_492 | 5 | 493.0 | Topological domain | Extracellular |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000332149 | - | 2 | 14 | 1_84 | 5 | 493.0 | Topological domain | Cytoplasmic |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000398585 | - | 2 | 14 | 106_492 | 42 | 530.0 | Topological domain | Extracellular |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000398585 | - | 2 | 14 | 1_84 | 42 | 530.0 | Topological domain | Cytoplasmic |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000458356 | - | 2 | 14 | 106_492 | 5 | 493.0 | Topological domain | Extracellular |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000458356 | - | 2 | 14 | 1_84 | 5 | 493.0 | Topological domain | Cytoplasmic |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000332149 | - | 2 | 14 | 85_105 | 5 | 493.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000398585 | - | 2 | 14 | 85_105 | 42 | 530.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
| Hgene | TMPRSS2 | chr21:42870046 | chr21:37705944 | ENST00000458356 | - | 2 | 14 | 85_105 | 5 | 493.0 | Transmembrane | Helical%3B Signal-anchor for type II membrane protein |
Top |
Fusion Gene Sequence for TMPRSS2-MORC3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >92395_92395_1_TMPRSS2-MORC3_TMPRSS2_chr21_42870046_ENST00000332149_MORC3_chr21_37705944_ENST00000400485_length(transcript)=4281nt_BP=150nt CGAGTAGGCGCGAGCTAAGCAGGAGGCGGAGGCGGAGGCGGAGGGCGAGGGGCGGGGAGCGCCGCCTGGAGCGCGGCAGGTCATATTGAA CATTCCAGATACCTATCATTACTCGATGCTGTTGATAACAGCAAGATGGCTTTGAACTCACTTTGCCCGAAGTTTTTACATACAAATTCT ACTAGTCACACCTGGCCATTCAGTGCAGTTGCTGAATTAATAGATAATGCTTATGATCCTGATGTGAACGCTAAACAAATATGGATTGAC AAAACAGTGATAAATGACCATATATGCTTGACATTCACCGACAATGGGAATGGTATGACTTCTGATAAATTACATAAAATGCTAAGCTTT GGCTTCAGTGACAAAGTCACCATGAATGGTCATGTCCCAGTTGGATTATATGGGAATGGCTTCAAGTCGGGTTCTATGCGTCTGGGTAAA GACGCAATCGTTTTTACCAAAAATGGAGAAAGCATGAGCGTGGGCCTTTTGTCTCAGACCTACTTGGAAGTCATAAAAGCGGAGCATGTT GTTGTTCCAATAGTGGCATTCAACAAGCACCGACAGATGATTAATTTAGCAGAATCAAAAGCCAGCCTTGCTGCAATTCTGGAACATTCT CTGTTTTCCACGGAACAGAAGTTACTGGCAGAACTTGATGCTATTATAGGCAAGAAGGGGACGAGGATCATCATTTGGAATCTTAGAAGC TACAAAAATGCAACAGAGTTCGATTTTGAAAAGGATAAATATGATATCAGAATTCCCGAGGATTTAGATGAGATAACAGGGAAGAAGGGG TACAAGAAGCAGGAAAGGATGGACCAGATTGCCCCTGAGAGTGACTATTCCCTGAGGGCTTATTGCAGTATATTATATCTAAAGCCAAGA ATGCAGATCATCCTACGTGGACAGAAAGTGAAGACACAGCTGGTTTCGAAGAGTCTTGCCTACATCGAACGTGATGTTTATCGACCAAAA TTTTTATCTAAAACAGTGAGAATTACCTTTGGATTCAACTGCAGAAATAAAGATCATTATGGGATAATGATGTATCACAGAAATAGACTC ATCAAAGCTTATGAAAAAGTTGGATGTCAGTTAAGGGCAAACAACATGGGTGTTGGAGTGGTTGGAATTATAGAGTGTAATTTCCTTAAG CCAACTCATAATAAACAAGATTTCGACTATACTAATGAGTACAGACTTACAATAACAGCACTAGGAGAAAAGCTGAATGATTACTGGAAT GAAATGAAAGTGAAGAAAAATACAGAATATCCTCTAAATTTGCCAGTTGAAGATATACAGAAGCGTCCTGATCAGACATGGGTTCAGTGT GATGCCTGTCTAAAGTGGCGGAAATTACCTGATGGGATGGATCAACTTCCTGAAAAATGGTATTGCTCCAATAACCCTGACCCACAGTTC AGAAATTGTGAGGTTCCAGAAGAACCTGAAGATGAGGATTTGGTACATCCCACTTATGAAAAAACCTACAAAAAGACCAACAAGGAAAAA TTCAGGATCAGACAACCGGAAATGATCCCTCGGATTAATGCTGAACTGTTGTTTCGGCCAACTGCTCTTTCAACTCCAAGCTTTTCTTCT CCTAAGGAAAGTGTTCCAAGAAGACATCTTTCAGAAGGAACAAATTCTTATGCGACAAGACTTCTAAATAATCATCAAGTTCCACCTCAG TCTGAACCTGAGAGCAACAGCTTGAAACGGAGACTTTCTACTCGTTCCTCAATTTTGAATGCAAAGAATCGGAGATTGAGTAGTCAGTTT GAAAATTCAGTTTATAAAGGTGATGATGATGATGAAGATGTCATCATCTTAGAAGAAAACAGTACCCCCAAACCTGCAGTAGATCATGAT ATTGACATGAAATCAGAACAGAGTCACGTTGAGCAAGGTGGTGTTCAGGTTGAGTTTGTGGGTGACAGTGAACCTTGTGGCCAGACTGGT TCAACAAGCACCTCATCATCCCGATGCGACCAGGGAAATACTGCAGCTACCCAGACTGAAGTACCAAGTTTAGTTGTTAAAAAAGAAGAA ACTGTTGAAGACGAGATAGACGTAAGAAATGATGCAGTGATTCTGCCCTCCTGTGTAGAAGCTGAAGCAAAGATACATGAAACCCAGGAA ACCACCGATAAATCTGCAGATGATGCAGGCTGCCAATTACAAGAACTGAGAAACCAGCTACTCCTTGTCACTGAGGAAAAAGAGAATTAT AAAAGACAGTGTCATATGTTTACTGATCAAATCAAAGTGTTACAACAGAGGATACTAGAAATGAATGACAAGTATGTTAAGAAAGAAACT TGCCATCAGTCCACTGAAACCGATGCTGTATTTTTACTTGAAAGTATTAATGGCAAATCTGAAAGTCCAGACCATATGGTATCTCAGTAT CAGCAAGCTTTGGAAGAAATAGAAAGGCTGAAAAAACAATGTAGTGCTTTGCAACATGTAAAGGCTGAATGCAGCCAGTGTTCCAATAAT GAGAGTAAAAGTGAAATGGATGAGATGGCTGTGCAGCTTGACGATGTGTTTAGACAACTGGACAAATGCAGTATTGAGAGGGACCAGTAT AAAAGTGAGGTTGAATTGCTGGAAATGGAAAAGTCACAAATCCGTTCACAGTGTGAAGAACTCAAAACTGAAGTAGAACAGTTAAAATCT ACAAATCAACAGACGGCAACAGATGTTTCAACATCAAGTAACATTGAGGAGTCTGTAAATCATATGGATGGAGAAAGCCTCAAACTCCGA TCTCTTCGAGTTAACGTAGGACAACTGCTGGCTATGATTGTGCCTGATCTTGATCTTCAGCAAGTGAATTACGATGTTGATGTAGTTGAT GAGATTTTAGGACAAGTTGTTGAACAAATGAGTGAAATCAGTAGTACTTAAAGTATATGTTATGTAAGATAAAATATTTGCTCAATTCTT TTGGTTGTACAGCTTTCAAAATATAATTAATTTTGTTTTATAGATATGATAGGCAACAGACTGAAAACCATAATCTTTACTGTATTCTAT GCATTCAAATGTGGTCACAAATATTGTGGACACATTATCTTATGTTTTGAAATACCTGTGAATTGTTGGCATTGAGCAGCTGAAGCTAAC TCATGACTCTGTTTTGAATGTAAATATTTGTAATTAAGCCTGCACATATTTTTTTATTGCCCTAGAGTACTCAAGTGTTTTTCACCAAGA GCTTTTCAGGTTGCCCCTAAGCTTTGTGCAATTTTTTCTGGTTCCCCAAAGTGTATTTTTCTCTAAGTCGAGGGCTATGCCATAATACAA ATGGAAATGTTACCTTTGATTTTCTTATAAAGGAGTTTAAATAGGATTTTTAAATAATGTAGTAACACTCCTGATACAACTCTGGTTATA AGTGAATTGAGCATTAATGTTTCTTTTGTATAAATTCCTGTCCTGAAATATTTTATTCATGAAAATAAGGTAAGCAAAAACCAACTCCAT TTTGCCAGGATTTTGTTGTGCTGAGATTGCCAATCACGGTTAAACACAAAGTGTTTTTATAGAATGAAGAAACAGTCTTTAACAGAAAAA AGGTATTGAAATATTAAATGGCCACCAAGTTTACTTTGAAGCCCATTTTTGGCTGTTTAGTCAGCATGAAGTGGGCATGATAATTTTTTA ATATTTCTTTTTGTGAAATTTCCTGTACAGCCTTTTGTAGGATTACTACAGGTTAATGTGAGTTGAGGAAGACAGTCTTTCTCAAACAAT ACTACATACCTTTTATTATATTACAAACCTAAGTGTTTTATATCCTAGAGTCAAGGAACAAATTTCCTTAGGATTATAGTATTACATGCC ATAAAATACTATGCTTTATTGGTCCCATGTTTTGTGCAATTTTAAAGAGATGGCTTTCTATTAAGTATAAACTATGTATATATAAGAACC ATATTTTCCACAACTAAAATGTGCATTATTTTTTCCAAAGTTTTATCATTGCTATTTATTTTTACCTTTGTTTTTGAACATTCAATCCGT TCATTTTGTATGTATGCTTAATACGTGTCGGTCATATACAGTATTGAATTTTTACTGTATAGTAATTCTGGAAAGAGCAAATAAATGAAG ATTGTTTTTATTTGCCTGATAAAGTAATTGAAAGTGTATTTTTGGTATGAAGCTGGTTTTCTGTCACAATTGTAATTTCCCAAATTTTAA >92395_92395_1_TMPRSS2-MORC3_TMPRSS2_chr21_42870046_ENST00000332149_MORC3_chr21_37705944_ENST00000400485_length(amino acids)=931AA_BP=5 MALNSLCPKFLHTNSTSHTWPFSAVAELIDNAYDPDVNAKQIWIDKTVINDHICLTFTDNGNGMTSDKLHKMLSFGFSDKVTMNGHVPVG LYGNGFKSGSMRLGKDAIVFTKNGESMSVGLLSQTYLEVIKAEHVVVPIVAFNKHRQMINLAESKASLAAILEHSLFSTEQKLLAELDAI IGKKGTRIIIWNLRSYKNATEFDFEKDKYDIRIPEDLDEITGKKGYKKQERMDQIAPESDYSLRAYCSILYLKPRMQIILRGQKVKTQLV SKSLAYIERDVYRPKFLSKTVRITFGFNCRNKDHYGIMMYHRNRLIKAYEKVGCQLRANNMGVGVVGIIECNFLKPTHNKQDFDYTNEYR LTITALGEKLNDYWNEMKVKKNTEYPLNLPVEDIQKRPDQTWVQCDACLKWRKLPDGMDQLPEKWYCSNNPDPQFRNCEVPEEPEDEDLV HPTYEKTYKKTNKEKFRIRQPEMIPRINAELLFRPTALSTPSFSSPKESVPRRHLSEGTNSYATRLLNNHQVPPQSEPESNSLKRRLSTR SSILNAKNRRLSSQFENSVYKGDDDDEDVIILEENSTPKPAVDHDIDMKSEQSHVEQGGVQVEFVGDSEPCGQTGSTSTSSSRCDQGNTA ATQTEVPSLVVKKEETVEDEIDVRNDAVILPSCVEAEAKIHETQETTDKSADDAGCQLQELRNQLLLVTEEKENYKRQCHMFTDQIKVLQ QRILEMNDKYVKKETCHQSTETDAVFLLESINGKSESPDHMVSQYQQALEEIERLKKQCSALQHVKAECSQCSNNESKSEMDEMAVQLDD VFRQLDKCSIERDQYKSEVELLEMEKSQIRSQCEELKTEVEQLKSTNQQTATDVSTSSNIEESVNHMDGESLKLRSLRVNVGQLLAMIVP -------------------------------------------------------------- >92395_92395_2_TMPRSS2-MORC3_TMPRSS2_chr21_42870046_ENST00000398585_MORC3_chr21_37705944_ENST00000400485_length(transcript)=4318nt_BP=187nt ACCAGGGTCCCGGCTCGGGGTCCGGGCTGGGGAGGGGAACCTGGGCGCCTGGGACCCGCCGATGCCCCCTGCCCCGCCCGGAGGTGAAAG CGGGTGTGAGGAGCGCGGCGCGGCAGGTCATATTGAACATTCCAGATACCTATCATTACTCGATGCTGTTGATAACAGCAAGATGGCTTT GAACTCACTTTGCCCGAAGTTTTTACATACAAATTCTACTAGTCACACCTGGCCATTCAGTGCAGTTGCTGAATTAATAGATAATGCTTA TGATCCTGATGTGAACGCTAAACAAATATGGATTGACAAAACAGTGATAAATGACCATATATGCTTGACATTCACCGACAATGGGAATGG TATGACTTCTGATAAATTACATAAAATGCTAAGCTTTGGCTTCAGTGACAAAGTCACCATGAATGGTCATGTCCCAGTTGGATTATATGG GAATGGCTTCAAGTCGGGTTCTATGCGTCTGGGTAAAGACGCAATCGTTTTTACCAAAAATGGAGAAAGCATGAGCGTGGGCCTTTTGTC TCAGACCTACTTGGAAGTCATAAAAGCGGAGCATGTTGTTGTTCCAATAGTGGCATTCAACAAGCACCGACAGATGATTAATTTAGCAGA ATCAAAAGCCAGCCTTGCTGCAATTCTGGAACATTCTCTGTTTTCCACGGAACAGAAGTTACTGGCAGAACTTGATGCTATTATAGGCAA GAAGGGGACGAGGATCATCATTTGGAATCTTAGAAGCTACAAAAATGCAACAGAGTTCGATTTTGAAAAGGATAAATATGATATCAGAAT TCCCGAGGATTTAGATGAGATAACAGGGAAGAAGGGGTACAAGAAGCAGGAAAGGATGGACCAGATTGCCCCTGAGAGTGACTATTCCCT GAGGGCTTATTGCAGTATATTATATCTAAAGCCAAGAATGCAGATCATCCTACGTGGACAGAAAGTGAAGACACAGCTGGTTTCGAAGAG TCTTGCCTACATCGAACGTGATGTTTATCGACCAAAATTTTTATCTAAAACAGTGAGAATTACCTTTGGATTCAACTGCAGAAATAAAGA TCATTATGGGATAATGATGTATCACAGAAATAGACTCATCAAAGCTTATGAAAAAGTTGGATGTCAGTTAAGGGCAAACAACATGGGTGT TGGAGTGGTTGGAATTATAGAGTGTAATTTCCTTAAGCCAACTCATAATAAACAAGATTTCGACTATACTAATGAGTACAGACTTACAAT AACAGCACTAGGAGAAAAGCTGAATGATTACTGGAATGAAATGAAAGTGAAGAAAAATACAGAATATCCTCTAAATTTGCCAGTTGAAGA TATACAGAAGCGTCCTGATCAGACATGGGTTCAGTGTGATGCCTGTCTAAAGTGGCGGAAATTACCTGATGGGATGGATCAACTTCCTGA AAAATGGTATTGCTCCAATAACCCTGACCCACAGTTCAGAAATTGTGAGGTTCCAGAAGAACCTGAAGATGAGGATTTGGTACATCCCAC TTATGAAAAAACCTACAAAAAGACCAACAAGGAAAAATTCAGGATCAGACAACCGGAAATGATCCCTCGGATTAATGCTGAACTGTTGTT TCGGCCAACTGCTCTTTCAACTCCAAGCTTTTCTTCTCCTAAGGAAAGTGTTCCAAGAAGACATCTTTCAGAAGGAACAAATTCTTATGC GACAAGACTTCTAAATAATCATCAAGTTCCACCTCAGTCTGAACCTGAGAGCAACAGCTTGAAACGGAGACTTTCTACTCGTTCCTCAAT TTTGAATGCAAAGAATCGGAGATTGAGTAGTCAGTTTGAAAATTCAGTTTATAAAGGTGATGATGATGATGAAGATGTCATCATCTTAGA AGAAAACAGTACCCCCAAACCTGCAGTAGATCATGATATTGACATGAAATCAGAACAGAGTCACGTTGAGCAAGGTGGTGTTCAGGTTGA GTTTGTGGGTGACAGTGAACCTTGTGGCCAGACTGGTTCAACAAGCACCTCATCATCCCGATGCGACCAGGGAAATACTGCAGCTACCCA GACTGAAGTACCAAGTTTAGTTGTTAAAAAAGAAGAAACTGTTGAAGACGAGATAGACGTAAGAAATGATGCAGTGATTCTGCCCTCCTG TGTAGAAGCTGAAGCAAAGATACATGAAACCCAGGAAACCACCGATAAATCTGCAGATGATGCAGGCTGCCAATTACAAGAACTGAGAAA CCAGCTACTCCTTGTCACTGAGGAAAAAGAGAATTATAAAAGACAGTGTCATATGTTTACTGATCAAATCAAAGTGTTACAACAGAGGAT ACTAGAAATGAATGACAAGTATGTTAAGAAAGAAACTTGCCATCAGTCCACTGAAACCGATGCTGTATTTTTACTTGAAAGTATTAATGG CAAATCTGAAAGTCCAGACCATATGGTATCTCAGTATCAGCAAGCTTTGGAAGAAATAGAAAGGCTGAAAAAACAATGTAGTGCTTTGCA ACATGTAAAGGCTGAATGCAGCCAGTGTTCCAATAATGAGAGTAAAAGTGAAATGGATGAGATGGCTGTGCAGCTTGACGATGTGTTTAG ACAACTGGACAAATGCAGTATTGAGAGGGACCAGTATAAAAGTGAGGTTGAATTGCTGGAAATGGAAAAGTCACAAATCCGTTCACAGTG TGAAGAACTCAAAACTGAAGTAGAACAGTTAAAATCTACAAATCAACAGACGGCAACAGATGTTTCAACATCAAGTAACATTGAGGAGTC TGTAAATCATATGGATGGAGAAAGCCTCAAACTCCGATCTCTTCGAGTTAACGTAGGACAACTGCTGGCTATGATTGTGCCTGATCTTGA TCTTCAGCAAGTGAATTACGATGTTGATGTAGTTGATGAGATTTTAGGACAAGTTGTTGAACAAATGAGTGAAATCAGTAGTACTTAAAG TATATGTTATGTAAGATAAAATATTTGCTCAATTCTTTTGGTTGTACAGCTTTCAAAATATAATTAATTTTGTTTTATAGATATGATAGG CAACAGACTGAAAACCATAATCTTTACTGTATTCTATGCATTCAAATGTGGTCACAAATATTGTGGACACATTATCTTATGTTTTGAAAT ACCTGTGAATTGTTGGCATTGAGCAGCTGAAGCTAACTCATGACTCTGTTTTGAATGTAAATATTTGTAATTAAGCCTGCACATATTTTT TTATTGCCCTAGAGTACTCAAGTGTTTTTCACCAAGAGCTTTTCAGGTTGCCCCTAAGCTTTGTGCAATTTTTTCTGGTTCCCCAAAGTG TATTTTTCTCTAAGTCGAGGGCTATGCCATAATACAAATGGAAATGTTACCTTTGATTTTCTTATAAAGGAGTTTAAATAGGATTTTTAA ATAATGTAGTAACACTCCTGATACAACTCTGGTTATAAGTGAATTGAGCATTAATGTTTCTTTTGTATAAATTCCTGTCCTGAAATATTT TATTCATGAAAATAAGGTAAGCAAAAACCAACTCCATTTTGCCAGGATTTTGTTGTGCTGAGATTGCCAATCACGGTTAAACACAAAGTG TTTTTATAGAATGAAGAAACAGTCTTTAACAGAAAAAAGGTATTGAAATATTAAATGGCCACCAAGTTTACTTTGAAGCCCATTTTTGGC TGTTTAGTCAGCATGAAGTGGGCATGATAATTTTTTAATATTTCTTTTTGTGAAATTTCCTGTACAGCCTTTTGTAGGATTACTACAGGT TAATGTGAGTTGAGGAAGACAGTCTTTCTCAAACAATACTACATACCTTTTATTATATTACAAACCTAAGTGTTTTATATCCTAGAGTCA AGGAACAAATTTCCTTAGGATTATAGTATTACATGCCATAAAATACTATGCTTTATTGGTCCCATGTTTTGTGCAATTTTAAAGAGATGG CTTTCTATTAAGTATAAACTATGTATATATAAGAACCATATTTTCCACAACTAAAATGTGCATTATTTTTTCCAAAGTTTTATCATTGCT ATTTATTTTTACCTTTGTTTTTGAACATTCAATCCGTTCATTTTGTATGTATGCTTAATACGTGTCGGTCATATACAGTATTGAATTTTT ACTGTATAGTAATTCTGGAAAGAGCAAATAAATGAAGATTGTTTTTATTTGCCTGATAAAGTAATTGAAAGTGTATTTTTGGTATGAAGC >92395_92395_2_TMPRSS2-MORC3_TMPRSS2_chr21_42870046_ENST00000398585_MORC3_chr21_37705944_ENST00000400485_length(amino acids)=975AA_BP=49 MGAWDPPMPPAPPGGESGCEERGAAGHIEHSRYLSLLDAVDNSKMALNSLCPKFLHTNSTSHTWPFSAVAELIDNAYDPDVNAKQIWIDK TVINDHICLTFTDNGNGMTSDKLHKMLSFGFSDKVTMNGHVPVGLYGNGFKSGSMRLGKDAIVFTKNGESMSVGLLSQTYLEVIKAEHVV VPIVAFNKHRQMINLAESKASLAAILEHSLFSTEQKLLAELDAIIGKKGTRIIIWNLRSYKNATEFDFEKDKYDIRIPEDLDEITGKKGY KKQERMDQIAPESDYSLRAYCSILYLKPRMQIILRGQKVKTQLVSKSLAYIERDVYRPKFLSKTVRITFGFNCRNKDHYGIMMYHRNRLI KAYEKVGCQLRANNMGVGVVGIIECNFLKPTHNKQDFDYTNEYRLTITALGEKLNDYWNEMKVKKNTEYPLNLPVEDIQKRPDQTWVQCD ACLKWRKLPDGMDQLPEKWYCSNNPDPQFRNCEVPEEPEDEDLVHPTYEKTYKKTNKEKFRIRQPEMIPRINAELLFRPTALSTPSFSSP KESVPRRHLSEGTNSYATRLLNNHQVPPQSEPESNSLKRRLSTRSSILNAKNRRLSSQFENSVYKGDDDDEDVIILEENSTPKPAVDHDI DMKSEQSHVEQGGVQVEFVGDSEPCGQTGSTSTSSSRCDQGNTAATQTEVPSLVVKKEETVEDEIDVRNDAVILPSCVEAEAKIHETQET TDKSADDAGCQLQELRNQLLLVTEEKENYKRQCHMFTDQIKVLQQRILEMNDKYVKKETCHQSTETDAVFLLESINGKSESPDHMVSQYQ QALEEIERLKKQCSALQHVKAECSQCSNNESKSEMDEMAVQLDDVFRQLDKCSIERDQYKSEVELLEMEKSQIRSQCEELKTEVEQLKST -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TMPRSS2-MORC3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TMPRSS2-MORC3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TMPRSS2-MORC3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
