
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TPGS2-DERA (FusionGDB2 ID:93316) |
Fusion Gene Summary for TPGS2-DERA |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TPGS2-DERA | Fusion gene ID: 93316 | Hgene | Tgene | Gene symbol | TPGS2 | DERA | Gene ID | 25941 | 51071 |
| Gene name | tubulin polyglutamylase complex subunit 2 | deoxyribose-phosphate aldolase | |
| Synonyms | C18orf10|HMFN0601|L17|PGs2 | CGI-26|DEOC | |
| Cytomap | 18q12.2 | 12p12.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | tubulin polyglutamylase complex subunit 2 | deoxyribose-phosphate aldolase2-deoxy-D-ribose 5-phosphate aldolase2-deoxyribose-5-phosphate aldolase homologdeoxyriboaldolasedeoxyribose-phosphate aldolase (putative)phosphodeoxyriboaldolaseputative deoxyribose-phosphate aldolase | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | Q68CL5 | Q9Y315 | |
| Ensembl transtripts involved in fusion gene | ENST00000334295, ENST00000587129, ENST00000589049, ENST00000590842, ENST00000593035, ENST00000383056, ENST00000590652, | ENST00000532573, ENST00000428559, ENST00000526530, ENST00000532964, | |
| Fusion gene scores | * DoF score | 3 X 3 X 3=27 | 10 X 7 X 6=420 |
| # samples | 3 | 9 | |
| ** MAII score | log2(3/27*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(9/420*10)=-2.22239242133645 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TPGS2 [Title/Abstract] AND DERA [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TPGS2(34385337)-DERA(16185476), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | DERA | GO:0046121 | deoxyribonucleoside catabolic process | 9226884 |
 Fusion gene breakpoints across TPGS2 (5'-gene) Fusion gene breakpoints across TPGS2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
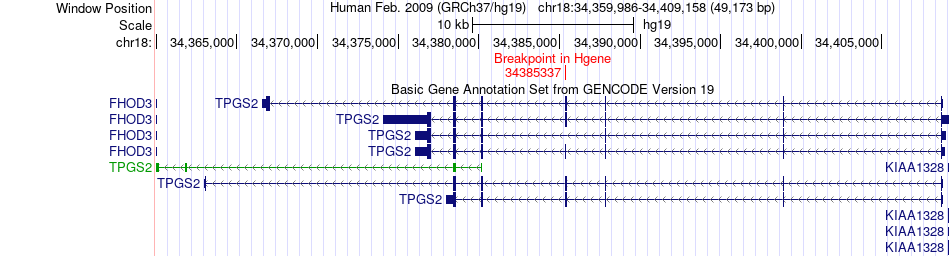 |
 Fusion gene breakpoints across DERA (3'-gene) Fusion gene breakpoints across DERA (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
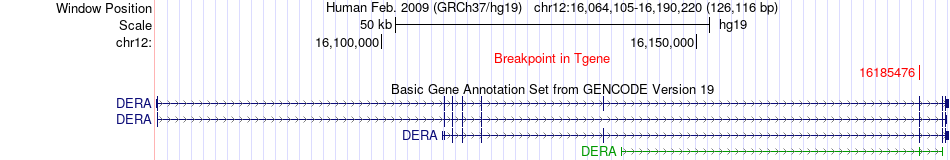 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-A48C-01A | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
Top |
Fusion Gene ORF analysis for TPGS2-DERA |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000334295 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| 5CDS-3UTR | ENST00000587129 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| 5CDS-3UTR | ENST00000589049 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| 5CDS-3UTR | ENST00000590842 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| 5CDS-3UTR | ENST00000593035 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000334295 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000334295 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000334295 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000587129 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000587129 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000587129 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000589049 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000589049 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000589049 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000590842 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000590842 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000590842 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000593035 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000593035 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| In-frame | ENST00000593035 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3CDS | ENST00000383056 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3CDS | ENST00000383056 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3CDS | ENST00000383056 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3CDS | ENST00000590652 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3CDS | ENST00000590652 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3CDS | ENST00000590652 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3UTR | ENST00000383056 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
| intron-3UTR | ENST00000590652 | ENST00000532573 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000587129 | TPGS2 | chr18 | 34385337 | - | ENST00000428559 | DERA | chr12 | 16185476 | + | 1279 | 419 | 37 | 738 | 233 |
| ENST00000587129 | TPGS2 | chr18 | 34385337 | - | ENST00000532964 | DERA | chr12 | 16185476 | + | 744 | 419 | 37 | 738 | 233 |
| ENST00000587129 | TPGS2 | chr18 | 34385337 | - | ENST00000526530 | DERA | chr12 | 16185476 | + | 1273 | 419 | 37 | 738 | 233 |
| ENST00000590842 | TPGS2 | chr18 | 34385337 | - | ENST00000428559 | DERA | chr12 | 16185476 | + | 1317 | 457 | 75 | 776 | 233 |
| ENST00000590842 | TPGS2 | chr18 | 34385337 | - | ENST00000532964 | DERA | chr12 | 16185476 | + | 782 | 457 | 75 | 776 | 233 |
| ENST00000590842 | TPGS2 | chr18 | 34385337 | - | ENST00000526530 | DERA | chr12 | 16185476 | + | 1311 | 457 | 75 | 776 | 233 |
| ENST00000334295 | TPGS2 | chr18 | 34385337 | - | ENST00000428559 | DERA | chr12 | 16185476 | + | 1670 | 810 | 305 | 1129 | 274 |
| ENST00000334295 | TPGS2 | chr18 | 34385337 | - | ENST00000532964 | DERA | chr12 | 16185476 | + | 1135 | 810 | 305 | 1129 | 274 |
| ENST00000334295 | TPGS2 | chr18 | 34385337 | - | ENST00000526530 | DERA | chr12 | 16185476 | + | 1664 | 810 | 305 | 1129 | 274 |
| ENST00000593035 | TPGS2 | chr18 | 34385337 | - | ENST00000428559 | DERA | chr12 | 16185476 | + | 1333 | 473 | 73 | 792 | 239 |
| ENST00000593035 | TPGS2 | chr18 | 34385337 | - | ENST00000532964 | DERA | chr12 | 16185476 | + | 798 | 473 | 73 | 792 | 239 |
| ENST00000593035 | TPGS2 | chr18 | 34385337 | - | ENST00000526530 | DERA | chr12 | 16185476 | + | 1327 | 473 | 73 | 792 | 239 |
| ENST00000589049 | TPGS2 | chr18 | 34385337 | - | ENST00000428559 | DERA | chr12 | 16185476 | + | 1256 | 396 | 14 | 715 | 233 |
| ENST00000589049 | TPGS2 | chr18 | 34385337 | - | ENST00000532964 | DERA | chr12 | 16185476 | + | 721 | 396 | 14 | 715 | 233 |
| ENST00000589049 | TPGS2 | chr18 | 34385337 | - | ENST00000526530 | DERA | chr12 | 16185476 | + | 1250 | 396 | 14 | 715 | 233 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000587129 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.001021793 | 0.99897826 |
| ENST00000587129 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.002468711 | 0.9975313 |
| ENST00000587129 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.001036742 | 0.99896324 |
| ENST00000590842 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.001081446 | 0.99891853 |
| ENST00000590842 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.002558696 | 0.9974413 |
| ENST00000590842 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.001118404 | 0.99888164 |
| ENST00000334295 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.001997565 | 0.99800247 |
| ENST00000334295 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.003791056 | 0.9962089 |
| ENST00000334295 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.002045445 | 0.9979545 |
| ENST00000593035 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.002141755 | 0.9978582 |
| ENST00000593035 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.003806382 | 0.99619365 |
| ENST00000593035 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.002084774 | 0.9979152 |
| ENST00000589049 | ENST00000428559 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.000984607 | 0.99901533 |
| ENST00000589049 | ENST00000532964 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.002393704 | 0.9976063 |
| ENST00000589049 | ENST00000526530 | TPGS2 | chr18 | 34385337 | - | DERA | chr12 | 16185476 | + | 0.001002364 | 0.9989976 |
Top |
Fusion Genomic Features for TPGS2-DERA |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TPGS2 | chr18 | 34385336 | - | DERA | chr12 | 16185475 | + | 9.04E-05 | 0.99990964 |
| TPGS2 | chr18 | 34385336 | - | DERA | chr12 | 16185475 | + | 9.04E-05 | 0.99990964 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
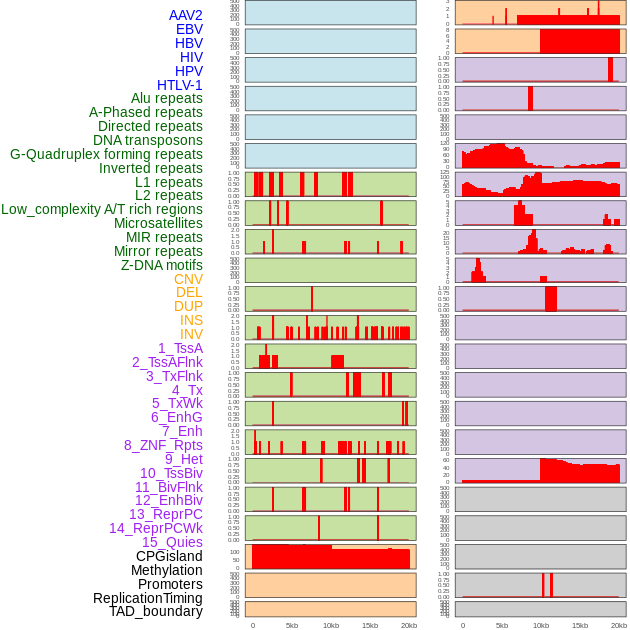 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
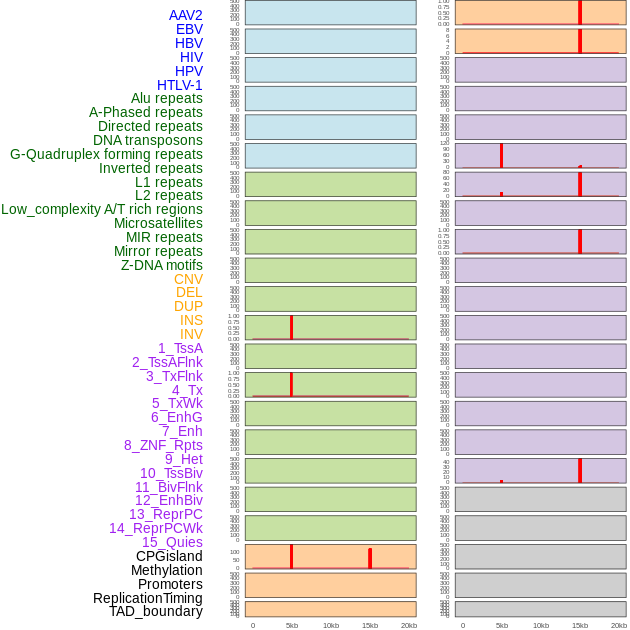 |
Top |
Fusion Protein Features for TPGS2-DERA |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:34385337/chr12:16185476) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| TPGS2 | DERA |
| FUNCTION: Catalyzes a reversible aldol reaction between acetaldehyde and D-glyceraldehyde 3-phosphate to generate 2-deoxy-D-ribose 5-phosphate. Participates in stress granule (SG) assembly. May allow ATP production from extracellular deoxyinosine in conditions of energy deprivation. {ECO:0000269|PubMed:25229427}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for TPGS2-DERA |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >93316_93316_1_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000334295_DERA_chr12_16185476_ENST00000428559_length(transcript)=1670nt_BP=810nt GTCCCGGCTCCAGAACGCCGCGGCACTGGGGCGGGAGGGGCCCGCCACGTCCGCCATCTTGAAACAACCGCCACTCGGGGCGTCGCGCCT GGGCCGCCACGCCTCGGCCTGCAGGCTCGGTCCCCATTGGCCGCACCACGCAGAGGCCCGCCCCCGCCTGCGCGGCCGGCCCCTGCGGGG CGAGAGGGTGGGGGCGAAGGGGAAGCTACGTCCCGGAGGTGCGGTGTGGGGCACCGGGCGGGGCCGCGGGAACCGGCGCCCCACGGAGCT GCTGCTGTCAGACCAACCCCGGGCCCCCATCATCACTGCGCCGCGCTTTCAGGCGCCGAGAACTACCGTTCCCGGCATGCCATGAAATTG GCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCC CGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGA CCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACT TTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATTAACAGCA TCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACACATGAAG GATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATT TCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAA AGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACC ATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTT TAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTAC CGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTG CACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAA AACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGA TTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTG >93316_93316_1_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000334295_DERA_chr12_16185476_ENST00000428559_length(amino acids)=274AA_BP=168 MRRAFRRRELPFPACHEIGLGAEAGSGPPPAPAARESRSRAMEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHM ISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPLGSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKE TVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHD -------------------------------------------------------------- >93316_93316_2_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000334295_DERA_chr12_16185476_ENST00000526530_length(transcript)=1664nt_BP=810nt GTCCCGGCTCCAGAACGCCGCGGCACTGGGGCGGGAGGGGCCCGCCACGTCCGCCATCTTGAAACAACCGCCACTCGGGGCGTCGCGCCT GGGCCGCCACGCCTCGGCCTGCAGGCTCGGTCCCCATTGGCCGCACCACGCAGAGGCCCGCCCCCGCCTGCGCGGCCGGCCCCTGCGGGG CGAGAGGGTGGGGGCGAAGGGGAAGCTACGTCCCGGAGGTGCGGTGTGGGGCACCGGGCGGGGCCGCGGGAACCGGCGCCCCACGGAGCT GCTGCTGTCAGACCAACCCCGGGCCCCCATCATCACTGCGCCGCGCTTTCAGGCGCCGAGAACTACCGTTCCCGGCATGCCATGAAATTG GCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCC CGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGA CCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACT TTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATTAACAGCA TCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACACATGAAG GATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATT TCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAA AGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACC ATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTT TAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTAC CGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTG CACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAA AACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGA TTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTG >93316_93316_2_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000334295_DERA_chr12_16185476_ENST00000526530_length(amino acids)=274AA_BP=168 MRRAFRRRELPFPACHEIGLGAEAGSGPPPAPAARESRSRAMEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHM ISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPLGSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKE TVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHD -------------------------------------------------------------- >93316_93316_3_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000334295_DERA_chr12_16185476_ENST00000532964_length(transcript)=1135nt_BP=810nt GTCCCGGCTCCAGAACGCCGCGGCACTGGGGCGGGAGGGGCCCGCCACGTCCGCCATCTTGAAACAACCGCCACTCGGGGCGTCGCGCCT GGGCCGCCACGCCTCGGCCTGCAGGCTCGGTCCCCATTGGCCGCACCACGCAGAGGCCCGCCCCCGCCTGCGCGGCCGGCCCCTGCGGGG CGAGAGGGTGGGGGCGAAGGGGAAGCTACGTCCCGGAGGTGCGGTGTGGGGCACCGGGCGGGGCCGCGGGAACCGGCGCCCCACGGAGCT GCTGCTGTCAGACCAACCCCGGGCCCCCATCATCACTGCGCCGCGCTTTCAGGCGCCGAGAACTACCGTTCCCGGCATGCCATGAAATTG GCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCC CGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGA CCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACT TTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATTAACAGCA TCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACACATGAAG GATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATT TCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAA AGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACC >93316_93316_3_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000334295_DERA_chr12_16185476_ENST00000532964_length(amino acids)=274AA_BP=168 MRRAFRRRELPFPACHEIGLGAEAGSGPPPAPAARESRSRAMEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHM ISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPLGSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKE TVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDSLAWLSLVKEELGDEWLKPELFRIGASTLLSDIERQIYHHVTGRYAAYHD -------------------------------------------------------------- >93316_93316_4_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000587129_DERA_chr12_16185476_ENST00000428559_length(transcript)=1279nt_BP=419nt ACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGA GAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCA TATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGAC ATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATTAACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTC CATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAA AGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTT TAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCC AGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCA TGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAA TTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCA TCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAA AAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCC TAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCA TCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTA >93316_93316_4_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000587129_DERA_chr12_16185476_ENST00000428559_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_5_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000587129_DERA_chr12_16185476_ENST00000526530_length(transcript)=1273nt_BP=419nt ACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGA GAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCA TATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGAC ATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATTAACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTC CATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAA AGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTT TAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCC AGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCA TGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAA TTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCA TCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAA AAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCC TAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCA TCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAAATTTCTGACATTGTCATATACAACGATGAATATCATTA >93316_93316_5_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000587129_DERA_chr12_16185476_ENST00000526530_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_6_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000587129_DERA_chr12_16185476_ENST00000532964_length(transcript)=744nt_BP=419nt ACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGA GAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCA TATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGAC ATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATTAACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTC CATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAA AGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTT TAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCC AGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCA >93316_93316_6_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000587129_DERA_chr12_16185476_ENST00000532964_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_7_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000589049_DERA_chr12_16185476_ENST00000428559_length(transcript)=1256nt_BP=396nt CGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGC GCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAA AGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGC ACATCATTCCACTGGGAAGCATGGCAATTAACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCAC CCACTCTGGCAGACCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCC CGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCA GTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCA GTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCA GTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCA GTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAG GCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATG GTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCT CGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAG >93316_93316_7_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000589049_DERA_chr12_16185476_ENST00000428559_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_8_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000589049_DERA_chr12_16185476_ENST00000526530_length(transcript)=1250nt_BP=396nt CGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGC GCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAA AGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGC ACATCATTCCACTGGGAAGCATGGCAATTAACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCAC CCACTCTGGCAGACCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCC CGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCA GTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCA GTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCA GTCACCAGTTCCAGAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCA GTAGGTAACTGGCATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAG GCACATCTGAAATGATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATG GTAGCTTTGATAACATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCT CGCAAGCGCTGCTGTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAG >93316_93316_8_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000589049_DERA_chr12_16185476_ENST00000526530_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_9_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000589049_DERA_chr12_16185476_ENST00000532964_length(transcript)=721nt_BP=396nt CGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGC GCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAA AGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGC ACATCATTCCACTGGGAAGCATGGCAATTAACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCAC CCACTCTGGCAGACCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCC CGGTAGCTATAGTAATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCA GTGCAAAGGATTCCCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCA GTACTCTGCTCTCGGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCA >93316_93316_9_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000589049_DERA_chr12_16185476_ENST00000532964_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_10_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000590842_DERA_chr12_16185476_ENST00000428559_length(transcript)=1317nt_BP=457nt GAAATTGGCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCA TCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACT GAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTG AAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATT AACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACA CATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATT AGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCT CTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAG ATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGA CAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAA TTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGA AGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGG AGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAG TCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAA >93316_93316_10_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000590842_DERA_chr12_16185476_ENST00000428559_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_11_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000590842_DERA_chr12_16185476_ENST00000526530_length(transcript)=1311nt_BP=457nt GAAATTGGCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCA TCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACT GAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTG AAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATT AACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACA CATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATT AGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCT CTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAG ATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCAGAAAAGTTCTTTACGA CAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGCATTCCTCTCTTTAAAA TTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAATGATCTTAATTACTAGA AGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAACATCAAATTCTAAGGG AGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCTGTAACGACTTCAGGAG TCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAATTGTTGTCCTAATGAA >93316_93316_11_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000590842_DERA_chr12_16185476_ENST00000526530_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_12_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000590842_DERA_chr12_16185476_ENST00000532964_length(transcript)=782nt_BP=457nt GAAATTGGCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGAATCGCGGTCGCGAGCCATGGAGGAGGAGGCA TCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCACGCGCATCCTAGAATCTTCCCCAGGTGTGACT GAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACAAAAGAATAACTGTGTGATGCCTGAAGATGTG AAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGAGCACATCATTCCACTGGGAAGCATGGCAATT AACAGCATCTCAAAACTGACTCAGCTCACCCAGTCTTCCATGTATTCACTTCCTAATGCACCCACTCTGGCAGACCTGGAGGACGATACA CATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGTAATGCTGCGGGCCATT AGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTCCCTTGCTTGGCTCTCT CTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTCGGACATTGAGAGGCAG >93316_93316_12_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000590842_DERA_chr12_16185476_ENST00000532964_length(amino acids)=233AA_BP=127 MEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHMISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDEHIIPL GSMAINSISKLTQLTQSSMYSLPNAPTLADLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGIRSAKDS -------------------------------------------------------------- >93316_93316_13_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000593035_DERA_chr12_16185476_ENST00000428559_length(transcript)=1333nt_BP=473nt ACCGGGCGGGGCCGCGGGAACCGGCGCCCCACGGAGCTGCTGCTGTCAGACCAACCCCGGGCCCCCATCATCACTGCGCCGCGCTTTCAG GCGCCGAGAACTACCGTTCCCGGCATGCCATGAAATTGGCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGA ATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCAC GCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACA AAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGA CCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGT AATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTC CCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTC GGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCA GAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGC ATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAAT GATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAA CATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCT GTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAA >93316_93316_13_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000593035_DERA_chr12_16185476_ENST00000428559_length(amino acids)=239AA_BP=133 MRRAFRRRELPFPACHEIGLGAEAGSGPPPAPAARESRSRAMEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHM ISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDDLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGI -------------------------------------------------------------- >93316_93316_14_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000593035_DERA_chr12_16185476_ENST00000526530_length(transcript)=1327nt_BP=473nt ACCGGGCGGGGCCGCGGGAACCGGCGCCCCACGGAGCTGCTGCTGTCAGACCAACCCCGGGCCCCCATCATCACTGCGCCGCGCTTTCAG GCGCCGAGAACTACCGTTCCCGGCATGCCATGAAATTGGCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGA ATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCAC GCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACA AAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGA CCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGT AATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTC CCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTC GGACATTGAGAGGCAGATTTACCATCATGTGACTGGAAGATATGCAGCTTATCATGATCTTCCAATGTCTTAAATCAGTCACCAGTTCCA GAAAAGTTCTTTACGACAATGTTTAAAAATTATTTTTCTACGTAATTGCTAAAATTATTTAATTAAAAAATTGGGCAGTAGGTAACTGGC ATTCCTCTCTTTAAAATTTCTACCGAACTTAATGGAATGGAAAAAGCAAACTCATCCACATGTGGTACTCATTTCAGGCACATCTGAAAT GATCTTAATTACTAGAAGATCTGCACTATTAACTTTGTGAAGAGTTTCTCCTAAAAACTTTAAGTAAAATGTTAATGGTAGCTTTGATAA CATCAAATTCTAAGGGAGAAAAAAACAATATTAAACCGCCCAAGCAGTGTGCCCTAGCAGAGGAAAATGCAACATCTCGCAAGCGCTGCT GTAACGACTTCAGGAGTCACTGATTCAGCACTAATTTCCTGCTGTGAAAACTCATCTTTCATTTTTGCCGTGGATAGGCGCTTTTATTAA >93316_93316_14_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000593035_DERA_chr12_16185476_ENST00000526530_length(amino acids)=239AA_BP=133 MRRAFRRRELPFPACHEIGLGAEAGSGPPPAPAARESRSRAMEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHM ISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDDLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGI -------------------------------------------------------------- >93316_93316_15_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000593035_DERA_chr12_16185476_ENST00000532964_length(transcript)=798nt_BP=473nt ACCGGGCGGGGCCGCGGGAACCGGCGCCCCACGGAGCTGCTGCTGTCAGACCAACCCCGGGCCCCCATCATCACTGCGCCGCGCTTTCAG GCGCCGAGAACTACCGTTCCCGGCATGCCATGAAATTGGCCTCGGCGCTGAGGCGGGGTCCGGCCCTCCACCCGCTCCCGCCGCGCGCGA ATCGCGGTCGCGAGCCATGGAGGAGGAGGCATCGTCCCCGGGGCTGGGCTGCAGCAAGCCGCACCTGGAGAAGCTGACCCTGGGCATCAC GCGCATCCTAGAATCTTCCCCAGGTGTGACTGAGGTGACCATCATAGAAAAGCCTCCTGCTGAACGTCATATGATTTCTTCCTGGGAACA AAAGAATAACTGTGTGATGCCTGAAGATGTGAAGAACTTTTACCTGATGACCAATGGCTTCCACATGACATGGAGTGTGAAGCTGGATGA CCTGGAGGACGATACACATGAAGGATCAGATTTTATTAAGACCTCTACTGGAAAAGAAACAGTAAATGCCACCTTCCCGGTAGCTATAGT AATGCTGCGGGCCATTAGAGATTTCTTCTGGAAAACTGGAAACAAGATAGGGTTTAAACCAGCAGGAGGCATCCGCAGTGCAAAGGATTC CCTTGCTTGGCTCTCTCTTGTAAAGGAGGAGCTTGGAGATGAGTGGCTGAAGCCAGAACTCTTTCGAATAGGTGCCAGTACTCTGCTCTC >93316_93316_15_TPGS2-DERA_TPGS2_chr18_34385337_ENST00000593035_DERA_chr12_16185476_ENST00000532964_length(amino acids)=239AA_BP=133 MRRAFRRRELPFPACHEIGLGAEAGSGPPPAPAARESRSRAMEEEASSPGLGCSKPHLEKLTLGITRILESSPGVTEVTIIEKPPAERHM ISSWEQKNNCVMPEDVKNFYLMTNGFHMTWSVKLDDLEDDTHEGSDFIKTSTGKETVNATFPVAIVMLRAIRDFFWKTGNKIGFKPAGGI -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TPGS2-DERA |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TPGS2-DERA |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TPGS2-DERA |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
