
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TRIOBP-SEPT9 (FusionGDB2 ID:94191) |
Fusion Gene Summary for TRIOBP-SEPT9 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TRIOBP-SEPT9 | Fusion gene ID: 94191 | Hgene | Tgene | Gene symbol | TRIOBP | SEPT9 | Gene ID | 11078 | 10801 |
| Gene name | TRIO and F-actin binding protein | septin 9 | |
| Synonyms | DFNB28|HRIHFB2122|TAP68|TARA|dJ37E16.4 | AF17q25|MSF|MSF1|NAPB|PNUTL4|SEPT9|SINT1|SeptD1 | |
| Cytomap | 22q13.1 | 17q25.3 | |
| Type of gene | protein-coding | protein-coding | |
| Description | TRIO and F-actin-binding proteinprotein Taratara-like proteintrio-associated repeat on actin | septin-9MLL septin-like fusion protein MSF-AOv/Br septinovarian/breast septinseptin D1 | |
| Modification date | 20200313 | 20200328 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000403663, ENST00000406386, ENST00000407319, | ENST00000592481, ENST00000427180, ENST00000541152, ENST00000590917, ENST00000591088, ENST00000592951, ENST00000329047, ENST00000423034, ENST00000427177, ENST00000427674, ENST00000431235, ENST00000449803, ENST00000585930, ENST00000588690, ENST00000590294, ENST00000591198, ENST00000592420, | |
| Fusion gene scores | * DoF score | 10 X 9 X 7=630 | 28 X 12 X 9=3024 |
| # samples | 9 | 27 | |
| ** MAII score | log2(9/630*10)=-2.8073549220576 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(27/3024*10)=-3.48542682717024 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TRIOBP [Title/Abstract] AND SEPT9 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TRIOBP(38147835)-SEPT9(75478226), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TRIOBP-SEPT9 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TRIOBP | GO:1900026 | positive regulation of substrate adhesion-dependent cell spreading | 11148140 |
 Fusion gene breakpoints across TRIOBP (5'-gene) Fusion gene breakpoints across TRIOBP (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
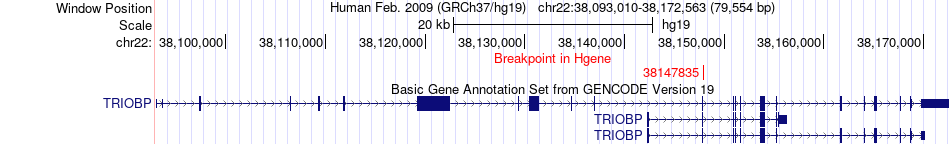 |
 Fusion gene breakpoints across SEPT9 (3'-gene) Fusion gene breakpoints across SEPT9 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
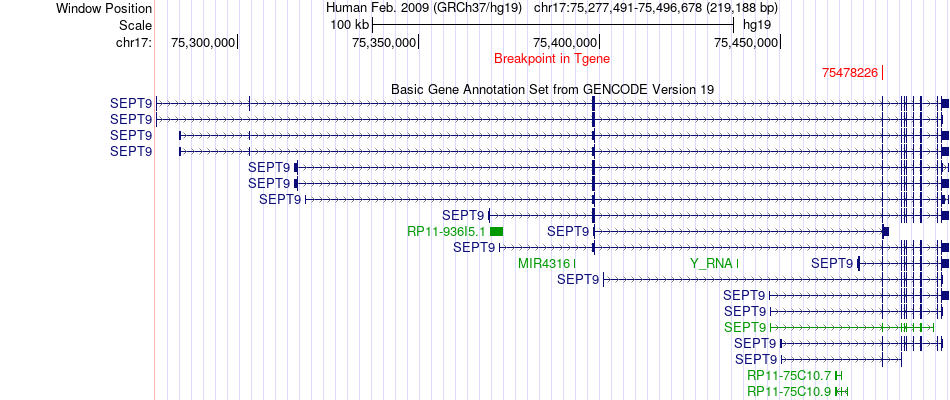 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | STAD | TCGA-CD-8531-01A | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
Top |
Fusion Gene ORF analysis for TRIOBP-SEPT9 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000403663 | ENST00000592481 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-3UTR | ENST00000406386 | ENST00000592481 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-3UTR | ENST00000407319 | ENST00000592481 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000403663 | ENST00000427180 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000403663 | ENST00000541152 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000403663 | ENST00000590917 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000403663 | ENST00000591088 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000403663 | ENST00000592951 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000406386 | ENST00000427180 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000406386 | ENST00000541152 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000406386 | ENST00000590917 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000406386 | ENST00000591088 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000406386 | ENST00000592951 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000407319 | ENST00000427180 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000407319 | ENST00000541152 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000407319 | ENST00000590917 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000407319 | ENST00000591088 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| 5CDS-5UTR | ENST00000407319 | ENST00000592951 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000329047 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000423034 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000427177 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000427674 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000431235 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000449803 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000585930 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000588690 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000590294 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000403663 | ENST00000591198 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000329047 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000423034 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000427177 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000427674 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000431235 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000449803 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000585930 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000588690 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000590294 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000406386 | ENST00000591198 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000329047 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000423034 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000427177 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000427674 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000431235 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000449803 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000585930 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000588690 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000590294 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| Frame-shift | ENST00000407319 | ENST00000591198 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| In-frame | ENST00000403663 | ENST00000592420 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| In-frame | ENST00000406386 | ENST00000592420 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
| In-frame | ENST00000407319 | ENST00000592420 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000406386 | TRIOBP | chr22 | 38147835 | + | ENST00000592420 | SEPT9 | chr17 | 75478226 | + | 7619 | 5634 | 231 | 5792 | 1853 |
| ENST00000407319 | TRIOBP | chr22 | 38147835 | + | ENST00000592420 | SEPT9 | chr17 | 75478226 | + | 2235 | 250 | 18 | 671 | 217 |
| ENST00000403663 | TRIOBP | chr22 | 38147835 | + | ENST00000592420 | SEPT9 | chr17 | 75478226 | + | 2230 | 245 | 13 | 666 | 217 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000406386 | ENST00000592420 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + | 0.021659762 | 0.9783402 |
| ENST00000407319 | ENST00000592420 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + | 0.33663622 | 0.6633638 |
| ENST00000403663 | ENST00000592420 | TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478226 | + | 0.293539 | 0.706461 |
Top |
Fusion Genomic Features for TRIOBP-SEPT9 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478225 | + | 8.58E-06 | 0.9999914 |
| TRIOBP | chr22 | 38147835 | + | SEPT9 | chr17 | 75478225 | + | 8.58E-06 | 0.9999914 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
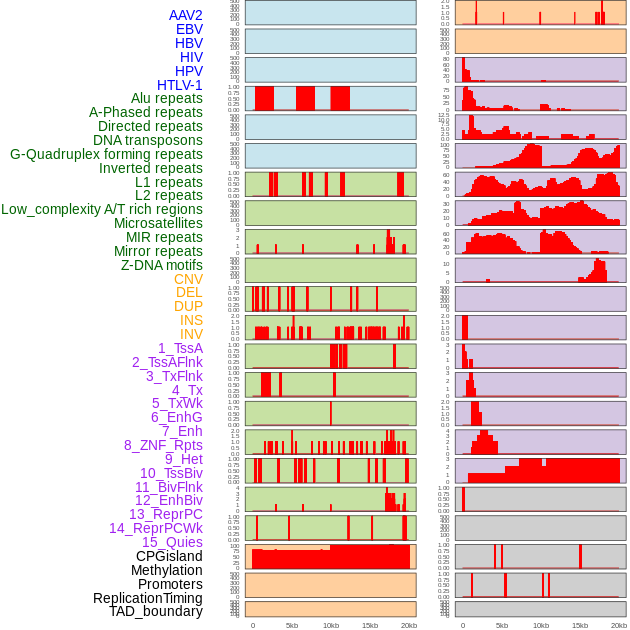 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
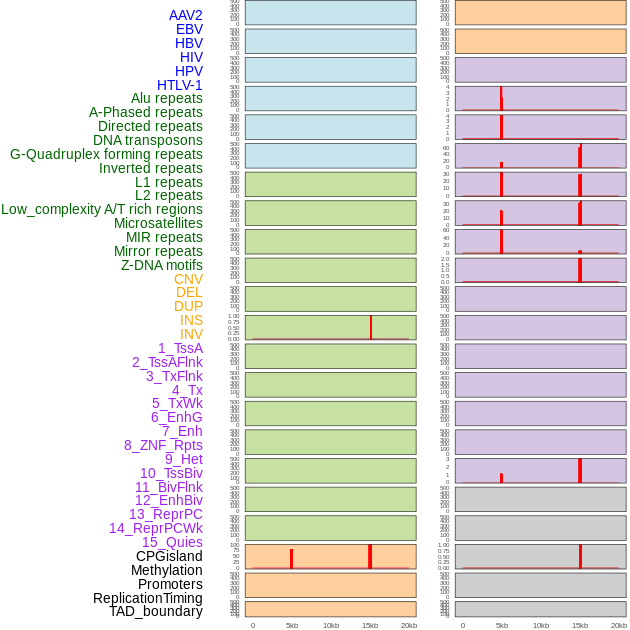 |
Top |
Fusion Protein Features for TRIOBP-SEPT9 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr22:38147835/chr17:75478226) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000329047 | 1 | 11 | 295_567 | 222 | 569.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000423034 | 1 | 11 | 295_567 | 233 | 580.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427177 | 2 | 12 | 295_567 | 240 | 587.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427180 | 0 | 10 | 295_567 | 128 | 475.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427674 | 1 | 11 | 295_567 | 76 | 423.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000431235 | 2 | 12 | 295_567 | 76 | 423.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000449803 | 2 | 12 | 295_567 | 76 | 423.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000541152 | 0 | 10 | 295_567 | 0 | 336.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000585930 | 0 | 10 | 295_567 | 16 | 363.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000588690 | 1 | 12 | 295_567 | 76 | 512.6666666666666 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000590294 | 1 | 12 | 295_567 | 222 | 354.3333333333333 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591088 | 0 | 10 | 295_567 | 0 | 336.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591198 | 1 | 11 | 295_567 | 221 | 568.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000592951 | 0 | 10 | 295_567 | 0 | 336.0 | Domain | Septin-type G | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000329047 | 1 | 11 | 445_453 | 222 | 569.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000423034 | 1 | 11 | 445_453 | 233 | 580.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427177 | 2 | 12 | 445_453 | 240 | 587.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427180 | 0 | 10 | 445_453 | 128 | 475.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427674 | 1 | 11 | 445_453 | 76 | 423.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000431235 | 2 | 12 | 445_453 | 76 | 423.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000449803 | 2 | 12 | 445_453 | 76 | 423.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000541152 | 0 | 10 | 445_453 | 0 | 336.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000585930 | 0 | 10 | 445_453 | 16 | 363.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000588690 | 1 | 12 | 445_453 | 76 | 512.6666666666666 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000590294 | 1 | 12 | 445_453 | 222 | 354.3333333333333 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591088 | 0 | 10 | 445_453 | 0 | 336.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591198 | 1 | 11 | 445_453 | 221 | 568.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000592951 | 0 | 10 | 445_453 | 0 | 336.0 | Nucleotide binding | GTP | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000329047 | 1 | 11 | 305_312 | 222 | 569.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000329047 | 1 | 11 | 362_365 | 222 | 569.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000329047 | 1 | 11 | 444_447 | 222 | 569.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000423034 | 1 | 11 | 305_312 | 233 | 580.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000423034 | 1 | 11 | 362_365 | 233 | 580.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000423034 | 1 | 11 | 444_447 | 233 | 580.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427177 | 2 | 12 | 305_312 | 240 | 587.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427177 | 2 | 12 | 362_365 | 240 | 587.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427177 | 2 | 12 | 444_447 | 240 | 587.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427180 | 0 | 10 | 305_312 | 128 | 475.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427180 | 0 | 10 | 362_365 | 128 | 475.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427180 | 0 | 10 | 444_447 | 128 | 475.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427674 | 1 | 11 | 305_312 | 76 | 423.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427674 | 1 | 11 | 362_365 | 76 | 423.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000427674 | 1 | 11 | 444_447 | 76 | 423.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000431235 | 2 | 12 | 305_312 | 76 | 423.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000431235 | 2 | 12 | 362_365 | 76 | 423.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000431235 | 2 | 12 | 444_447 | 76 | 423.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000449803 | 2 | 12 | 305_312 | 76 | 423.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000449803 | 2 | 12 | 362_365 | 76 | 423.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000449803 | 2 | 12 | 444_447 | 76 | 423.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000541152 | 0 | 10 | 305_312 | 0 | 336.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000541152 | 0 | 10 | 362_365 | 0 | 336.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000541152 | 0 | 10 | 444_447 | 0 | 336.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000585930 | 0 | 10 | 305_312 | 16 | 363.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000585930 | 0 | 10 | 362_365 | 16 | 363.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000585930 | 0 | 10 | 444_447 | 16 | 363.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000588690 | 1 | 12 | 305_312 | 76 | 512.6666666666666 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000588690 | 1 | 12 | 362_365 | 76 | 512.6666666666666 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000588690 | 1 | 12 | 444_447 | 76 | 512.6666666666666 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000590294 | 1 | 12 | 305_312 | 222 | 354.3333333333333 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000590294 | 1 | 12 | 362_365 | 222 | 354.3333333333333 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000590294 | 1 | 12 | 444_447 | 222 | 354.3333333333333 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591088 | 0 | 10 | 305_312 | 0 | 336.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591088 | 0 | 10 | 362_365 | 0 | 336.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591088 | 0 | 10 | 444_447 | 0 | 336.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591198 | 1 | 11 | 305_312 | 221 | 568.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591198 | 1 | 11 | 362_365 | 221 | 568.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000591198 | 1 | 11 | 444_447 | 221 | 568.0 | Region | G4 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000592951 | 0 | 10 | 305_312 | 0 | 336.0 | Region | G1 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000592951 | 0 | 10 | 362_365 | 0 | 336.0 | Region | G3 motif | |
| Tgene | SEPT9 | chr22:38147835 | chr17:75478226 | ENST00000592951 | 0 | 10 | 444_447 | 0 | 336.0 | Region | G4 motif |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TRIOBP | chr22:38147835 | chr17:75478226 | ENST00000406386 | + | 12 | 24 | 2062_2247 | 1793 | 3262.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TRIOBP | chr22:38147835 | chr17:75478226 | ENST00000406386 | + | 12 | 24 | 2281_2361 | 1793 | 3262.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TRIOBP | chr22:38147835 | chr17:75478226 | ENST00000407319 | + | 2 | 8 | 2062_2247 | 80 | 432.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TRIOBP | chr22:38147835 | chr17:75478226 | ENST00000407319 | + | 2 | 8 | 2281_2361 | 80 | 432.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TRIOBP | chr22:38147835 | chr17:75478226 | ENST00000406386 | + | 12 | 24 | 1778_1887 | 1793 | 3262.0 | Domain | PH |
| Hgene | TRIOBP | chr22:38147835 | chr17:75478226 | ENST00000407319 | + | 2 | 8 | 1778_1887 | 80 | 432.0 | Domain | PH |
Top |
Fusion Gene Sequence for TRIOBP-SEPT9 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >94191_94191_1_TRIOBP-SEPT9_TRIOBP_chr22_38147835_ENST00000403663_SEPT9_chr17_75478226_ENST00000592420_length(transcript)=2230nt_BP=245nt GTTTTATGGGCGGATGGAAGGGGCCGGGGCAGCGTCGGGGAAAGGAAGGGCCGGAGGCGCGGCGGCGGGCGGCCGAGAGGGGCGGCGGCG GCGGCGGCGGCGGGGTTCCCGCGCCGCGGAGCCCGGCCCGAGAGCCGCGTCCACGTTCCTGCCTCCTGCTCCCGCCGCCCTGGGGCGCCG CCATGACGCCCGATCTGCTCAACTTCAAGAAGGGATGGATGTCGATCTTGGACGAGCCTGGAGAGCCACTGAGGCGGCTCCCAGCTGCGT TGGCGACATGGCCGACACCCCCAGAGATGCCGGGCTCAAGCAGGCGCCTGCATCACGGAACGAGAAGGCCCCGGTGGACTTCGGCTACGT GGGGATTGACTCCATCCTGGAGCAGATGCGCCGGAAGGCCATGAAGCAGGGCTTCGAGTTCAACATCATGGTGGTCGGTGAGTCCTCACC TTGCATGCCACTCAACCAATGCCGGAAACCAGCCTCAGCCCCCAGGGCGGCCCACAAGCCTTTGGGCTTGGTCTTCCCTTCTGTAAAATG GGGCAGCAACCCTGACCTCAAGGACGGATTGCCAGTGACATTGCGTGTGCATGCATGCGCCTGGGAGGAGCCCACAGCCTCCAGCGGCTC CTCAGAGGGGCCTGTCCTGCACATCCAGGGCCCCTGAGATGACTTTGGGGAGCCCAGGCCGGCCTGGAGCAGGTCCTGATGAGTGGTCGC TGCCTCTGAGCAATGACGTCAGCTTGTGCAGAGGGTTCTGTGGCAGCTGTCCCCTCACTGTTGCTCCAGCCTGGCCCTCCAGGCTCCCCA CCGCACCCCACCGCACCCCGGCCAGAGGCTTCCCAGCAGCTCAGCTTCAGCTCTGCCCTGGAATTGGGGCCTCGTGGCTTCCACACCCCC TGGTGGCCCCGGAGAGCTGGAGGTCACCACGGGAGGTCGTCGATCAGCACGACCTGGGCAGGGTGAGGGGGCCAGGCTGGAGTCTACAGT GGGCCGCCTGTTGTTTCCAAAGCCCCCCAGGTCCCATTCAAACCTCACAGGGACCCTGTGAAGGGGGCGTCCCGTTTTCCAGGCAGAAGC GAGGCTCAGGGAAGGCTGCCAAGGTTCCTTAGCTCACAGGGCAGAGAACCGCAGCCACCGTTTTGACTCCCAGAACTGTGTTCTGCCCAC TCCACGGGGTCCAGGGAGGGGTCTGGGGAAGTGCCCCCGCCAGGTCACACCCTAAACAAAGGTGAGCTTGCGGGGTTTCTGCAAGGTGTC TGGGCACCTCTCAGGCCGGGCCTGTTTCTCTGACCTTTGCCAGCCGTGCACGTCCTTCCTGCCCAGCTCTGTGGACGCAAACCCACACCT GCCAGCCCGACACCTTTCTTTCCTCTTCTGCACTGATCTGGGCCCTGGCTCCCCTGGGGTCTTGGTGTCCTCTCTGCTTCCTGTAGCCTC ATGGGACCCGGCTGCTCATGGCCCTGCTGTGCTCACAGCTGCCTGGAAGCTGGCAGCCTGCTTCAAGGGAGGGCGGAGGTGTGGGGTCCC CTCCTACGGCCCCCACCCTGTTCTGCCTGTTGTGGGGGACGTGCTTGTACCCTGAGGATTCAGGGCTGCCTCCCGGGAAGATGGCTGGAG AGAAAGAGAGGGGAGAGGCCCAAAGGCAGTGGCCAGGCTGGGACTCATCTCTGTGGGCTACGGAGGGGCTGCCCTGCTCCCTAGCCAGGG ACAGGGTGAGAAGGACCTGGGAACCTCTGCGTCCCAGCCATGGCACAGGCCTTCTTCCGGAGCTTTGGCCCCAAGGAGAGGGAGAGGCAG CCCAGCAGCCCTCGCCTTCCGTGGCTGCCACTGTGACAGCCTTGCCCTCTGGGAATGGTGCGCAGGAGGCCACTTCACCTGTCTGGGCCT CTTCGCCCTTTCAACCCTGAGAAGAACAGTGGTCCTGATTCCCCACAAAATCACTCCCTCCCAGGGATGCCGAGTGAGGAACAAGACGTG GGGCAGCTCTGCTCTCCTCCCTCCTCCCTCCCATGGCAGCTGTGCCCACCTGCAGAGGTGCAGGCAAGGTGGAGGGGCACCTGTTTGGAG GTCCACAGACCCCGGGGAGAATCCCAGCTTTGTCCTAACTCAGCGGGGTGAGGCTTGTGTGTCTTCCCCTGGGCTCCTCCTCTGTGAGCC >94191_94191_1_TRIOBP-SEPT9_TRIOBP_chr22_38147835_ENST00000403663_SEPT9_chr17_75478226_ENST00000592420_length(amino acids)=217AA_BP=72 MEGAGAASGKGRAGGAAAGGREGRRRRRRRGSRAAEPGPRAASTFLPPAPAALGRRHDARSAQLQEGMDVDLGRAWRATEAAPSCVGDMA DTPRDAGLKQAPASRNEKAPVDFGYVGIDSILEQMRRKAMKQGFEFNIMVVGESSPCMPLNQCRKPASAPRAAHKPLGLVFPSVKWGSNP -------------------------------------------------------------- >94191_94191_2_TRIOBP-SEPT9_TRIOBP_chr22_38147835_ENST00000406386_SEPT9_chr17_75478226_ENST00000592420_length(transcript)=7619nt_BP=5634nt ATCGCTCTGATTCATTGTTAAAATAAAGAGGCCATGACCCTGGCACTCCCACCCCGGCAGCCCCTGTGCGTGGAAACCCAGCCAAGGTCT GATGATGGAGGAGCCTTGGCCCTGGCTGTCCCAGGGGAGGAGGTGAAATTCCTCAGCTCTCCACCAAGATTGGCCACAAAAGCCTGATCC CCTGGAACACAGCAGGCCTCACATAGACGGTCAGCCATTGGATCATAGGAACTGCCCTGGCCTGACTCACCCAATATGGAGGAGGTGCCT GGGGATGCCCTGTGTGAACACTTTGAGGCCAACATACTTACCCAGAACCGCTGTCAAAACTGCTTCCACCCTGAGGAGGCCCATGGAGCA AGATACCAGGAGCTCAGGAGCCCTTCAGGTGCTGAGGTGCCCTACTGCGACCTGCCTCGATGTCCACCTGCCCCTGAGGACCCACTCAGC GCCTCAACCTCCGGCTGCCAGTCTGTGGTGGACCCAGGCCTCAGGCCAGGGCCCAAGAGGGGCCCATCCCCCTCAGCAGGGCTCCCAGAA GAGGGTCCCACAGCTGCCCCCAGGAGCAGGAGCCGGGAGCTTGAGGCAGTACCCTATCTGGAGGGCCTGACCACTTCCTTGTGTGGCAGC TGCAACGAGGACCCCGGCTCTGACCCCACCTCCAGCCCTGACTCCGCCACCCCTGATGATACCAGCAACTCGTCCTCTGTGGACTGGGAC ACTGTTGAGAGGCAGGAGGAGGAGGCCCCCAGCTGGGACGAGCTCGCAGTGATGATCCCGAGGAGGCCTCGGGAGGGGCCGAGAGCTGAC AGCTCCCAAAGGGCTCCGTCTCTCCTCACCAGGTCCCCTGTGGGAGGAGATGCTGCAGGCCAGAAAAAGGAGGACACCGGCGGTGGGGGC CGGAGCGCAGGACAGCACTGGGCAAGGCTCCGGGGAGAAAGCGGGTTGTCCCTGGAGCGGCACCGGTCAACACTGACCCAGGCTTCCTCC ATGACACCACACAGTGGACCTCGAAGCACCACGTCTCAGGCTTCTCCTGCCCAAAGGGACACTGCTCAGGCTGCCTCTACACGTGAAATC CCCAGAGCCTCCTCTCCCCATCGAATCACCCAAAGGGACACCTCCAGGGCCTCATCCACCCAACAGGAAATCTCCAGGGCCTCATCCACC CAACAGGAAACCTCCAGGGCCTCATCCACCCAAGAGGACACCCCTAGGGCCTCATCCACCCAAGAGGACACCCCCAGGGCCTCATCTACA CAGTGGAACACCCCCAGAGCTTCCTCTCCCTCACGAAGCACCCAACTGGATAACCCCAGAACCTCTTCTACCCAGCAGGACAACCCCCAA ACTTCTTTTCCTACTTGTACTCCCCAGCGGGAAAACCCCAGGACACCCTGTGTCCAGCAGGACGATCCCAGAGCCTCCTCTCCCAACAGA ACCACTCAACGAGAGAATTCCAGAACATCCTGTGCCCAGCGGGACAATCCCAAAGCCTCCAGAACCTCCTCTCCCAATAGAGCCACACGA GACAACCCCAGAACATCCTGCGCCCAGCGGGACAATCCCAGAGCCTCCTCTCCCAGTAGAGCTACACGAGACAACCCCACAACATCCTGT GCCCAGCGGGACAATCCCAGAGCCTCCAGAACCTCCTCTCCCAATAGAGCCACACGAGACAACCCCAGAACATCCTGTGCCCAGCGGGAC AATCCCAGAGCCTCCTCTCCCAGTAGAGCTACACGAGACAACCCCACAACATCCTGTGCCCAGCGGGACAATCCCAGAGCCTCCAGAACC TCCTCTCCCAATAGAGCCACACGAGACAACCCCAGAACATCCTGCGCCCAGCGGGACAATCCCAGAGCCTCCTCTCCCAATAGAGCTGCA CGAGACAACCCCACAACATCCTGTGCCCAGCGGGACAATCCCAGAGCCTCCAGAACCTCCTCTCCCAATAGAGCCACACGAGACAACCCC AGAACATCCTGTGCCCAGCGGGACAATCCCAGAGCCTCCTCTCCCAATAGAGCTACACGAGACAACCCCACAACATCCTGTGCCCAGCGG GACAATCCCAGAGCCTCCAGAACCTCCTCTCCCAATAGAGCCACACGAGATAACCCCAGAACATCCTGTGCCCAGCGGGACAATCCCAGA GCCTCCTCTCCCAACAGAACCACCCAACAAGACAGCCCCAGAACATCCTGTGCCCGACGGGACGATCCCAGAGCCTCCTCTCCTAACAGA ACCATCCAACAAGAGAACCCCAGAACATCCTGTGCCCTACGGGACAATCCCAGAGCCTCCTCTCCCAGCAGAACCATCCAACAAGAGAAC CCCAGAACATCCTGTGCCCAACGGGACGATCCCAGAGCCTCCTCTCCTAACAGAACCACCCAACAAGAGAACCCCAGAACATCCTGTGCC CGACGGGACAATCCCAGAGCCTCCTCTCGCAACAGAACCATCCAGCGAGACAACCCCAGAACATCCTGTGCCCAGCGGGACAATCCCAGA GCCTCCTCTCCTAACAGAACCATCCAACAAGAGAACCTCAGAACATCCTGTACCCGACAGGACAATCCCAGGACCTCCTCTCCCAATAGA GCCACACGAGACAACCCCAGAACATCCTGTGCCCAGCGGGACAATCTCAGAGCCTCCTCTCCCATCAGAGCCACCCAACAGGACAACCCC AGAACTTGTATTCAACAGAACATCCCCAGATCATCTTCTACCCAACAAGACAACCCTAAAACCTCTTGTACCAAACGAGATAACCTCAGA CCCACTTGTACACAGCGGGACCGCACACAGTCCTTTTCCTTTCAACGAGACAACCCTGGAACCTCCTCATCTCAATGCTGCACCCAAAAG GAGAATCTGAGACCATCATCTCCCCACCGCTCCACTCAATGGAACAATCCCAGGAATTCATCTCCCCATCGTACTAACAAAGACATCCCC TGGGCCTCGTTTCCCCTCCGGCCAACTCAGAGTGATGGTCCCCGAACCTCTTCCCCATCTCGCTCCAAGCAAAGCGAGGTTCCCTGGGCA TCCATCGCCCTCCGGCCAACCCAAGGTGACAGGCCTCAGACATCCTCTCCCAGCAGGCCAGCCCAGCATGACCCACCCCAGTCCTCCTTT GGCCCCACCCAGTACAACTTGCCATCCCGGGCCACCTCTTCCTCCCATAACCCAGGCCACCAGAGCACCTCCCGAACTTCCTCACCTGTG TACCCCGCTGCCTATGGGGCTCCCCTGACCTCTCCTGAGCCCTCCCAGCCTCCATGTGCTGTGTGCATTGGGCACCGGGATGCCCCTCGA GCCTCTTCGCCCCCTCGCTATTTGCAGCACGACCCCTTCCCCTTCTTCCCAGAGCCCCGCGCCCCTGAGAGTGAACCGCCCCACCACGAG CCTCCCTATATACCACCTGCTGTGTGCATTGGACACCGAGATGCCCCCCGGGCGTCCTCGCCCCCCCGCCACACCCAATTTGACCCCTTC CCCTTCCTCCCAGACACATCAGATGCCGAGCATCAGTGTCAGTCCCCCCAACACGAGCCCCTTCAGCTCCCTGCACCTGTGTGTATTGGG TACCGAGATGCACCCCGGGCCTCCTCCCCACCACGCCAGGCCCCAGAGCCTTCCCTCTTATTCCAGGACCTCCCCAGGGCCAGCACAGAG AGCCTTGTCCCTTCCATGGACTCTCTGCACGAGTGCCCCCACATCCCCACCCCTGTGTGCATTGGGCACCGGGATGCACCCTCCTTCTCA TCCCCACCACGCCAGGCTCCTGAGCCATCCCTCTTCTTCCAGGATCCCCCTGGAACTAGTATGGAGAGCCTGGCCCCCTCCACTGACTCT CTGCATGGCTCCCCAGTGCTGATCCCCCAAGTGTGCATCGGGCACCGGGATGCACCCCGAGCCTCCTCCCCACCCCGCCACCCACCCAGT GACCTAGCGTTCCTGGCACCCTCACCTTCACCGGGCAGCTCTGGGGGCTCCCGGGGCTCAGCGCCTCCCGGGGAGACCAGGCACAACTTG GAGCGGGAGGAGTACACTGTGCTGGCCGACCTGCCCCCACCCAGGAGGCTGGCCCAGAGACAGCCAGGGCCCCAGGCGCAGTGCAGCAGC GGGGGCCGCACCCACAGCCCTGGCCGTGCAGAGGTGGAGCGCCTCTTCGGGCAAGAGCGCAGGAAGTCCGAGGCAGCGGGGGCCTTCCAG GCCCAGGACGAGGGACGGTCACAGCAGCCCAGCCAAGGCCAGAGCCAACTTCTCCGAAGACAGTCCAGCCCTGCCCCCAGCAGGCAGGTG ACCATGCTCCCTGCCAAACAGGCAGAACTGACCCGGCGGAGCCAAGCAGAGCCCCCTCATCCTTGGAGTCCTGAGAAGAGACCTGAGGGA GATCGGCAGCTCCAGGGGTCCCCGCTGCCCCCCAGGACATCAGCCAGGACCCCTGAGAGGGAGCTGCGGACACAGAGACCTCTGGAGAGT GGCCAAGCAGGCCCAAGACAGCCTCTGGGGGTGTGGCAGAGTCAGGAGGAACCGCCAGGGTCCCAGGGCCCTCATAGACACCTAGAAAGG AGCTGGAGCAGCCAGGAGGGAGGCCTGGGCCCTGGGGGCTGGTGGGGATGTGGAGAGCCCAGCCTGGGGGCAGCCAAAGCCCCGGAGGGA GCATGGGGGGGCACTTCCAGGGAGTACAAGGAGAGCTGGGGGCAGCCAGAGGCCTGGGAGGAGAAGCCCACTCATGAGCTCCCCAGAGAA CTAGGAAAGAGAAGCCCACTCACGAGCCCCCCTGAGAACTGGGGAGGCCCCGCAGAGTCCTCACAATCCTGGCACTCTGGGACACCCACT GCTGTGGGCTGGGGGGCAGAGGGAGCGTGTCCATACCCGCGTGGCTCTGAGAGGCGACCCGAGCTTGACTGGAGGGATCTGCTTGGCCTT CTCCGGGCACCAGGAGAGGGGGTCTGGGCCCGTGTCCCCAGCCTGGACTGGGAGGGCCTCTTGGAGCTCCTGCAGGCCAGGCTGCCCCGC AAGGACCCAGCTGGACACAGGGATGACCTGGCCAGGGCTTTAGGGCCAGAGCTGGGTCCCCCAGGCACAAACGATGTCCCTGAGCAGGAG TCACACAGCCAGCCAGAAGGCTGGGCCGAGGCCACCCCAGTCAATGGACACAGCCCCGCACTGCAGTCCCAGAGCCCGGTCCAGCTGCCC AGCCCTGCCTGCACCTCCACCCAGTGGCCAAAGATCAAAGTGACAAGAGGACCAGCGACCGCAACTCTGGCAGGCCTGGAGCAGACGGGC CCCCTGGGGAGCAGGAGCACTGCGAAGGGCCCCAGCTTGCCAGAGCTGCAGTTCCAACCAGAGGAGCCTGAGGAGTCAGAACCAAGCAGA GGCCAAGACCCCCTGACTGACCAGAAGCAGGCAGACTCGGCAGACAAGAGGCCAGCAGAGGGCAAGGCTGGGAGCCCGCTCAAGGGCCGA CTGGTGACCTCATGGCGGATGCCCGGGGACCGGCCCACGCTGTTCAATCCGTTCCTGCTGTCTCTGGGGGTCCTCAGGTGGCGAAGGCCC GATCTGCTCAACTTCAAGAAGGGATGGATGTCGATCTTGGACGAGCCTGGAGAGCCACTGAGGCGGCTCCCAGCTGCGTTGGCGACATGG CCGACACCCCCAGAGATGCCGGGCTCAAGCAGGCGCCTGCATCACGGAACGAGAAGGCCCCGGTGGACTTCGGCTACGTGGGGATTGACT CCATCCTGGAGCAGATGCGCCGGAAGGCCATGAAGCAGGGCTTCGAGTTCAACATCATGGTGGTCGGTGAGTCCTCACCTTGCATGCCAC TCAACCAATGCCGGAAACCAGCCTCAGCCCCCAGGGCGGCCCACAAGCCTTTGGGCTTGGTCTTCCCTTCTGTAAAATGGGGCAGCAACC CTGACCTCAAGGACGGATTGCCAGTGACATTGCGTGTGCATGCATGCGCCTGGGAGGAGCCCACAGCCTCCAGCGGCTCCTCAGAGGGGC CTGTCCTGCACATCCAGGGCCCCTGAGATGACTTTGGGGAGCCCAGGCCGGCCTGGAGCAGGTCCTGATGAGTGGTCGCTGCCTCTGAGC AATGACGTCAGCTTGTGCAGAGGGTTCTGTGGCAGCTGTCCCCTCACTGTTGCTCCAGCCTGGCCCTCCAGGCTCCCCACCGCACCCCAC CGCACCCCGGCCAGAGGCTTCCCAGCAGCTCAGCTTCAGCTCTGCCCTGGAATTGGGGCCTCGTGGCTTCCACACCCCCTGGTGGCCCCG GAGAGCTGGAGGTCACCACGGGAGGTCGTCGATCAGCACGACCTGGGCAGGGTGAGGGGGCCAGGCTGGAGTCTACAGTGGGCCGCCTGT TGTTTCCAAAGCCCCCCAGGTCCCATTCAAACCTCACAGGGACCCTGTGAAGGGGGCGTCCCGTTTTCCAGGCAGAAGCGAGGCTCAGGG AAGGCTGCCAAGGTTCCTTAGCTCACAGGGCAGAGAACCGCAGCCACCGTTTTGACTCCCAGAACTGTGTTCTGCCCACTCCACGGGGTC CAGGGAGGGGTCTGGGGAAGTGCCCCCGCCAGGTCACACCCTAAACAAAGGTGAGCTTGCGGGGTTTCTGCAAGGTGTCTGGGCACCTCT CAGGCCGGGCCTGTTTCTCTGACCTTTGCCAGCCGTGCACGTCCTTCCTGCCCAGCTCTGTGGACGCAAACCCACACCTGCCAGCCCGAC ACCTTTCTTTCCTCTTCTGCACTGATCTGGGCCCTGGCTCCCCTGGGGTCTTGGTGTCCTCTCTGCTTCCTGTAGCCTCATGGGACCCGG CTGCTCATGGCCCTGCTGTGCTCACAGCTGCCTGGAAGCTGGCAGCCTGCTTCAAGGGAGGGCGGAGGTGTGGGGTCCCCTCCTACGGCC CCCACCCTGTTCTGCCTGTTGTGGGGGACGTGCTTGTACCCTGAGGATTCAGGGCTGCCTCCCGGGAAGATGGCTGGAGAGAAAGAGAGG GGAGAGGCCCAAAGGCAGTGGCCAGGCTGGGACTCATCTCTGTGGGCTACGGAGGGGCTGCCCTGCTCCCTAGCCAGGGACAGGGTGAGA AGGACCTGGGAACCTCTGCGTCCCAGCCATGGCACAGGCCTTCTTCCGGAGCTTTGGCCCCAAGGAGAGGGAGAGGCAGCCCAGCAGCCC TCGCCTTCCGTGGCTGCCACTGTGACAGCCTTGCCCTCTGGGAATGGTGCGCAGGAGGCCACTTCACCTGTCTGGGCCTCTTCGCCCTTT CAACCCTGAGAAGAACAGTGGTCCTGATTCCCCACAAAATCACTCCCTCCCAGGGATGCCGAGTGAGGAACAAGACGTGGGGCAGCTCTG CTCTCCTCCCTCCTCCCTCCCATGGCAGCTGTGCCCACCTGCAGAGGTGCAGGCAAGGTGGAGGGGCACCTGTTTGGAGGTCCACAGACC CCGGGGAGAATCCCAGCTTTGTCCTAACTCAGCGGGGTGAGGCTTGTGTGTCTTCCCCTGGGCTCCTCCTCTGTGAGCCTTAGCTTCCTC >94191_94191_2_TRIOBP-SEPT9_TRIOBP_chr22_38147835_ENST00000406386_SEPT9_chr17_75478226_ENST00000592420_length(amino acids)=1853AA_BP=1801 MPWPDSPNMEEVPGDALCEHFEANILTQNRCQNCFHPEEAHGARYQELRSPSGAEVPYCDLPRCPPAPEDPLSASTSGCQSVVDPGLRPG PKRGPSPSAGLPEEGPTAAPRSRSRELEAVPYLEGLTTSLCGSCNEDPGSDPTSSPDSATPDDTSNSSSVDWDTVERQEEEAPSWDELAV MIPRRPREGPRADSSQRAPSLLTRSPVGGDAAGQKKEDTGGGGRSAGQHWARLRGESGLSLERHRSTLTQASSMTPHSGPRSTTSQASPA QRDTAQAASTREIPRASSPHRITQRDTSRASSTQQEISRASSTQQETSRASSTQEDTPRASSTQEDTPRASSTQWNTPRASSPSRSTQLD NPRTSSTQQDNPQTSFPTCTPQRENPRTPCVQQDDPRASSPNRTTQRENSRTSCAQRDNPKASRTSSPNRATRDNPRTSCAQRDNPRASS PSRATRDNPTTSCAQRDNPRASRTSSPNRATRDNPRTSCAQRDNPRASSPSRATRDNPTTSCAQRDNPRASRTSSPNRATRDNPRTSCAQ RDNPRASSPNRAARDNPTTSCAQRDNPRASRTSSPNRATRDNPRTSCAQRDNPRASSPNRATRDNPTTSCAQRDNPRASRTSSPNRATRD NPRTSCAQRDNPRASSPNRTTQQDSPRTSCARRDDPRASSPNRTIQQENPRTSCALRDNPRASSPSRTIQQENPRTSCAQRDDPRASSPN RTTQQENPRTSCARRDNPRASSRNRTIQRDNPRTSCAQRDNPRASSPNRTIQQENLRTSCTRQDNPRTSSPNRATRDNPRTSCAQRDNLR ASSPIRATQQDNPRTCIQQNIPRSSSTQQDNPKTSCTKRDNLRPTCTQRDRTQSFSFQRDNPGTSSSQCCTQKENLRPSSPHRSTQWNNP RNSSPHRTNKDIPWASFPLRPTQSDGPRTSSPSRSKQSEVPWASIALRPTQGDRPQTSSPSRPAQHDPPQSSFGPTQYNLPSRATSSSHN PGHQSTSRTSSPVYPAAYGAPLTSPEPSQPPCAVCIGHRDAPRASSPPRYLQHDPFPFFPEPRAPESEPPHHEPPYIPPAVCIGHRDAPR ASSPPRHTQFDPFPFLPDTSDAEHQCQSPQHEPLQLPAPVCIGYRDAPRASSPPRQAPEPSLLFQDLPRASTESLVPSMDSLHECPHIPT PVCIGHRDAPSFSSPPRQAPEPSLFFQDPPGTSMESLAPSTDSLHGSPVLIPQVCIGHRDAPRASSPPRHPPSDLAFLAPSPSPGSSGGS RGSAPPGETRHNLEREEYTVLADLPPPRRLAQRQPGPQAQCSSGGRTHSPGRAEVERLFGQERRKSEAAGAFQAQDEGRSQQPSQGQSQL LRRQSSPAPSRQVTMLPAKQAELTRRSQAEPPHPWSPEKRPEGDRQLQGSPLPPRTSARTPERELRTQRPLESGQAGPRQPLGVWQSQEE PPGSQGPHRHLERSWSSQEGGLGPGGWWGCGEPSLGAAKAPEGAWGGTSREYKESWGQPEAWEEKPTHELPRELGKRSPLTSPPENWGGP AESSQSWHSGTPTAVGWGAEGACPYPRGSERRPELDWRDLLGLLRAPGEGVWARVPSLDWEGLLELLQARLPRKDPAGHRDDLARALGPE LGPPGTNDVPEQESHSQPEGWAEATPVNGHSPALQSQSPVQLPSPACTSTQWPKIKVTRGPATATLAGLEQTGPLGSRSTAKGPSLPELQ FQPEEPEESEPSRGQDPLTDQKQADSADKRPAEGKAGSPLKGRLVTSWRMPGDRPTLFNPFLLSLGVLRWRRPDLLNFKKGWMSILDEPG -------------------------------------------------------------- >94191_94191_3_TRIOBP-SEPT9_TRIOBP_chr22_38147835_ENST00000407319_SEPT9_chr17_75478226_ENST00000592420_length(transcript)=2235nt_BP=250nt AAAAAGTTTTATGGGCGGATGGAAGGGGCCGGGGCAGCGTCGGGGAAAGGAAGGGCCGGAGGCGCGGCGGCGGGCGGCCGAGAGGGGCGG CGGCGGCGGCGGCGGCGGGGTTCCCGCGCCGCGGAGCCCGGCCCGAGAGCCGCGTCCACGTTCCTGCCTCCTGCTCCCGCCGCCCTGGGG CGCCGCCATGACGCCCGATCTGCTCAACTTCAAGAAGGGATGGATGTCGATCTTGGACGAGCCTGGAGAGCCACTGAGGCGGCTCCCAGC TGCGTTGGCGACATGGCCGACACCCCCAGAGATGCCGGGCTCAAGCAGGCGCCTGCATCACGGAACGAGAAGGCCCCGGTGGACTTCGGC TACGTGGGGATTGACTCCATCCTGGAGCAGATGCGCCGGAAGGCCATGAAGCAGGGCTTCGAGTTCAACATCATGGTGGTCGGTGAGTCC TCACCTTGCATGCCACTCAACCAATGCCGGAAACCAGCCTCAGCCCCCAGGGCGGCCCACAAGCCTTTGGGCTTGGTCTTCCCTTCTGTA AAATGGGGCAGCAACCCTGACCTCAAGGACGGATTGCCAGTGACATTGCGTGTGCATGCATGCGCCTGGGAGGAGCCCACAGCCTCCAGC GGCTCCTCAGAGGGGCCTGTCCTGCACATCCAGGGCCCCTGAGATGACTTTGGGGAGCCCAGGCCGGCCTGGAGCAGGTCCTGATGAGTG GTCGCTGCCTCTGAGCAATGACGTCAGCTTGTGCAGAGGGTTCTGTGGCAGCTGTCCCCTCACTGTTGCTCCAGCCTGGCCCTCCAGGCT CCCCACCGCACCCCACCGCACCCCGGCCAGAGGCTTCCCAGCAGCTCAGCTTCAGCTCTGCCCTGGAATTGGGGCCTCGTGGCTTCCACA CCCCCTGGTGGCCCCGGAGAGCTGGAGGTCACCACGGGAGGTCGTCGATCAGCACGACCTGGGCAGGGTGAGGGGGCCAGGCTGGAGTCT ACAGTGGGCCGCCTGTTGTTTCCAAAGCCCCCCAGGTCCCATTCAAACCTCACAGGGACCCTGTGAAGGGGGCGTCCCGTTTTCCAGGCA GAAGCGAGGCTCAGGGAAGGCTGCCAAGGTTCCTTAGCTCACAGGGCAGAGAACCGCAGCCACCGTTTTGACTCCCAGAACTGTGTTCTG CCCACTCCACGGGGTCCAGGGAGGGGTCTGGGGAAGTGCCCCCGCCAGGTCACACCCTAAACAAAGGTGAGCTTGCGGGGTTTCTGCAAG GTGTCTGGGCACCTCTCAGGCCGGGCCTGTTTCTCTGACCTTTGCCAGCCGTGCACGTCCTTCCTGCCCAGCTCTGTGGACGCAAACCCA CACCTGCCAGCCCGACACCTTTCTTTCCTCTTCTGCACTGATCTGGGCCCTGGCTCCCCTGGGGTCTTGGTGTCCTCTCTGCTTCCTGTA GCCTCATGGGACCCGGCTGCTCATGGCCCTGCTGTGCTCACAGCTGCCTGGAAGCTGGCAGCCTGCTTCAAGGGAGGGCGGAGGTGTGGG GTCCCCTCCTACGGCCCCCACCCTGTTCTGCCTGTTGTGGGGGACGTGCTTGTACCCTGAGGATTCAGGGCTGCCTCCCGGGAAGATGGC TGGAGAGAAAGAGAGGGGAGAGGCCCAAAGGCAGTGGCCAGGCTGGGACTCATCTCTGTGGGCTACGGAGGGGCTGCCCTGCTCCCTAGC CAGGGACAGGGTGAGAAGGACCTGGGAACCTCTGCGTCCCAGCCATGGCACAGGCCTTCTTCCGGAGCTTTGGCCCCAAGGAGAGGGAGA GGCAGCCCAGCAGCCCTCGCCTTCCGTGGCTGCCACTGTGACAGCCTTGCCCTCTGGGAATGGTGCGCAGGAGGCCACTTCACCTGTCTG GGCCTCTTCGCCCTTTCAACCCTGAGAAGAACAGTGGTCCTGATTCCCCACAAAATCACTCCCTCCCAGGGATGCCGAGTGAGGAACAAG ACGTGGGGCAGCTCTGCTCTCCTCCCTCCTCCCTCCCATGGCAGCTGTGCCCACCTGCAGAGGTGCAGGCAAGGTGGAGGGGCACCTGTT TGGAGGTCCACAGACCCCGGGGAGAATCCCAGCTTTGTCCTAACTCAGCGGGGTGAGGCTTGTGTGTCTTCCCCTGGGCTCCTCCTCTGT >94191_94191_3_TRIOBP-SEPT9_TRIOBP_chr22_38147835_ENST00000407319_SEPT9_chr17_75478226_ENST00000592420_length(amino acids)=217AA_BP=72 MEGAGAASGKGRAGGAAAGGREGRRRRRRRGSRAAEPGPRAASTFLPPAPAALGRRHDARSAQLQEGMDVDLGRAWRATEAAPSCVGDMA DTPRDAGLKQAPASRNEKAPVDFGYVGIDSILEQMRRKAMKQGFEFNIMVVGESSPCMPLNQCRKPASAPRAAHKPLGLVFPSVKWGSNP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TRIOBP-SEPT9 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TRIOBP-SEPT9 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TRIOBP-SEPT9 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
