
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TRIP10-C1orf186 (FusionGDB2 ID:94227) |
Fusion Gene Summary for TRIP10-C1orf186 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TRIP10-C1orf186 | Fusion gene ID: 94227 | Hgene | Tgene | Gene symbol | TRIP10 | C1orf186 | Gene ID | 9322 | 440712 |
| Gene name | thyroid hormone receptor interactor 10 | regulator of hemoglobinization and erythroid cell expansion | |
| Synonyms | CIP4|HSTP|STOT|STP|TRIP-10 | C1orf186 | |
| Cytomap | 19p13.3 | 1q32.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | cdc42-interacting protein 4TR-interacting protein 10protein Felicsalt tolerant proteinsalt toleratorthyroid receptor-interacting protein 10 | regulator of hemoglobinization and erythroid cell expansion proteinregulator of human erythroid cell expansionregulator of human erythroid cell expansion proteinuncharacterized protein C1orf186 | |
| Modification date | 20200320 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000313244, ENST00000313285, ENST00000596758, ENST00000600428, ENST00000596543, | ENST00000469540, ENST00000331555, | |
| Fusion gene scores | * DoF score | 1 X 1 X 1=1 | 4 X 4 X 4=64 |
| # samples | 1 | 4 | |
| ** MAII score | log2(1/1*10)=3.32192809488736 | log2(4/64*10)=-0.678071905112638 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TRIP10 [Title/Abstract] AND C1orf186 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TRIP10(6750644)-C1orf186(206241676), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across TRIP10 (5'-gene) Fusion gene breakpoints across TRIP10 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
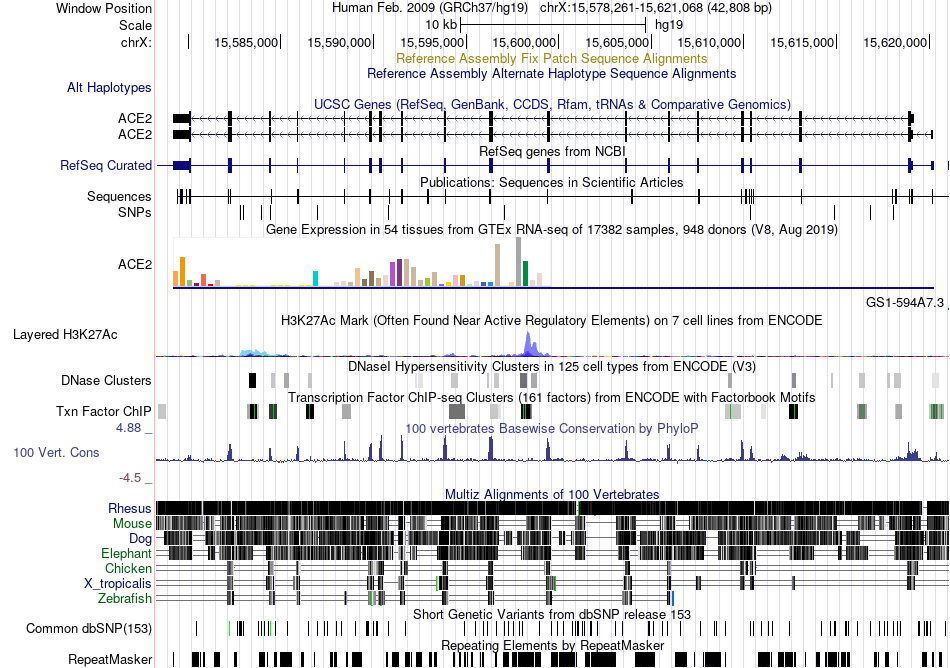 |
 Fusion gene breakpoints across C1orf186 (3'-gene) Fusion gene breakpoints across C1orf186 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
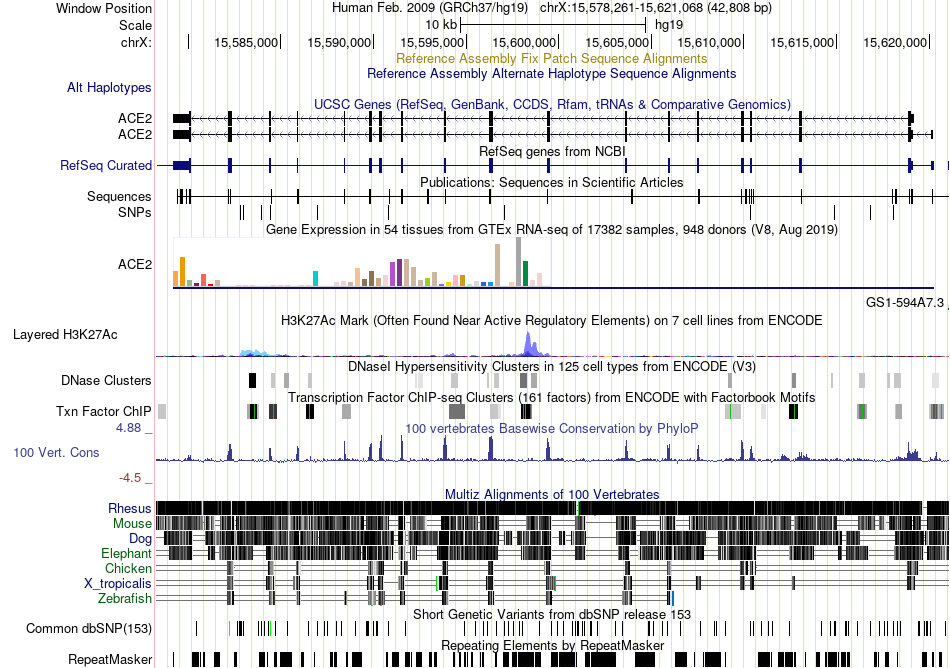 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-D1-A3DG | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
Top |
Fusion Gene ORF analysis for TRIP10-C1orf186 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000313244 | ENST00000469540 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| 5CDS-intron | ENST00000313285 | ENST00000469540 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| 5CDS-intron | ENST00000596758 | ENST00000469540 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| 5CDS-intron | ENST00000600428 | ENST00000469540 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| In-frame | ENST00000313244 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| In-frame | ENST00000313285 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| In-frame | ENST00000596758 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| In-frame | ENST00000600428 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| intron-3CDS | ENST00000596543 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
| intron-intron | ENST00000596543 | ENST00000469540 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000600428 | TRIP10 | chr19 | 6750644 | + | ENST00000331555 | C1orf186 | chr1 | 206241676 | - | 2725 | 1811 | 385 | 2217 | 610 |
| ENST00000313285 | TRIP10 | chr19 | 6750644 | + | ENST00000331555 | C1orf186 | chr1 | 206241676 | - | 2485 | 1571 | 82 | 1977 | 631 |
| ENST00000313244 | TRIP10 | chr19 | 6750644 | + | ENST00000331555 | C1orf186 | chr1 | 206241676 | - | 2606 | 1692 | 35 | 2098 | 687 |
| ENST00000596758 | TRIP10 | chr19 | 6750644 | + | ENST00000331555 | C1orf186 | chr1 | 206241676 | - | 2403 | 1489 | 0 | 1895 | 631 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000600428 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - | 0.003496806 | 0.99650323 |
| ENST00000313285 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - | 0.005826753 | 0.9941732 |
| ENST00000313244 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - | 0.009077699 | 0.9909223 |
| ENST00000596758 | ENST00000331555 | TRIP10 | chr19 | 6750644 | + | C1orf186 | chr1 | 206241676 | - | 0.005531153 | 0.9944688 |
Top |
Fusion Genomic Features for TRIP10-C1orf186 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
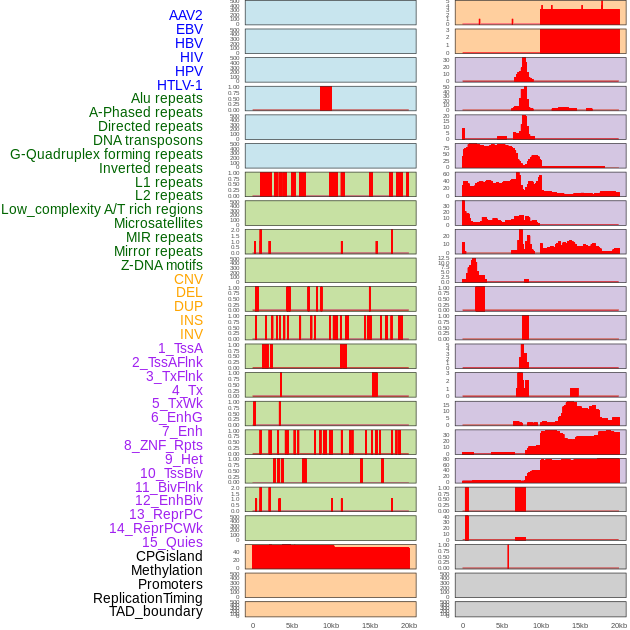 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TRIP10-C1orf186 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:6750644/chr1:206241676) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 388_481 | 552 | 602.0 | Coiled coil | . |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 67_259 | 552 | 602.0 | Coiled coil | . |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 388_481 | 496 | 546.0 | Coiled coil | . |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 67_259 | 496 | 546.0 | Coiled coil | . |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 1_264 | 552 | 602.0 | Domain | F-BAR |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 393_470 | 552 | 602.0 | Domain | REM-1 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 1_264 | 496 | 546.0 | Domain | F-BAR |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 393_470 | 496 | 546.0 | Domain | REM-1 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 1_117 | 552 | 602.0 | Region | Required for translocation to the plasma membrane in response to insulin |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 1_117 | 496 | 546.0 | Region | Required for translocation to the plasma membrane in response to insulin |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 540_601 | 552 | 602.0 | Domain | SH3 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 540_601 | 496 | 546.0 | Domain | SH3 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 538_601 | 552 | 602.0 | Region | Note=Required for podosome formation |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 538_601 | 496 | 546.0 | Region | Note=Required for podosome formation |
| Tgene | C1orf186 | chr19:6750644 | chr1:206241676 | ENST00000331555 | 2 | 6 | 9_29 | 37 | 173.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for TRIP10-C1orf186 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >94227_94227_1_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000313244_C1orf186_chr1_206241676_ENST00000331555_length(transcript)=2606nt_BP=1692nt GCGGTGGTGGCTGCGGCGGCGGCGGCGGGAGCAGCATGGATTGGGGCACTGAGCTGTGGGATCAGTTCGAGGTGCTCGAGCGCCACACGC AGTGGGGGCTGGACCTGTTGGACAGATATGTAAAGTTCGTGAAAGAACGCACCGAAGTGGAACAGGCTTACGCCAAACAACTGCGGAGCC TGGTGAAAAAATATCTGCCCAAGAGACCTGCCAAGGATGATCCTGAGTCCAAATTCAGCCAGCAACAGTCCTTCGTACAGATTCTCCAGG AGGTGAATGACTTTGCAGGCCAGCGGGAGCTGGTGGCTGAGAACCTCAGTGTCCGTGTATGTCTTGAGCTGACCAAGTACTCACAAGAGA TGAAACAGGAGAGGAAGATGCACTTCCAAGAAGGGCGGCGGGCCCAGCAGCAGCTGGAAAATGGCTTTAAACAGCTGGAGAATAGTAAGC GTAAATTTGAGCGGGACTGCCGGGAGGCAGAGAAGGCAGCCCAGACTGCTGAACGGCTAGACCAGGATATCAACGCCACCAAGGCTGATG TGGAGAAGGCCAAGCAGCAAGCCCACCTTCGGAGTCACATGGCCGAAGAAAGCAAAAACGAATATGCGGCTCAACTGCAGCGCTTCAACC GAGACCAAGCCCACTTCTATTTTTCACAGATGCCCCAGATATTCGATAAGCTCCAAGACATGGATGAACGCAGGGCCACCCGCCTGGGTG CCGGGTATGGGCTCCTGTCGGAGGCCGAGCTGGAGGTGGTGCCCATAATAGCCAAGTGCTTGGAGGGCATGAAGGTGGCTGCAAATGCTG TGGATCCCAAGAACGACTCCCACGTCCTTATAGAGCTGCACAAGTCAGGTTTTGCCCGCCCGGGCGACGTGGAATTCGAGGACTTCAGCC AGCCCATGAACCGTGCACCCTCCGACAGCAGTCTGGGCACCCCCTCGGATGGACGGCCTGAACTCCGAGGCCCGGGTCGCAGCCGCACCA AGCGCTGGCCTTTTGGCAAGAAGAACAAGCCTCGCCCCCCACCCCTCTCCCCCCTGGGGGGCCCCGTACCCTCGGCATTGCCTAACGGAC CCCCGTCCCCCCGCTCCGGCCGTGACCCCTTGGCCATACTGAGCGAGATCAGTAAGTCGGTCAAACCGAGGCTAGCATCCTTCCGCAGCC TTCGAGGCAGCCGTGGGACAGTGGTGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGCAGTTGGAAG AACGCAGTCGTGAACTTCAGAAGGAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTCAGATGGGGG ACCCCGCCAGCTTGGAGCCCCAGATCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGGCGTGGCTGG CAGAAGCTGAAAGTCGAGTCCTTAGCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAG ACAGCAGCAGCAACAGCGCATCACAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCATTTACACGG AGTTTGATGAGGATTTCGAGGAGGAACCCACATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGCCCACAAGAGTGAACAGA TACTGAAAGCGGCCAGTCTCCAGGTTCCCAGGCCCAGCCCTGGCCACCATCATCCACCTGCTGTCAAAGAGATGAAGGAGACTCAGACAG AGAGAGACATCCCAATGTCTGATTCCCTTTACAGGCATGACAGCGACACACCCTCAGATAGCTTGGATAGCTCCTGCAGTTCGCCTCCTG CCTGCCAGGCCACAGAGGATGTGGATTACACACAAGTCGTCTTTTCTGACCCTGGAGAACTAAAAAATGACTCCCCGCTGGACTATGAGA ACATAAAGGAAATCACAGATTATGTCAATGTCAATCCAGAAAGACACAAGCCCAGTTTCTGGTATTTTGTCAACCCTGCTCTGTCTGAGC CAGCGGAATATGATCAAGTGGCCATGTGAATTCCAAATATTTTTAATGGGGTCCAGTTCTCTATGGATTCTTACATTTAATTTGTAGGGA AATGCCATTTTTCCCCCTTAAACAAGGCATGGGGCTCACAAGTCTATGGAGACAGGCCAAAAAGAATGTGGAGAAGAAAACTGATAAATA CACAGAGGTCCTCAAGACCCATGGACTCCTGGTCTGTACCCAAAAAAGCTGTTCGTTCCTCAAAAACAAAAACAAGGCTTGGCTGGGAAA ACAGGCCAATGCCCCGGCAAGAAAGGTTGAGATCAGATGTTAGGAAGAACTTTCAGGTAAAGTATGAGAACTATGGAGTCCATCAGCAGA GATAGTAGTGAAGTCTCTCCCCAGGGAAAATTTTAAAAAGGTTGAATCAGCTGTTGTAGAGTTCTATTTGGCAATCTCATGGTTAAATGA >94227_94227_1_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000313244_C1orf186_chr1_206241676_ENST00000331555_length(amino acids)=687AA_BP=552 MDWGTELWDQFEVLERHTQWGLDLLDRYVKFVKERTEVEQAYAKQLRSLVKKYLPKRPAKDDPESKFSQQQSFVQILQEVNDFAGQRELV AENLSVRVCLELTKYSQEMKQERKMHFQEGRRAQQQLENGFKQLENSKRKFERDCREAEKAAQTAERLDQDINATKADVEKAKQQAHLRS HMAEESKNEYAAQLQRFNRDQAHFYFSQMPQIFDKLQDMDERRATRLGAGYGLLSEAELEVVPIIAKCLEGMKVAANAVDPKNDSHVLIE LHKSGFARPGDVEFEDFSQPMNRAPSDSSLGTPSDGRPELRGPGRSRTKRWPFGKKNKPRPPPLSPLGGPVPSALPNGPPSPRSGRDPLA ILSEISKSVKPRLASFRSLRGSRGTVVTEDFSHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQMGDPASLEPQIAET LSNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTS PIGHCVAIYHFEAHKSEQILKAASLQVPRPSPGHHHPPAVKEMKETQTERDIPMSDSLYRHDSDTPSDSLDSSCSSPPACQATEDVDYTQ -------------------------------------------------------------- >94227_94227_2_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000313285_C1orf186_chr1_206241676_ENST00000331555_length(transcript)=2485nt_BP=1571nt GAGTCTCCCCGGGGAGGGCGGCGGGCGGCGGGCGGCGGGGACCGGGTGCGGTGGTGGCTGCGGCGGCGGCGGCGGGAGCAGCATGGATTG GGGCACTGAGCTGTGGGATCAGTTCGAGGTGCTCGAGCGCCACACGCAGTGGGGGCTGGACCTGTTGGACAGATATGTAAAGTTCGTGAA AGAACGCACCGAAGTGGAACAGGCTTACGCCAAACAACTGCGGAGCCTGGTGAAAAAATATCTGCCCAAGAGACCTGCCAAGGATGATCC TGAGTCCAAATTCAGCCAGCAACAGTCCTTCGTACAGATTCTCCAGGAGGTGAATGACTTTGCAGGCCAGCGGGAGCTGGTGGCTGAGAA CCTCAGTGTCCGTGTATGTCTTGAGCTGACCAAGTACTCACAAGAGATGAAACAGGAGAGGAAGATGCACTTCCAAGAAGGGCGGCGGGC CCAGCAGCAGCTGGAAAATGGCTTTAAACAGCTGGAGAATAGTAAGCGTAAATTTGAGCGGGACTGCCGGGAGGCAGAGAAGGCAGCCCA GACTGCTGAACGGCTAGACCAGGATATCAACGCCACCAAGGCTGATGTGGAGAAGGCCAAGCAGCAAGCCCACCTTCGGAGTCACATGGC CGAAGAAAGCAAAAACGAATATGCGGCTCAACTGCAGCGCTTCAACCGAGACCAAGCCCACTTCTATTTTTCACAGATGCCCCAGATATT CGATAAGCTCCAAGACATGGATGAACGCAGGGCCACCCGCCTGGGTGCCGGGTATGGGCTCCTGTCGGAGGCCGAGCTGGAGGTGGTGCC CATAATAGCCAAGTGCTTGGAGGGCATGAAGGTGGCTGCAAATGCTGTGGATCCCAAGAACGACTCCCACGTCCTTATAGAGCTGCACAA GTCAGGTTTTGCCCGCCCGGGCGACGTGGAATTCGAGGACTTCAGCCAGCCCATGAACCGTGCACCCTCCGACAGCAGTCTGGGCACCCC CTCGGATGGACGGCCTGAACTCCGAGGCCCGGGTCGCAGCCGCACCAAGCGCTGGCCTTTTGGCAAGAAGAACAAGACAGTGGTGACCGA GGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAGGAGGTTGA CCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAGATCGCTGA AACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTAGCAACCG GGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCACAGGACAC CAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAGGAACCCAC ATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGCCCACAAGAGTGAACAGATACTGAAAGCGGCCAGTCTCCAGGTTCCCAG GCCCAGCCCTGGCCACCATCATCCACCTGCTGTCAAAGAGATGAAGGAGACTCAGACAGAGAGAGACATCCCAATGTCTGATTCCCTTTA CAGGCATGACAGCGACACACCCTCAGATAGCTTGGATAGCTCCTGCAGTTCGCCTCCTGCCTGCCAGGCCACAGAGGATGTGGATTACAC ACAAGTCGTCTTTTCTGACCCTGGAGAACTAAAAAATGACTCCCCGCTGGACTATGAGAACATAAAGGAAATCACAGATTATGTCAATGT CAATCCAGAAAGACACAAGCCCAGTTTCTGGTATTTTGTCAACCCTGCTCTGTCTGAGCCAGCGGAATATGATCAAGTGGCCATGTGAAT TCCAAATATTTTTAATGGGGTCCAGTTCTCTATGGATTCTTACATTTAATTTGTAGGGAAATGCCATTTTTCCCCCTTAAACAAGGCATG GGGCTCACAAGTCTATGGAGACAGGCCAAAAAGAATGTGGAGAAGAAAACTGATAAATACACAGAGGTCCTCAAGACCCATGGACTCCTG GTCTGTACCCAAAAAAGCTGTTCGTTCCTCAAAAACAAAAACAAGGCTTGGCTGGGAAAACAGGCCAATGCCCCGGCAAGAAAGGTTGAG ATCAGATGTTAGGAAGAACTTTCAGGTAAAGTATGAGAACTATGGAGTCCATCAGCAGAGATAGTAGTGAAGTCTCTCCCCAGGGAAAAT TTTAAAAAGGTTGAATCAGCTGTTGTAGAGTTCTATTTGGCAATCTCATGGTTAAATGACTTCCCTTTGAGCTCTTTAATTATTGGCAAT >94227_94227_2_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000313285_C1orf186_chr1_206241676_ENST00000331555_length(amino acids)=631AA_BP=496 MDWGTELWDQFEVLERHTQWGLDLLDRYVKFVKERTEVEQAYAKQLRSLVKKYLPKRPAKDDPESKFSQQQSFVQILQEVNDFAGQRELV AENLSVRVCLELTKYSQEMKQERKMHFQEGRRAQQQLENGFKQLENSKRKFERDCREAEKAAQTAERLDQDINATKADVEKAKQQAHLRS HMAEESKNEYAAQLQRFNRDQAHFYFSQMPQIFDKLQDMDERRATRLGAGYGLLSEAELEVVPIIAKCLEGMKVAANAVDPKNDSHVLIE LHKSGFARPGDVEFEDFSQPMNRAPSDSSLGTPSDGRPELRGPGRSRTKRWPFGKKNKTVVTEDFSHLPPEQQRKRLQQQLEERSRELQK EVDQREALKKMKDVYEKTPQMGDPASLEPQIAETLSNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSAS QDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSPIGHCVAIYHFEAHKSEQILKAASLQVPRPSPGHHHPPAVKEMKETQTERDIPMSD SLYRHDSDTPSDSLDSSCSSPPACQATEDVDYTQVVFSDPGELKNDSPLDYENIKEITDYVNVNPERHKPSFWYFVNPALSEPAEYDQVA -------------------------------------------------------------- >94227_94227_3_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000596758_C1orf186_chr1_206241676_ENST00000331555_length(transcript)=2403nt_BP=1489nt ATGGATTGGGGCACTGAGCTGTGGGATCAGTTCGAGGTGCTCGAGCGCCACACGCAGTGGGGGCTGGACCTGTTGGACAGATATGTAAAG TTCGTGAAAGAACGCACCGAAGTGGAACAGGCTTACGCCAAACAACTGCGGAGCCTGGTGAAAAAATATCTGCCCAAGAGACCTGCCAAG GATGATCCTGAGTCCAAATTCAGCCAGCAACAGTCCTTCGTACAGATTCTCCAGGAGGTGAATGACTTTGCAGGCCAGCGGGAGCTGGTG GCTGAGAACCTCAGTGTCCGTGTATGTCTTGAGCTGACCAAGTACTCACAAGAGATGAAACAGGAGAGGAAGATGCACTTCCAAGAAGGG CGGCGGGCCCAGCAGCAGCTGGAAAATGGCTTTAAACAGCTGGAGAATAGTAAGCGTAAATTTGAGCGGGACTGCCGGGAGGCAGAGAAG GCAGCCCAGACTGCTGAACGGCTAGACCAGGATATCAACGCCACCAAGGCTGATGTGGAGAAGGCCAAGCAGCAAGCCCACCTTCGGAGT CACATGGCCGAAGAAAGCAAAAACGAATATGCGGCTCAACTGCAGCGCTTCAACCGAGACCAAGCCCACTTCTATTTTTCACAGATGCCC CAGATATTCGATAAGCTCCAAGACATGGATGAACGCAGGGCCACCCGCCTGGGTGCCGGGTATGGGCTCCTGTCGGAGGCCGAGCTGGAG GTGGTGCCCATAATAGCCAAGTGCTTGGAGGGCATGAAGGTGGCTGCAAATGCTGTGGATCCCAAGAACGACTCCCACGTCCTTATAGAG CTGCACAAGTCAGGTTTTGCCCGCCCGGGCGACGTGGAATTCGAGGACTTCAGCCAGCCCATGAACCGTGCACCCTCCGACAGCAGTCTG GGCACCCCCTCGGATGGACGGCCTGAACTCCGAGGCCCGGGTCGCAGCCGCACCAAGCGCTGGCCTTTTGGCAAGAAGAACAAGACAGTG GTGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCAGCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAG GAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGATGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAG ATCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATTGGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTT AGCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCCTCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCA CAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGAAGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAG GAACCCACATCCCCCATAGGTCACTGTGTGGCCATCTACCACTTTGAAGCCCACAAGAGTGAACAGATACTGAAAGCGGCCAGTCTCCAG GTTCCCAGGCCCAGCCCTGGCCACCATCATCCACCTGCTGTCAAAGAGATGAAGGAGACTCAGACAGAGAGAGACATCCCAATGTCTGAT TCCCTTTACAGGCATGACAGCGACACACCCTCAGATAGCTTGGATAGCTCCTGCAGTTCGCCTCCTGCCTGCCAGGCCACAGAGGATGTG GATTACACACAAGTCGTCTTTTCTGACCCTGGAGAACTAAAAAATGACTCCCCGCTGGACTATGAGAACATAAAGGAAATCACAGATTAT GTCAATGTCAATCCAGAAAGACACAAGCCCAGTTTCTGGTATTTTGTCAACCCTGCTCTGTCTGAGCCAGCGGAATATGATCAAGTGGCC ATGTGAATTCCAAATATTTTTAATGGGGTCCAGTTCTCTATGGATTCTTACATTTAATTTGTAGGGAAATGCCATTTTTCCCCCTTAAAC AAGGCATGGGGCTCACAAGTCTATGGAGACAGGCCAAAAAGAATGTGGAGAAGAAAACTGATAAATACACAGAGGTCCTCAAGACCCATG GACTCCTGGTCTGTACCCAAAAAAGCTGTTCGTTCCTCAAAAACAAAAACAAGGCTTGGCTGGGAAAACAGGCCAATGCCCCGGCAAGAA AGGTTGAGATCAGATGTTAGGAAGAACTTTCAGGTAAAGTATGAGAACTATGGAGTCCATCAGCAGAGATAGTAGTGAAGTCTCTCCCCA GGGAAAATTTTAAAAAGGTTGAATCAGCTGTTGTAGAGTTCTATTTGGCAATCTCATGGTTAAATGACTTCCCTTTGAGCTCTTTAATTA >94227_94227_3_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000596758_C1orf186_chr1_206241676_ENST00000331555_length(amino acids)=631AA_BP=496 MDWGTELWDQFEVLERHTQWGLDLLDRYVKFVKERTEVEQAYAKQLRSLVKKYLPKRPAKDDPESKFSQQQSFVQILQEVNDFAGQRELV AENLSVRVCLELTKYSQEMKQERKMHFQEGRRAQQQLENGFKQLENSKRKFERDCREAEKAAQTAERLDQDINATKADVEKAKQQAHLRS HMAEESKNEYAAQLQRFNRDQAHFYFSQMPQIFDKLQDMDERRATRLGAGYGLLSEAELEVVPIIAKCLEGMKVAANAVDPKNDSHVLIE LHKSGFARPGDVEFEDFSQPMNRAPSDSSLGTPSDGRPELRGPGRSRTKRWPFGKKNKTVVTEDFSHLPPEQQRKRLQQQLEERSRELQK EVDQREALKKMKDVYEKTPQMGDPASLEPQIAETLSNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSAS QDTKESSEEPPSEESQDTPIYTEFDEDFEEEPTSPIGHCVAIYHFEAHKSEQILKAASLQVPRPSPGHHHPPAVKEMKETQTERDIPMSD SLYRHDSDTPSDSLDSSCSSPPACQATEDVDYTQVVFSDPGELKNDSPLDYENIKEITDYVNVNPERHKPSFWYFVNPALSEPAEYDQVA -------------------------------------------------------------- >94227_94227_4_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000600428_C1orf186_chr1_206241676_ENST00000331555_length(transcript)=2725nt_BP=1811nt AATGCAGCGGGAGCAGACAAGGGCCACAAAGCATTAAATAAACTACCTTTCCCAGTATCCCCAAGGACGGACTTCCTCTTCTTGAGCCAG GGATGTGGCTCCCAGCCAGAGCTGAGGCTTTTGGTAAACACACGCATCTGTTCAGGCCTGACCTGTTTTCCTAATCTTCCTGGTCCACAG AGAGGCCTAAAGCTGGCCTCAGGTTAAAGTTTGACCCTAGGATGACCTATGACCCCAGGCTGAGACCCCAGCCTTAGGCTAAGCCTTTTA TTCCAGAGTGAGGCAATTGGAGCCCCACCCCCAGATTCCAGGCTAGACCGGACTCCGGGCTGAACTCTGACCCCAGGATCAGTTCGAGGT GCTCGAGCGCCACACGCAGTGGGGGCTGGACCTGTTGGACAGATATGTAAAGTTCGTGAAAGAACGCACCGAAGTGGAACAGGCTTACGC CAAACAACTGCGGAGCCTGGTGAAAAAATATCTGCCCAAGAGACCTGCCAAGGATGATCCTGAGTCCAAATTCAGCCAGCAACAGTCCTT CGTACAGATTCTCCAGGAGGTGAATGACTTTGCAGGCCAGCGGGAGCTGGTGGCTGAGAACCTCAGTGTCCGTGTATGTCTTGAGCTGAC CAAGTACTCACAAGAGATGAAACAGGAGAGGAAGATGCACTTCCAAGAAGGGCGGCGGGCCCAGCAGCAGCTGGAAAATGGCTTTAAACA GCTGGAGAATAGTAAGCGTAAATTTGAGCGGGACTGCCGGGAGGCAGAGAAGGCAGCCCAGACTGCTGAACGGCTAGACCAGGATATCAA CGCCACCAAGGCTGATGTGGAGAAGGCCAAGCAGCAAGCCCACCTTCGGAGTCACATGGCCGAAGAAAGCAAAAACGAATATGCGGCTCA ACTGCAGCGCTTCAACCGAGACCAAGCCCACTTCTATTTTTCACAGATGCCCCAGATATTCGATAAGCTCCAAGACATGGATGAACGCAG GGCCACCCGCCTGGGTGCCGGGTATGGGCTCCTGTCGGAGGCCGAGCTGGAGGTGGTGCCCATAATAGCCAAGTGCTTGGAGGGCATGAA GGTGGCTGCAAATGCTGTGGATCCCAAGAACGACTCCCACGTCCTTATAGAGCTGCACAAGTCAGGTTTTGCCCGCCCGGGCGACGTGGA ATTCGAGGACTTCAGCCAGCCCATGAACCGTGCACCCTCCGACAGCAGTCTGGGCACCCCCTCGGATGGACGGCCTGAACTCCGAGGCCC GGGTCGCAGCCGCACCAAGCGCTGGCCTTTTGGCAAGAAGAACAAGACAGTGGTGACCGAGGATTTTAGCCACTTGCCCCCAGAGCAGCA GCGAAAACGGCTTCAACAGCAGTTGGAAGAACGCAGTCGTGAACTTCAGAAGGAGGTTGACCAGAGGGAAGCCCTAAAGAAAATGAAGGA TGTCTATGAGAAGACACCTCAGATGGGGGACCCCGCCAGCTTGGAGCCCCAGATCGCTGAAACCCTGAGCAACATTGAACGGCTGAAATT GGAAGTGCAGAAGTATGAGGCGTGGCTGGCAGAAGCTGAAAGTCGAGTCCTTAGCAACCGGGGAGACAGCCTGAGCCGGCACGCCCGGCC TCCCGACCCCCCCGCTAGCGCCCCGCCAGACAGCAGCAGCAACAGCGCATCACAGGACACCAAGGAGAGCTCTGAAGAGCCTCCCTCAGA AGAGAGCCAGGACACCCCCATTTACACGGAGTTTGATGAGGATTTCGAGGAGGAACCCACATCCCCCATAGGTCACTGTGTGGCCATCTA CCACTTTGAAGCCCACAAGAGTGAACAGATACTGAAAGCGGCCAGTCTCCAGGTTCCCAGGCCCAGCCCTGGCCACCATCATCCACCTGC TGTCAAAGAGATGAAGGAGACTCAGACAGAGAGAGACATCCCAATGTCTGATTCCCTTTACAGGCATGACAGCGACACACCCTCAGATAG CTTGGATAGCTCCTGCAGTTCGCCTCCTGCCTGCCAGGCCACAGAGGATGTGGATTACACACAAGTCGTCTTTTCTGACCCTGGAGAACT AAAAAATGACTCCCCGCTGGACTATGAGAACATAAAGGAAATCACAGATTATGTCAATGTCAATCCAGAAAGACACAAGCCCAGTTTCTG GTATTTTGTCAACCCTGCTCTGTCTGAGCCAGCGGAATATGATCAAGTGGCCATGTGAATTCCAAATATTTTTAATGGGGTCCAGTTCTC TATGGATTCTTACATTTAATTTGTAGGGAAATGCCATTTTTCCCCCTTAAACAAGGCATGGGGCTCACAAGTCTATGGAGACAGGCCAAA AAGAATGTGGAGAAGAAAACTGATAAATACACAGAGGTCCTCAAGACCCATGGACTCCTGGTCTGTACCCAAAAAAGCTGTTCGTTCCTC AAAAACAAAAACAAGGCTTGGCTGGGAAAACAGGCCAATGCCCCGGCAAGAAAGGTTGAGATCAGATGTTAGGAAGAACTTTCAGGTAAA GTATGAGAACTATGGAGTCCATCAGCAGAGATAGTAGTGAAGTCTCTCCCCAGGGAAAATTTTAAAAAGGTTGAATCAGCTGTTGTAGAG TTCTATTTGGCAATCTCATGGTTAAATGACTTCCCTTTGAGCTCTTTAATTATTGGCAATAAACAACTTCTTTAAAAGTTTTAAATAAAA >94227_94227_4_TRIP10-C1orf186_TRIP10_chr19_6750644_ENST00000600428_C1orf186_chr1_206241676_ENST00000331555_length(amino acids)=610AA_BP=475 MDLLDRYVKFVKERTEVEQAYAKQLRSLVKKYLPKRPAKDDPESKFSQQQSFVQILQEVNDFAGQRELVAENLSVRVCLELTKYSQEMKQ ERKMHFQEGRRAQQQLENGFKQLENSKRKFERDCREAEKAAQTAERLDQDINATKADVEKAKQQAHLRSHMAEESKNEYAAQLQRFNRDQ AHFYFSQMPQIFDKLQDMDERRATRLGAGYGLLSEAELEVVPIIAKCLEGMKVAANAVDPKNDSHVLIELHKSGFARPGDVEFEDFSQPM NRAPSDSSLGTPSDGRPELRGPGRSRTKRWPFGKKNKTVVTEDFSHLPPEQQRKRLQQQLEERSRELQKEVDQREALKKMKDVYEKTPQM GDPASLEPQIAETLSNIERLKLEVQKYEAWLAEAESRVLSNRGDSLSRHARPPDPPASAPPDSSSNSASQDTKESSEEPPSEESQDTPIY TEFDEDFEEEPTSPIGHCVAIYHFEAHKSEQILKAASLQVPRPSPGHHHPPAVKEMKETQTERDIPMSDSLYRHDSDTPSDSLDSSCSSP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TRIP10-C1orf186 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 293_537 | 552.3333333333334 | 602.0 | CDC42 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 487_541 | 552.3333333333334 | 602.0 | DNM2 and WASL |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 546_601 | 552.3333333333334 | 602.0 | ARHGAP17%2C DAAM1%2C DIAPH1 and DIAPH2 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 546_601 | 496.3333333333333 | 546.0 | ARHGAP17%2C DAAM1%2C DIAPH1 and DIAPH2 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 293_537 | 496.3333333333333 | 546.0 | CDC42 |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 529_601 | 552.3333333333334 | 602.0 | DNM1 and WASL |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 529_601 | 496.3333333333333 | 546.0 | DNM1 and WASL |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 487_541 | 496.3333333333333 | 546.0 | DNM2 and WASL |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 293_601 | 552.3333333333334 | 602.0 | PDE6G |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 293_601 | 496.3333333333333 | 546.0 | PDE6G |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313244 | + | 14 | 15 | 544_601 | 552.3333333333334 | 602.0 | WAS |
| Hgene | TRIP10 | chr19:6750644 | chr1:206241676 | ENST00000313285 | + | 13 | 14 | 544_601 | 496.3333333333333 | 546.0 | WAS |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TRIP10-C1orf186 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TRIP10-C1orf186 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
