
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TSC1-PDGFRB (FusionGDB2 ID:94475) |
Fusion Gene Summary for TSC1-PDGFRB |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TSC1-PDGFRB | Fusion gene ID: 94475 | Hgene | Tgene | Gene symbol | TSC1 | PDGFRB | Gene ID | 7248 | 5159 |
| Gene name | TSC complex subunit 1 | platelet derived growth factor receptor beta | |
| Synonyms | LAM|TSC | CD140B|IBGC4|IMF1|JTK12|KOGS|PDGFR|PDGFR-1|PDGFR1|PENTT | |
| Cytomap | 9q34.13 | 5q32 | |
| Type of gene | protein-coding | protein-coding | |
| Description | hamartintruncated hemartintuberous sclerosis 1 protein | platelet-derived growth factor receptor betaActivated tyrosine kinase PDGFRBCD140 antigen-like family member BNDEL1-PDGFRBPDGF-R-betaPDGFR-betabeta-type platelet-derived growth factor receptorplatelet-derived growth factor receptor 1platelet-deriv | |
| Modification date | 20200313 | 20200329 | |
| UniProtAcc | . | P09619 | |
| Ensembl transtripts involved in fusion gene | ENST00000298552, ENST00000440111, ENST00000545250, ENST00000403810, ENST00000475903, | ENST00000523456, ENST00000261799, | |
| Fusion gene scores | * DoF score | 5 X 7 X 6=210 | 28 X 26 X 6=4368 |
| # samples | 8 | 15 | |
| ** MAII score | log2(8/210*10)=-1.39231742277876 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/4368*10)=-4.86393845042397 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TSC1 [Title/Abstract] AND PDGFRB [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TSC1(135776976)-PDGFRB(149505142), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TSC1-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a CGC due to the frame-shifted ORF. TSC1-PDGFRB seems lost the major protein functional domain in Hgene partner, which is a tumor suppressor due to the frame-shifted ORF. TSC1-PDGFRB seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. TSC1-PDGFRB seems lost the major protein functional domain in Tgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. TSC1-PDGFRB seems lost the major protein functional domain in Tgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | TSC1 | GO:0006417 | regulation of translation | 17308101 |
| Hgene | TSC1 | GO:0032780 | negative regulation of ATPase activity | 29127155 |
| Hgene | TSC1 | GO:0032868 | response to insulin | 16996505 |
| Hgene | TSC1 | GO:0050821 | protein stabilization | 11175345 |
| Hgene | TSC1 | GO:0051492 | regulation of stress fiber assembly | 10806479 |
| Hgene | TSC1 | GO:0051894 | positive regulation of focal adhesion assembly | 10806479 |
| Hgene | TSC1 | GO:0090630 | activation of GTPase activity | 10806479 |
| Tgene | PDGFRB | GO:0007165 | signal transduction | 10821867 |
| Tgene | PDGFRB | GO:0010863 | positive regulation of phospholipase C activity | 1653029 |
| Tgene | PDGFRB | GO:0018108 | peptidyl-tyrosine phosphorylation | 1653029|2536956|2850496 |
| Tgene | PDGFRB | GO:0030335 | positive regulation of cell migration | 17470632 |
| Tgene | PDGFRB | GO:0032516 | positive regulation of phosphoprotein phosphatase activity | 7691811 |
| Tgene | PDGFRB | GO:0038091 | positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway | 17470632 |
| Tgene | PDGFRB | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity | 1314164 |
| Tgene | PDGFRB | GO:0046777 | protein autophosphorylation | 1314164|2536956|2850496 |
| Tgene | PDGFRB | GO:0048008 | platelet-derived growth factor receptor signaling pathway | 1314164|2536956 |
| Tgene | PDGFRB | GO:0060326 | cell chemotaxis | 2554309|17991872 |
 Fusion gene breakpoints across TSC1 (5'-gene) Fusion gene breakpoints across TSC1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
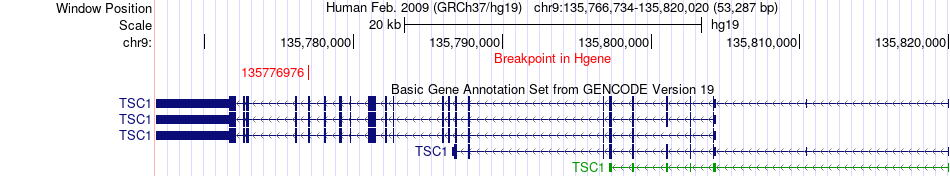 |
 Fusion gene breakpoints across PDGFRB (3'-gene) Fusion gene breakpoints across PDGFRB (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
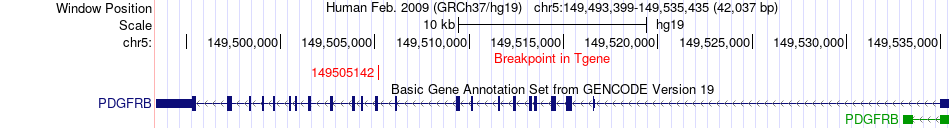 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChiTaRS5.0 | N/A | KM516092 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
Top |
Fusion Gene ORF analysis for TSC1-PDGFRB |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000298552 | ENST00000523456 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| 5CDS-intron | ENST00000440111 | ENST00000523456 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| 5CDS-intron | ENST00000545250 | ENST00000523456 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| Frame-shift | ENST00000298552 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| In-frame | ENST00000440111 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| In-frame | ENST00000545250 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| intron-3CDS | ENST00000403810 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| intron-3CDS | ENST00000475903 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| intron-intron | ENST00000403810 | ENST00000523456 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
| intron-intron | ENST00000475903 | ENST00000523456 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000545250 | TSC1 | chr9 | 135776976 | - | ENST00000261799 | PDGFRB | chr5 | 149505142 | - | 6005 | 2432 | 83 | 4078 | 1331 |
| ENST00000440111 | TSC1 | chr9 | 135776976 | - | ENST00000261799 | PDGFRB | chr5 | 149505142 | - | 6158 | 2585 | 83 | 4231 | 1382 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000545250 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - | 0.001822732 | 0.9981773 |
| ENST00000440111 | ENST00000261799 | TSC1 | chr9 | 135776976 | - | PDGFRB | chr5 | 149505142 | - | 0.001771916 | 0.99822813 |
Top |
Fusion Genomic Features for TSC1-PDGFRB |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
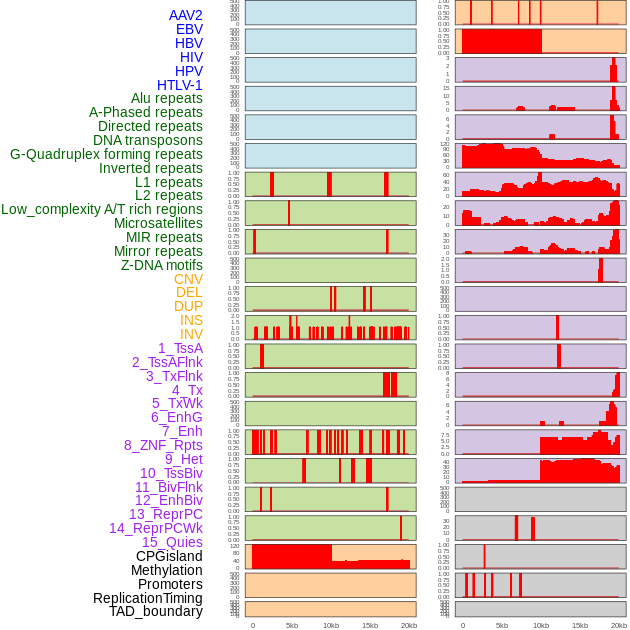 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TSC1-PDGFRB |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr9:135776976/chr5:149505142) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | PDGFRB |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Tyrosine-protein kinase that acts as cell-surface receptor for homodimeric PDGFB and PDGFD and for heterodimers formed by PDGFA and PDGFB, and plays an essential role in the regulation of embryonic development, cell proliferation, survival, differentiation, chemotaxis and migration. Plays an essential role in blood vessel development by promoting proliferation, migration and recruitment of pericytes and smooth muscle cells to endothelial cells. Plays a role in the migration of vascular smooth muscle cells and the formation of neointima at vascular injury sites. Required for normal development of the cardiovascular system. Required for normal recruitment of pericytes (mesangial cells) in the kidney glomerulus, and for normal formation of a branched network of capillaries in kidney glomeruli. Promotes rearrangement of the actin cytoskeleton and the formation of membrane ruffles. Binding of its cognate ligands - homodimeric PDGFB, heterodimers formed by PDGFA and PDGFB or homodimeric PDGFD -leads to the activation of several signaling cascades; the response depends on the nature of the bound ligand and is modulated by the formation of heterodimers between PDGFRA and PDGFRB. Phosphorylates PLCG1, PIK3R1, PTPN11, RASA1/GAP, CBL, SHC1 and NCK1. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate, mobilization of cytosolic Ca(2+) and the activation of protein kinase C. Phosphorylation of PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, leads to the activation of the AKT1 signaling pathway. Phosphorylation of SHC1, or of the C-terminus of PTPN11, creates a binding site for GRB2, resulting in the activation of HRAS, RAF1 and down-stream MAP kinases, including MAPK1/ERK2 and/or MAPK3/ERK1. Promotes phosphorylation and activation of SRC family kinases. Promotes phosphorylation of PDCD6IP/ALIX and STAM. Receptor signaling is down-regulated by protein phosphatases that dephosphorylate the receptor and its down-stream effectors, and by rapid internalization of the activated receptor. {ECO:0000269|PubMed:11297552, ECO:0000269|PubMed:11331881, ECO:0000269|PubMed:1314164, ECO:0000269|PubMed:1396585, ECO:0000269|PubMed:1653029, ECO:0000269|PubMed:1709159, ECO:0000269|PubMed:1846866, ECO:0000269|PubMed:20494825, ECO:0000269|PubMed:20529858, ECO:0000269|PubMed:21098708, ECO:0000269|PubMed:21679854, ECO:0000269|PubMed:21733313, ECO:0000269|PubMed:2554309, ECO:0000269|PubMed:26599395, ECO:0000269|PubMed:2835772, ECO:0000269|PubMed:2850496, ECO:0000269|PubMed:7685273, ECO:0000269|PubMed:7691811, ECO:0000269|PubMed:7692233, ECO:0000269|PubMed:8195171}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 600_962 | 558 | 1107.0 | Domain | Protein kinase | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 606_614 | 558 | 1107.0 | Nucleotide binding | ATP |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000298552 | - | 19 | 23 | 721_997 | 834 | 1165.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000440111 | - | 17 | 21 | 721_997 | 834 | 1165.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000545250 | - | 16 | 20 | 721_997 | 783 | 1114.0 | Coiled coil | Ontology_term=ECO:0000255 |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000298552 | - | 19 | 23 | 1034_1037 | 834 | 1165.0 | Compositional bias | Note=Poly-Gly |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000298552 | - | 19 | 23 | 1038_1043 | 834 | 1165.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000440111 | - | 17 | 21 | 1034_1037 | 834 | 1165.0 | Compositional bias | Note=Poly-Gly |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000440111 | - | 17 | 21 | 1038_1043 | 834 | 1165.0 | Compositional bias | Note=Poly-Ser |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000545250 | - | 16 | 20 | 1034_1037 | 783 | 1114.0 | Compositional bias | Note=Poly-Gly |
| Hgene | TSC1 | chr9:135776976 | chr5:149505142 | ENST00000545250 | - | 16 | 20 | 1038_1043 | 783 | 1114.0 | Compositional bias | Note=Poly-Ser |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 129_210 | 558 | 1107.0 | Domain | Note=Ig-like C2-type 2 | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 214_309 | 558 | 1107.0 | Domain | Note=Ig-like C2-type 3 | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 331_403 | 558 | 1107.0 | Domain | Note=Ig-like C2-type 4 | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 33_120 | 558 | 1107.0 | Domain | Note=Ig-like C2-type 1 | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 416_524 | 558 | 1107.0 | Domain | Note=Ig-like C2-type 5 | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 33_532 | 558 | 1107.0 | Topological domain | Extracellular | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 554_1106 | 558 | 1107.0 | Topological domain | Cytoplasmic | |
| Tgene | PDGFRB | chr9:135776976 | chr5:149505142 | ENST00000261799 | 10 | 23 | 533_553 | 558 | 1107.0 | Transmembrane | Helical |
Top |
Fusion Gene Sequence for TSC1-PDGFRB |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >94475_94475_1_TSC1-PDGFRB_TSC1_chr9_135776976_ENST00000440111_PDGFRB_chr5_149505142_ENST00000261799_length(transcript)=6158nt_BP=2585nt CAGTTTGGTAGTGGCCCCAATGAAGAACCTTCAGAACCTGTAGCACACGTCCTGGAGCCAGCACAGCGCCTTCGAGCGAGAGAATGGCCC AACAAGCAAATGTCGGGGAGCTTCTTGCCATGCTGGACTCCCCCATGCTGGGTGTGCGGGACGACGTGACAGCTGTCTTTAAAGAGAACC TCAATTCTGACCGTGGCCCTATGCTTGTAAACACCTTGGTGGATTATTACCTGGAAACCAGCTCTCAGCCGGCATTGCACATCCTGACCA CCTTGCAAGAGCCACATGACAAGCACCTCTTGGACAGGATTAACGAATATGTGGGCAAAGCCGCCACTCGTTTATCCATCCTCTCGTTAC TGGGTCATGTCATAAGACTGCAGCCATCTTGGAAGCATAAGCTCTCTCAAGCACCTCTTTTGCCTTCTTTACTAAAATGTCTCAAGATGG ACACTGACGTCGTTGTCCTCACAACAGGCGTCTTGGTGTTGATAACCATGCTACCAATGATTCCACAGTCTGGGAAACAGCATCTTCTTG ATTTCTTTGACATTTTTGGCCGTCTGTCATCATGGTGCCTGAAGAAACCAGGCCACGTGGCGGAAGTCTATCTCGTCCATCTCCATGCCA GTGTGTACGCACTCTTTCATCGCCTTTATGGAATGTACCCTTGCAACTTCGTCTCCTTTTTGCGTTCTCATTACAGTATGAAAGAAAACC TGGAGACTTTTGAAGAAGTGGTCAAGCCAATGATGGAGCATGTGCGAATTCATCCGGAATTAGTGACTGGATCCAAGGACCATGAACTGG ACCCTCGAAGGTGGAAGAGATTAGAAACTCATGATGTTGTGATCGAGTGTGCCAAAATCTCTCTGGATCCCACAGAAGCCTCATATGAAG ATGGCTATTCTGTGTCTCACCAAATCTCAGCCCGCTTTCCTCATCGTTCAGCCGATGTCACCACCAGCCCTTATGCTGACACACAGAATA GCTATGGGTGTGCTACTTCTACCCCTTACTCCACGTCTCGGCTGATGTTGTTAAATATGCCAGGGCAGCTACCTCAGACTCTGAGTTCCC CATCGACACGGCTGATAACTGAACCACCACAAGCTACTCTTTGGAGCCCATCTATGGTTTGTGGTATGACCACTCCTCCAACTTCTCCTG GAAATGTCCCACCTGATCTGTCACACCCTTACAGTAAAGTCTTTGGTACAACTGCAGGTGGAAAAGGAACTCCTCTGGGAACCCCAGCAA CCTCTCCTCCTCCAGCCCCACTCTGTCATTCGGATGACTACGTGCACATTTCACTCCCCCAGGCCACAGTCACACCCCCCAGGAAGGAAG AGAGAATGGATTCTGCAAGACCATGTCTACACAGACAACACCATCTTCTGAATGACAGAGGATCAGAAGAGCCACCTGGCAGCAAAGGTT CTGTCACTCTAAGTGATCTTCCAGGGTTTTTAGGTGATCTGGCCTCTGAAGAAGATAGTATTGAAAAAGATAAAGAAGAAGCTGCAATAT CTAGAGAACTTTCTGAGATCACCACAGCAGAGGCAGAGCCTGTGGTTCCTCGAGGAGGCTTTGACTCTCCCTTTTACCGAGACAGTCTCC CAGGTTCTCAGCGGAAGACCCACTCGGCAGCCTCCAGTTCTCAGGGCGCCAGCGTGAACCCTGAGCCTTTACACTCCTCCCTGGACAAGC TTGGGCCTGACACACCAAAGCAAGCCTTTACTCCCATAGACCTGCCCTGCGGCAGTGCTGATGAAAGCCCTGCGGGAGACAGGGAATGCC AGACTTCTTTGGAGACCAGTATCTTCACTCCCAGTCCTTGTAAAATTCCACCTCCGACGAGAGTGGGCTTTGGAAGCGGGCAGCCTCCCC CGTATGATCATCTTTTTGAGGTGGCATTGCCAAAGACAGCCCATCATTTTGTCATCAGGAAGACTGAGGAGCTGTTAAAGAAAGCAAAAG GAAACACAGAGGAAGATGGTGTGCCCTCTACCTCCCCAATGGAAGTGCTGGACAGACTGATACAGCAGGGAGCAGACGCGCACAGCAAGG AGCTGAACAAGTTGCCTTTACCCAGCAAGTCTGTCGACTGGACCCACTTTGGAGGCTCTCCTCCTTCAGATGAGATCCGCACCCTCCGAG ACCAGTTGCTTTTACTGCACAACCAGTTACTCTATGAGCGTTTTAAGAGGCAGCAGCATGCCCTCCGGAACAGGCGGCTCCTCCGCAAGG TGATCAAAGCAGCAGCTCTGGAGGAACATAATGCTGCCATGAAAGATCAGTTGAAGTTACAAGAGAAGGACATCCAGATGTGGAAGGTTA GTCTGCAGAAAGAACAAGCTAGATACAATCAGCTCCAGGAGCAGCGTGACACTATGGTAACCAAGCTCCACAGCCAGATCAGACAGCTGC AGCATGACCGAGAGGAATTCTACAACCAGAGCCAGGAATTACAGACGAAGCTGGAGGACTGCAGGAACATGATTGCGGAGCTGCGGATAG AACTGAAGAAGGCCAACAACAAGGTGTGTCACACTGAGCTGCTGCTCAGTCAGGTTTCCCAAAAGAAGCCACGTTACGAGATCCGATGGA AGGTGATTGAGTCTGTGAGCTCTGACGGCCATGAGTACATCTACGTGGACCCCATGCAGCTGCCCTATGACTCCACGTGGGAGCTGCCGC GGGACCAGCTTGTGCTGGGACGCACCCTCGGCTCTGGGGCCTTTGGGCAGGTGGTGGAGGCCACGGCTCATGGCCTGAGCCATTCTCAGG CCACGATGAAAGTGGCCGTCAAGATGCTTAAATCCACAGCCCGCAGCAGTGAGAAGCAAGCCCTTATGTCGGAGCTGAAGATCATGAGTC ACCTTGGGCCCCACCTGAACGTGGTCAACCTGTTGGGGGCCTGCACCAAAGGAGGACCCATCTATATCATCACTGAGTACTGCCGCTACG GAGACCTGGTGGACTACCTGCACCGCAACAAACACACCTTCCTGCAGCACCACTCCGACAAGCGCCGCCCGCCCAGCGCGGAGCTCTACA GCAATGCTCTGCCCGTTGGGCTCCCCCTGCCCAGCCATGTGTCCTTGACCGGGGAGAGCGACGGTGGCTACATGGACATGAGCAAGGACG AGTCGGTGGACTATGTGCCCATGCTGGACATGAAAGGAGACGTCAAATATGCAGACATCGAGTCCTCCAACTACATGGCCCCTTACGATA ACTACGTTCCCTCTGCCCCTGAGAGGACCTGCCGAGCAACTTTGATCAACGAGTCTCCAGTGCTAAGCTACATGGACCTCGTGGGCTTCA GCTACCAGGTGGCCAATGGCATGGAGTTTCTGGCCTCCAAGAACTGCGTCCACAGAGACCTGGCGGCTAGGAACGTGCTCATCTGTGAAG GCAAGCTGGTCAAGATCTGTGACTTTGGCCTGGCTCGAGACATCATGCGGGACTCGAATTACATCTCCAAAGGCAGCACCTTTTTGCCTT TAAAGTGGATGGCTCCGGAGAGCATCTTCAACAGCCTCTACACCACCCTGAGCGACGTGTGGTCCTTCGGGATCCTGCTCTGGGAGATCT TCACCTTGGGTGGCACCCCTTACCCAGAGCTGCCCATGAACGAGCAGTTCTACAATGCCATCAAACGGGGTTACCGCATGGCCCAGCCTG CCCATGCCTCCGACGAGATCTATGAGATCATGCAGAAGTGCTGGGAAGAGAAGTTTGAGATTCGGCCCCCCTTCTCCCAGCTGGTGCTGC TTCTCGAGAGACTGTTGGGCGAAGGTTACAAAAAGAAGTACCAGCAGGTGGATGAGGAGTTTCTGAGGAGTGACCACCCAGCCATCCTTC GGTCCCAGGCCCGCTTGCCTGGGTTCCATGGCCTCCGATCTCCCCTGGACACCAGCTCCGTCCTCTATACTGCCGTGCAGCCCAATGAGG GTGACAACGACTATATCATCCCCCTGCCTGACCCCAAACCCGAGGTTGCTGACGAGGGCCCACTGGAGGGTTCCCCCAGCCTAGCCAGCT CCACCCTGAATGAAGTCAACACCTCCTCAACCATCTCCTGTGACAGCCCCCTGGAGCCCCAGGACGAACCAGAGCCAGAGCCCCAGCTTG AGCTCCAGGTGGAGCCGGAGCCAGAGCTGGAACAGTTGCCGGATTCGGGGTGCCCTGCGCCTCGGGCGGAAGCAGAGGATAGCTTCCTGT AGGGGGCTGGCCCCTACCCTGCCCTGCCTGAAGCTCCCCCCCTGCCAGCACCCAGCATCTCCTGGCCTGGCCTGACCGGGCTTCCTGTCA GCCAGGCTGCCCTTATCAGCTGTCCCCTTCTGGAAGCTTTCTGCTCCTGACGTGTTGTGCCCCAAACCCTGGGGCTGGCTTAGGAGGCAA GAAAACTGCAGGGGCCGTGACCAGCCCTCTGCCTCCAGGGAGGCCAACTGACTCTGAGCCAGGGTTCCCCCAGGGAACTCAGTTTTCCCA TATGTAAAATGGGAAAGTTAGGCTTGATGACCCAGAATCTAGGATTCTCTCCCTGGCTGACAGGTGGGGAGACCGAATCCCTCCCTGGGA AGATTCTTGGAGTTACTGAGGTGGTAAATTAACTTTTTTCTGTTCAGCCAGCTACCCCTCAAGGAATCATAGCTCTCTCCTCGCACTTTT ATCCACCCAGGAGCTAGGGAAGAGACCCTAGCCTCCCTGGCTGCTGGCTGAGCTAGGGCCTAGCCTTGAGCAGTGTTGCCTCATCCAGAA GAAAGCCAGTCTCCTCCCTATGATGCCAGTCCCTGCGTTCCCTGGCCCGAGCTGGTCTGGGGCCATTAGGCAGCCTAATTAATGCTGGAG GCTGAGCCAAGTACAGGACACCCCCAGCCTGCAGCCCTTGCCCAGGGCACTTGGAGCACACGCAGCCATAGCAAGTGCCTGTGTCCCTGT CCTTCAGGCCCATCAGTCCTGGGGCTTTTTCTTTATCACCCTCAGTCTTAATCCATCCACCAGAGTCTAGAAGGCCAGACGGGCCCCGCA TCTGTGATGAGAATGTAAATGTGCCAGTGTGGAGTGGCCACGTGTGTGTGCCAGTATATGGCCCTGGCTCTGCATTGGACCTGCTATGAG GCTTTGGAGGAATCCCTCACCCTCTCTGGGCCTCAGTTTCCCCTTCAAAAAATGAATAAGTCGGACTTATTAACTCTGAGTGCCTTGCCA GCACTAACATTCTAGAGTATTCCAGGTGGTTGCACATTTGTCCAGATGAAGCAAGGCCATATACCCTAAACTTCCATCCTGGGGGTCAGC TGGGCTCCTGGGAGATTCCAGATCACACATCACACTCTGGGGACTCAGGAACCATGCCCCTTCCCCAGGCCCCCAGCAAGTCTCAAGAAC ACAGCTGCACAGGCCTTGACTTAGAGTGACAGCCGGTGTCCTGGAAAGCCCCCAGCAGCTGCCCCAGGGACATGGGAAGACCACGGGACC TCTTTCACTACCCACGATGACCTCCGGGGGTATCCTGGGCAAAAGGGACAAAGAGGGCAAATGAGATCACCTCCTGCAGCCCACCACTCC AGCACCTGTGCCGAGGTCTGCGTCGAAGACAGAATGGACAGTGAGGACAGTTATGTCTTGTAAAAGACAAGAAGCTTCAGATGGGTACCC CAAGAAGGATGTGAGAGGTGGGCGCTTTGGAGGTTTGCCCCTCACCCACCAGCTGCCCCATCCCTGAGGCAGCGCTCCATGGGGGTATGG TTTTGTCACTGCCCAGACCTAGCAGTGACATCTCATTGTCCCCAGCCCAGTGGGCATTGGAGGTGCCAGGGGAGTCAGGGTTGTAGCCAA GACGCCCCCGCACGGGGAGGGTTGGGAAGGGGGTGCAGGAAGCTCAACCCCTCTGGGCACCAACCCTGCATTGCAGGTTGGCACCTTACT TCCCTGGGATCCCCAGAGTTGGTCCAAGGAGGGAGAGTGGGTTCTCAATACGGTACCAAAGATATAATCACCTAGGTTTACAAATATTTT TAGGACTCACGTTAACTCACATTTATACAGCAGAAATGCTATTTTGTATGCTGTTGAGTTTTTCTATCTGTGTACTTTTTTTTAAGGGAA >94475_94475_1_TSC1-PDGFRB_TSC1_chr9_135776976_ENST00000440111_PDGFRB_chr5_149505142_ENST00000261799_length(amino acids)=1382AA_BP=834 MAQQANVGELLAMLDSPMLGVRDDVTAVFKENLNSDRGPMLVNTLVDYYLETSSQPALHILTTLQEPHDKHLLDRINEYVGKAATRLSIL SLLGHVIRLQPSWKHKLSQAPLLPSLLKCLKMDTDVVVLTTGVLVLITMLPMIPQSGKQHLLDFFDIFGRLSSWCLKKPGHVAEVYLVHL HASVYALFHRLYGMYPCNFVSFLRSHYSMKENLETFEEVVKPMMEHVRIHPELVTGSKDHELDPRRWKRLETHDVVIECAKISLDPTEAS YEDGYSVSHQISARFPHRSADVTTSPYADTQNSYGCATSTPYSTSRLMLLNMPGQLPQTLSSPSTRLITEPPQATLWSPSMVCGMTTPPT SPGNVPPDLSHPYSKVFGTTAGGKGTPLGTPATSPPPAPLCHSDDYVHISLPQATVTPPRKEERMDSARPCLHRQHHLLNDRGSEEPPGS KGSVTLSDLPGFLGDLASEEDSIEKDKEEAAISRELSEITTAEAEPVVPRGGFDSPFYRDSLPGSQRKTHSAASSSQGASVNPEPLHSSL DKLGPDTPKQAFTPIDLPCGSADESPAGDRECQTSLETSIFTPSPCKIPPPTRVGFGSGQPPPYDHLFEVALPKTAHHFVIRKTEELLKK AKGNTEEDGVPSTSPMEVLDRLIQQGADAHSKELNKLPLPSKSVDWTHFGGSPPSDEIRTLRDQLLLLHNQLLYERFKRQQHALRNRRLL RKVIKAAALEEHNAAMKDQLKLQEKDIQMWKVSLQKEQARYNQLQEQRDTMVTKLHSQIRQLQHDREEFYNQSQELQTKLEDCRNMIAEL RIELKKANNKVCHTELLLSQVSQKKPRYEIRWKVIESVSSDGHEYIYVDPMQLPYDSTWELPRDQLVLGRTLGSGAFGQVVEATAHGLSH SQATMKVAVKMLKSTARSSEKQALMSELKIMSHLGPHLNVVNLLGACTKGGPIYIITEYCRYGDLVDYLHRNKHTFLQHHSDKRRPPSAE LYSNALPVGLPLPSHVSLTGESDGGYMDMSKDESVDYVPMLDMKGDVKYADIESSNYMAPYDNYVPSAPERTCRATLINESPVLSYMDLV GFSYQVANGMEFLASKNCVHRDLAARNVLICEGKLVKICDFGLARDIMRDSNYISKGSTFLPLKWMAPESIFNSLYTTLSDVWSFGILLW EIFTLGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKFEIRPPFSQLVLLLERLLGEGYKKKYQQVDEEFLRSDHPA ILRSQARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADEGPLEGSPSLASSTLNEVNTSSTISCDSPLEPQDEPEPEP -------------------------------------------------------------- >94475_94475_2_TSC1-PDGFRB_TSC1_chr9_135776976_ENST00000545250_PDGFRB_chr5_149505142_ENST00000261799_length(transcript)=6005nt_BP=2432nt CAGTTTGGTAGTGGCCCCAATGAAGAACCTTCAGAACCTGTAGCACACGTCCTGGAGCCAGCACAGCGCCTTCGAGCGAGAGAATGGCCC AACAAGCAAATGTCGGGGAGCTTCTTGCCATGCTGGACTCCCCCATGCTGGGTGTGCGGGACGACGTGACAGCTGTCTTTAAAGAGAACC TCAATTCTGACCGTGGCCCTATGCTTGTAAACACCTTGGTGGATTATTACCTGGAAACCAGCTCTCAGCCGGCATTGCACATCCTGACCA CCTTGCAAGAGCCACATGACAAGATGGACACTGACGTCGTTGTCCTCACAACAGGCGTCTTGGTGTTGATAACCATGCTACCAATGATTC CACAGTCTGGGAAACAGCATCTTCTTGATTTCTTTGACATTTTTGGCCGTCTGTCATCATGGTGCCTGAAGAAACCAGGCCACGTGGCGG AAGTCTATCTCGTCCATCTCCATGCCAGTGTGTACGCACTCTTTCATCGCCTTTATGGAATGTACCCTTGCAACTTCGTCTCCTTTTTGC GTTCTCATTACAGTATGAAAGAAAACCTGGAGACTTTTGAAGAAGTGGTCAAGCCAATGATGGAGCATGTGCGAATTCATCCGGAATTAG TGACTGGATCCAAGGACCATGAACTGGACCCTCGAAGGTGGAAGAGATTAGAAACTCATGATGTTGTGATCGAGTGTGCCAAAATCTCTC TGGATCCCACAGAAGCCTCATATGAAGATGGCTATTCTGTGTCTCACCAAATCTCAGCCCGCTTTCCTCATCGTTCAGCCGATGTCACCA CCAGCCCTTATGCTGACACACAGAATAGCTATGGGTGTGCTACTTCTACCCCTTACTCCACGTCTCGGCTGATGTTGTTAAATATGCCAG GGCAGCTACCTCAGACTCTGAGTTCCCCATCGACACGGCTGATAACTGAACCACCACAAGCTACTCTTTGGAGCCCATCTATGGTTTGTG GTATGACCACTCCTCCAACTTCTCCTGGAAATGTCCCACCTGATCTGTCACACCCTTACAGTAAAGTCTTTGGTACAACTGCAGGTGGAA AAGGAACTCCTCTGGGAACCCCAGCAACCTCTCCTCCTCCAGCCCCACTCTGTCATTCGGATGACTACGTGCACATTTCACTCCCCCAGG CCACAGTCACACCCCCCAGGAAGGAAGAGAGAATGGATTCTGCAAGACCATGTCTACACAGACAACACCATCTTCTGAATGACAGAGGAT CAGAAGAGCCACCTGGCAGCAAAGGTTCTGTCACTCTAAGTGATCTTCCAGGGTTTTTAGGTGATCTGGCCTCTGAAGAAGATAGTATTG AAAAAGATAAAGAAGAAGCTGCAATATCTAGAGAACTTTCTGAGATCACCACAGCAGAGGCAGAGCCTGTGGTTCCTCGAGGAGGCTTTG ACTCTCCCTTTTACCGAGACAGTCTCCCAGGTTCTCAGCGGAAGACCCACTCGGCAGCCTCCAGTTCTCAGGGCGCCAGCGTGAACCCTG AGCCTTTACACTCCTCCCTGGACAAGCTTGGGCCTGACACACCAAAGCAAGCCTTTACTCCCATAGACCTGCCCTGCGGCAGTGCTGATG AAAGCCCTGCGGGAGACAGGGAATGCCAGACTTCTTTGGAGACCAGTATCTTCACTCCCAGTCCTTGTAAAATTCCACCTCCGACGAGAG TGGGCTTTGGAAGCGGGCAGCCTCCCCCGTATGATCATCTTTTTGAGGTGGCATTGCCAAAGACAGCCCATCATTTTGTCATCAGGAAGA CTGAGGAGCTGTTAAAGAAAGCAAAAGGAAACACAGAGGAAGATGGTGTGCCCTCTACCTCCCCAATGGAAGTGCTGGACAGACTGATAC AGCAGGGAGCAGACGCGCACAGCAAGGAGCTGAACAAGTTGCCTTTACCCAGCAAGTCTGTCGACTGGACCCACTTTGGAGGCTCTCCTC CTTCAGATGAGATCCGCACCCTCCGAGACCAGTTGCTTTTACTGCACAACCAGTTACTCTATGAGCGTTTTAAGAGGCAGCAGCATGCCC TCCGGAACAGGCGGCTCCTCCGCAAGGTGATCAAAGCAGCAGCTCTGGAGGAACATAATGCTGCCATGAAAGATCAGTTGAAGTTACAAG AGAAGGACATCCAGATGTGGAAGGTTAGTCTGCAGAAAGAACAAGCTAGATACAATCAGCTCCAGGAGCAGCGTGACACTATGGTAACCA AGCTCCACAGCCAGATCAGACAGCTGCAGCATGACCGAGAGGAATTCTACAACCAGAGCCAGGAATTACAGACGAAGCTGGAGGACTGCA GGAACATGATTGCGGAGCTGCGGATAGAACTGAAGAAGGCCAACAACAAGGTGTGTCACACTGAGCTGCTGCTCAGTCAGGTTTCCCAAA AGAAGCCACGTTACGAGATCCGATGGAAGGTGATTGAGTCTGTGAGCTCTGACGGCCATGAGTACATCTACGTGGACCCCATGCAGCTGC CCTATGACTCCACGTGGGAGCTGCCGCGGGACCAGCTTGTGCTGGGACGCACCCTCGGCTCTGGGGCCTTTGGGCAGGTGGTGGAGGCCA CGGCTCATGGCCTGAGCCATTCTCAGGCCACGATGAAAGTGGCCGTCAAGATGCTTAAATCCACAGCCCGCAGCAGTGAGAAGCAAGCCC TTATGTCGGAGCTGAAGATCATGAGTCACCTTGGGCCCCACCTGAACGTGGTCAACCTGTTGGGGGCCTGCACCAAAGGAGGACCCATCT ATATCATCACTGAGTACTGCCGCTACGGAGACCTGGTGGACTACCTGCACCGCAACAAACACACCTTCCTGCAGCACCACTCCGACAAGC GCCGCCCGCCCAGCGCGGAGCTCTACAGCAATGCTCTGCCCGTTGGGCTCCCCCTGCCCAGCCATGTGTCCTTGACCGGGGAGAGCGACG GTGGCTACATGGACATGAGCAAGGACGAGTCGGTGGACTATGTGCCCATGCTGGACATGAAAGGAGACGTCAAATATGCAGACATCGAGT CCTCCAACTACATGGCCCCTTACGATAACTACGTTCCCTCTGCCCCTGAGAGGACCTGCCGAGCAACTTTGATCAACGAGTCTCCAGTGC TAAGCTACATGGACCTCGTGGGCTTCAGCTACCAGGTGGCCAATGGCATGGAGTTTCTGGCCTCCAAGAACTGCGTCCACAGAGACCTGG CGGCTAGGAACGTGCTCATCTGTGAAGGCAAGCTGGTCAAGATCTGTGACTTTGGCCTGGCTCGAGACATCATGCGGGACTCGAATTACA TCTCCAAAGGCAGCACCTTTTTGCCTTTAAAGTGGATGGCTCCGGAGAGCATCTTCAACAGCCTCTACACCACCCTGAGCGACGTGTGGT CCTTCGGGATCCTGCTCTGGGAGATCTTCACCTTGGGTGGCACCCCTTACCCAGAGCTGCCCATGAACGAGCAGTTCTACAATGCCATCA AACGGGGTTACCGCATGGCCCAGCCTGCCCATGCCTCCGACGAGATCTATGAGATCATGCAGAAGTGCTGGGAAGAGAAGTTTGAGATTC GGCCCCCCTTCTCCCAGCTGGTGCTGCTTCTCGAGAGACTGTTGGGCGAAGGTTACAAAAAGAAGTACCAGCAGGTGGATGAGGAGTTTC TGAGGAGTGACCACCCAGCCATCCTTCGGTCCCAGGCCCGCTTGCCTGGGTTCCATGGCCTCCGATCTCCCCTGGACACCAGCTCCGTCC TCTATACTGCCGTGCAGCCCAATGAGGGTGACAACGACTATATCATCCCCCTGCCTGACCCCAAACCCGAGGTTGCTGACGAGGGCCCAC TGGAGGGTTCCCCCAGCCTAGCCAGCTCCACCCTGAATGAAGTCAACACCTCCTCAACCATCTCCTGTGACAGCCCCCTGGAGCCCCAGG ACGAACCAGAGCCAGAGCCCCAGCTTGAGCTCCAGGTGGAGCCGGAGCCAGAGCTGGAACAGTTGCCGGATTCGGGGTGCCCTGCGCCTC GGGCGGAAGCAGAGGATAGCTTCCTGTAGGGGGCTGGCCCCTACCCTGCCCTGCCTGAAGCTCCCCCCCTGCCAGCACCCAGCATCTCCT GGCCTGGCCTGACCGGGCTTCCTGTCAGCCAGGCTGCCCTTATCAGCTGTCCCCTTCTGGAAGCTTTCTGCTCCTGACGTGTTGTGCCCC AAACCCTGGGGCTGGCTTAGGAGGCAAGAAAACTGCAGGGGCCGTGACCAGCCCTCTGCCTCCAGGGAGGCCAACTGACTCTGAGCCAGG GTTCCCCCAGGGAACTCAGTTTTCCCATATGTAAAATGGGAAAGTTAGGCTTGATGACCCAGAATCTAGGATTCTCTCCCTGGCTGACAG GTGGGGAGACCGAATCCCTCCCTGGGAAGATTCTTGGAGTTACTGAGGTGGTAAATTAACTTTTTTCTGTTCAGCCAGCTACCCCTCAAG GAATCATAGCTCTCTCCTCGCACTTTTATCCACCCAGGAGCTAGGGAAGAGACCCTAGCCTCCCTGGCTGCTGGCTGAGCTAGGGCCTAG CCTTGAGCAGTGTTGCCTCATCCAGAAGAAAGCCAGTCTCCTCCCTATGATGCCAGTCCCTGCGTTCCCTGGCCCGAGCTGGTCTGGGGC CATTAGGCAGCCTAATTAATGCTGGAGGCTGAGCCAAGTACAGGACACCCCCAGCCTGCAGCCCTTGCCCAGGGCACTTGGAGCACACGC AGCCATAGCAAGTGCCTGTGTCCCTGTCCTTCAGGCCCATCAGTCCTGGGGCTTTTTCTTTATCACCCTCAGTCTTAATCCATCCACCAG AGTCTAGAAGGCCAGACGGGCCCCGCATCTGTGATGAGAATGTAAATGTGCCAGTGTGGAGTGGCCACGTGTGTGTGCCAGTATATGGCC CTGGCTCTGCATTGGACCTGCTATGAGGCTTTGGAGGAATCCCTCACCCTCTCTGGGCCTCAGTTTCCCCTTCAAAAAATGAATAAGTCG GACTTATTAACTCTGAGTGCCTTGCCAGCACTAACATTCTAGAGTATTCCAGGTGGTTGCACATTTGTCCAGATGAAGCAAGGCCATATA CCCTAAACTTCCATCCTGGGGGTCAGCTGGGCTCCTGGGAGATTCCAGATCACACATCACACTCTGGGGACTCAGGAACCATGCCCCTTC CCCAGGCCCCCAGCAAGTCTCAAGAACACAGCTGCACAGGCCTTGACTTAGAGTGACAGCCGGTGTCCTGGAAAGCCCCCAGCAGCTGCC CCAGGGACATGGGAAGACCACGGGACCTCTTTCACTACCCACGATGACCTCCGGGGGTATCCTGGGCAAAAGGGACAAAGAGGGCAAATG AGATCACCTCCTGCAGCCCACCACTCCAGCACCTGTGCCGAGGTCTGCGTCGAAGACAGAATGGACAGTGAGGACAGTTATGTCTTGTAA AAGACAAGAAGCTTCAGATGGGTACCCCAAGAAGGATGTGAGAGGTGGGCGCTTTGGAGGTTTGCCCCTCACCCACCAGCTGCCCCATCC CTGAGGCAGCGCTCCATGGGGGTATGGTTTTGTCACTGCCCAGACCTAGCAGTGACATCTCATTGTCCCCAGCCCAGTGGGCATTGGAGG TGCCAGGGGAGTCAGGGTTGTAGCCAAGACGCCCCCGCACGGGGAGGGTTGGGAAGGGGGTGCAGGAAGCTCAACCCCTCTGGGCACCAA CCCTGCATTGCAGGTTGGCACCTTACTTCCCTGGGATCCCCAGAGTTGGTCCAAGGAGGGAGAGTGGGTTCTCAATACGGTACCAAAGAT ATAATCACCTAGGTTTACAAATATTTTTAGGACTCACGTTAACTCACATTTATACAGCAGAAATGCTATTTTGTATGCTGTTGAGTTTTT >94475_94475_2_TSC1-PDGFRB_TSC1_chr9_135776976_ENST00000545250_PDGFRB_chr5_149505142_ENST00000261799_length(amino acids)=1331AA_BP=783 MAQQANVGELLAMLDSPMLGVRDDVTAVFKENLNSDRGPMLVNTLVDYYLETSSQPALHILTTLQEPHDKMDTDVVVLTTGVLVLITMLP MIPQSGKQHLLDFFDIFGRLSSWCLKKPGHVAEVYLVHLHASVYALFHRLYGMYPCNFVSFLRSHYSMKENLETFEEVVKPMMEHVRIHP ELVTGSKDHELDPRRWKRLETHDVVIECAKISLDPTEASYEDGYSVSHQISARFPHRSADVTTSPYADTQNSYGCATSTPYSTSRLMLLN MPGQLPQTLSSPSTRLITEPPQATLWSPSMVCGMTTPPTSPGNVPPDLSHPYSKVFGTTAGGKGTPLGTPATSPPPAPLCHSDDYVHISL PQATVTPPRKEERMDSARPCLHRQHHLLNDRGSEEPPGSKGSVTLSDLPGFLGDLASEEDSIEKDKEEAAISRELSEITTAEAEPVVPRG GFDSPFYRDSLPGSQRKTHSAASSSQGASVNPEPLHSSLDKLGPDTPKQAFTPIDLPCGSADESPAGDRECQTSLETSIFTPSPCKIPPP TRVGFGSGQPPPYDHLFEVALPKTAHHFVIRKTEELLKKAKGNTEEDGVPSTSPMEVLDRLIQQGADAHSKELNKLPLPSKSVDWTHFGG SPPSDEIRTLRDQLLLLHNQLLYERFKRQQHALRNRRLLRKVIKAAALEEHNAAMKDQLKLQEKDIQMWKVSLQKEQARYNQLQEQRDTM VTKLHSQIRQLQHDREEFYNQSQELQTKLEDCRNMIAELRIELKKANNKVCHTELLLSQVSQKKPRYEIRWKVIESVSSDGHEYIYVDPM QLPYDSTWELPRDQLVLGRTLGSGAFGQVVEATAHGLSHSQATMKVAVKMLKSTARSSEKQALMSELKIMSHLGPHLNVVNLLGACTKGG PIYIITEYCRYGDLVDYLHRNKHTFLQHHSDKRRPPSAELYSNALPVGLPLPSHVSLTGESDGGYMDMSKDESVDYVPMLDMKGDVKYAD IESSNYMAPYDNYVPSAPERTCRATLINESPVLSYMDLVGFSYQVANGMEFLASKNCVHRDLAARNVLICEGKLVKICDFGLARDIMRDS NYISKGSTFLPLKWMAPESIFNSLYTTLSDVWSFGILLWEIFTLGGTPYPELPMNEQFYNAIKRGYRMAQPAHASDEIYEIMQKCWEEKF EIRPPFSQLVLLLERLLGEGYKKKYQQVDEEFLRSDHPAILRSQARLPGFHGLRSPLDTSSVLYTAVQPNEGDNDYIIPLPDPKPEVADE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TSC1-PDGFRB |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TSC1-PDGFRB |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TSC1-PDGFRB |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
