
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TXNL1-LRP1B (FusionGDB2 ID:95587) |
Fusion Gene Summary for TXNL1-LRP1B |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TXNL1-LRP1B | Fusion gene ID: 95587 | Hgene | Tgene | Gene symbol | TXNL1 | LRP1B | Gene ID | 9352 | 53353 |
| Gene name | thioredoxin like 1 | LDL receptor related protein 1B | |
| Synonyms | HEL-S-114|TRP32|TXL-1|TXNL|Txl | LRP-1B|LRP-DIT|LRPDIT | |
| Cytomap | 18q21.31 | 2q22.1-q22.2 | |
| Type of gene | protein-coding | protein-coding | |
| Description | thioredoxin-like protein 132 kDa thioredoxin-related proteinepididymis secretory protein Li 114thioredoxin-related 32 kDa proteinthioredoxin-related protein 1 | low-density lipoprotein receptor-related protein 1BLRP-deleted in tumorslow density lipoprotein receptor related protein-deleted in tumorlow density lipoprotein receptor-related protein 1Blow density lipoprotein-related protein 1B (deleted in tumors) | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | Q9NZR2 | |
| Ensembl transtripts involved in fusion gene | ENST00000217515, ENST00000590954, ENST00000540155, ENST00000585497, | ENST00000486364, ENST00000389484, | |
| Fusion gene scores | * DoF score | 3 X 4 X 3=36 | 12 X 13 X 5=780 |
| # samples | 4 | 13 | |
| ** MAII score | log2(4/36*10)=0.15200309344505 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | log2(13/780*10)=-2.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TXNL1 [Title/Abstract] AND LRP1B [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TXNL1(54305574)-LRP1B(141299540), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | TXNL1-LRP1B seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. TXNL1-LRP1B seems lost the major protein functional domain in Tgene partner, which is a CGC due to the frame-shifted ORF. TXNL1-LRP1B seems lost the major protein functional domain in Tgene partner, which is a essential gene due to the frame-shifted ORF. TXNL1-LRP1B seems lost the major protein functional domain in Tgene partner, which is a tumor suppressor due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across TXNL1 (5'-gene) Fusion gene breakpoints across TXNL1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
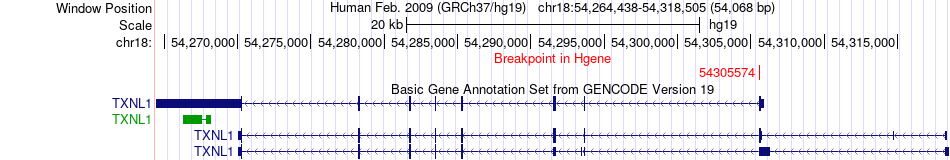 |
 Fusion gene breakpoints across LRP1B (3'-gene) Fusion gene breakpoints across LRP1B (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
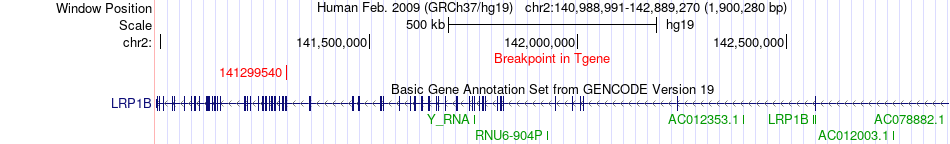 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | BLCA | TCGA-G2-A2EF-01A | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
Top |
Fusion Gene ORF analysis for TXNL1-LRP1B |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000217515 | ENST00000486364 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| 5CDS-intron | ENST00000590954 | ENST00000486364 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| 5UTR-3CDS | ENST00000540155 | ENST00000389484 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| 5UTR-intron | ENST00000540155 | ENST00000486364 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| Frame-shift | ENST00000590954 | ENST00000389484 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| In-frame | ENST00000217515 | ENST00000389484 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| intron-3CDS | ENST00000585497 | ENST00000389484 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
| intron-intron | ENST00000585497 | ENST00000486364 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000217515 | TXNL1 | chr18 | 54305574 | - | ENST00000389484 | LRP1B | chr2 | 141299540 | - | 8672 | 303 | 54 | 6908 | 2284 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000217515 | ENST00000389484 | TXNL1 | chr18 | 54305574 | - | LRP1B | chr2 | 141299540 | - | 0.000134682 | 0.9998653 |
Top |
Fusion Genomic Features for TXNL1-LRP1B |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for TXNL1-LRP1B |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr18:54305574/chr2:141299540) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LRP1B |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Potential cell surface proteins that bind and internalize ligands in the process of receptor-mediated endocytosis. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2464_2504 | 2398 | 4600.0 | Domain | EGF-like 10 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2509_2548 | 2398 | 4600.0 | Domain | LDL-receptor class A 11 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2551_2587 | 2398 | 4600.0 | Domain | LDL-receptor class A 12 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2590_2626 | 2398 | 4600.0 | Domain | LDL-receptor class A 13 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2629_2675 | 2398 | 4600.0 | Domain | LDL-receptor class A 14 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2681_2717 | 2398 | 4600.0 | Domain | LDL-receptor class A 15 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2719_2757 | 2398 | 4600.0 | Domain | LDL-receptor class A 16 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2760_2800 | 2398 | 4600.0 | Domain | LDL-receptor class A 17 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2804_2841 | 2398 | 4600.0 | Domain | LDL-receptor class A 18 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2844_2885 | 2398 | 4600.0 | Domain | LDL-receptor class A 19 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2890_2926 | 2398 | 4600.0 | Domain | LDL-receptor class A 20 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2927_2967 | 2398 | 4600.0 | Domain | EGF-like 11 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2968_3008 | 2398 | 4600.0 | Domain | EGF-like 12%3B calcium-binding | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3273_3314 | 2398 | 4600.0 | Domain | EGF-like 13 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3316_3353 | 2398 | 4600.0 | Domain | LDL-receptor class A 21 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3356_3392 | 2398 | 4600.0 | Domain | LDL-receptor class A 22 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3395_3432 | 2398 | 4600.0 | Domain | LDL-receptor class A 23 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3435_3472 | 2398 | 4600.0 | Domain | LDL-receptor class A 24 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3475_3511 | 2398 | 4600.0 | Domain | LDL-receptor class A 25 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3514_3550 | 2398 | 4600.0 | Domain | LDL-receptor class A 26 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3552_3588 | 2398 | 4600.0 | Domain | LDL-receptor class A 27 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3593_3629 | 2398 | 4600.0 | Domain | LDL-receptor class A 28 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3631_3668 | 2398 | 4600.0 | Domain | LDL-receptor class A 29 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3673_3711 | 2398 | 4600.0 | Domain | LDL-receptor class A 30 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3714_3752 | 2398 | 4600.0 | Domain | LDL-receptor class A 31 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3761_3797 | 2398 | 4600.0 | Domain | LDL-receptor class A 32 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3801_3843 | 2398 | 4600.0 | Domain | EGF-like 14 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3844_3881 | 2398 | 4600.0 | Domain | EGF-like 15 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4171_4208 | 2398 | 4600.0 | Domain | EGF-like 16 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4213_4249 | 2398 | 4600.0 | Domain | EGF-like 17 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4249_4285 | 2398 | 4600.0 | Domain | EGF-like 18 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4285_4321 | 2398 | 4600.0 | Domain | EGF-like 19 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4321_4357 | 2398 | 4600.0 | Domain | EGF-like 20 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4357_4392 | 2398 | 4600.0 | Domain | EGF-like 21 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4390_4427 | 2398 | 4600.0 | Domain | EGF-like 22 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4492_4495 | 2398 | 4600.0 | Motif | Endocytosis signal | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4559_4562 | 2398 | 4600.0 | Motif | Endocytosis signal | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2417_2459 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 25 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3055_3098 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 26 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3099_3141 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 27 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3142_3185 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 28 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3186_3224 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 29 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3225_3268 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 30 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3933_3980 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 31 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 3981_4038 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 32 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4039_4082 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 33 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4083_4127 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 34 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4468_4599 | 2398 | 4600.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 4445_4467 | 2398 | 4600.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TXNL1 | chr18:54305574 | chr2:141299540 | ENST00000217515 | - | 1 | 8 | 115_285 | 32 | 290.0 | Domain | PITH |
| Hgene | TXNL1 | chr18:54305574 | chr2:141299540 | ENST00000217515 | - | 1 | 8 | 2_109 | 32 | 290.0 | Domain | Note=Thioredoxin |
| Hgene | TXNL1 | chr18:54305574 | chr2:141299540 | ENST00000590954 | - | 3 | 10 | 115_285 | 32 | 290.0 | Domain | PITH |
| Hgene | TXNL1 | chr18:54305574 | chr2:141299540 | ENST00000590954 | - | 3 | 10 | 2_109 | 32 | 290.0 | Domain | Note=Thioredoxin |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1005_1043 | 2398 | 4600.0 | Domain | LDL-receptor class A 7 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1052_1089 | 2398 | 4600.0 | Domain | LDL-receptor class A 8 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1094_1132 | 2398 | 4600.0 | Domain | LDL-receptor class A 9 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1135_1174 | 2398 | 4600.0 | Domain | LDL-receptor class A 10 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 116_154 | 2398 | 4600.0 | Domain | EGF-like 1 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1174_1213 | 2398 | 4600.0 | Domain | EGF-like 5 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1214_1253 | 2398 | 4600.0 | Domain | EGF-like 6 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1527_1570 | 2398 | 4600.0 | Domain | EGF-like 7 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 155_194 | 2398 | 4600.0 | Domain | EGF-like 2%3B calcium-binding | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1834_1875 | 2398 | 4600.0 | Domain | EGF-like 8 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2143_2183 | 2398 | 4600.0 | Domain | EGF-like 9 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 31_70 | 2398 | 4600.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 471_517 | 2398 | 4600.0 | Domain | EGF-like 3 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 76_114 | 2398 | 4600.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 794_834 | 2398 | 4600.0 | Domain | EGF-like 4 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 844_882 | 2398 | 4600.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 885_923 | 2398 | 4600.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 926_963 | 2398 | 4600.0 | Domain | LDL-receptor class A 5 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 966_1003 | 2398 | 4600.0 | Domain | LDL-receptor class A 6 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1300_1346 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 8 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1347_1389 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 9 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1390_1436 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 10 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1437_1480 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 11 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1481_1522 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 12 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1618_1660 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 13 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1661_1704 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 14 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1705_1744 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 15 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1745_1787 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 16 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1922_1964 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 17 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 1965_2007 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 28 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2008_2051 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 19 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2052_2095 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 20 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2239_2280 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 21 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2281_2329 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 22 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2330_2374 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 23 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 2375_2416 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 24 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 295_337 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 1 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 338_381 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 2 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 382_425 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 3 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 568_610 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 4 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 611_656 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 5 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 657_706 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 6 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 707_750 | 2398 | 4600.0 | Repeat | Note=LDL-receptor class B 7 | |
| Tgene | LRP1B | chr18:54305574 | chr2:141299540 | ENST00000389484 | 42 | 91 | 25_4444 | 2398 | 4600.0 | Topological domain | Extracellular |
Top |
Fusion Gene Sequence for TXNL1-LRP1B |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >95587_95587_1_TXNL1-LRP1B_TXNL1_chr18_54305574_ENST00000217515_LRP1B_chr2_141299540_ENST00000389484_length(transcript)=8672nt_BP=303nt TCCCCCATCCCTCCCCGCCCCCTCGGCCGCTCCGCTCCCTGGCGTGACTCGGCACTGAGAGTCCCGGGAGAAGACTCGGCGGTCGCCACC TCTTCTGTCCAGGCCTCGGCCTTCCTGAGCATCTCTCCTTCCTCTCCCAGATCGTCTTCTCCTTCAGTTTCAAAGCCAGTGGCGTCGCGG CCACCCTGCCGGGCTCTCTGTGAGGATGGTGGGGGTGAAGCCCGTCGGGAGCGACCCGGATTTCCAGCCAGAGCTGAGCGGCGCGGGCTC CAGACTCGCCGTGGTCAAGTTCACCATGAGAGGGTGATAGTTAAATCTGGGCCAGGGACTTTCCTCAGTTTGGCTGTTTATGACAATTAT ATATTCTGGTCGGACTGGGGAAGAAGAGCTATACTGCGGTCCAACAAGTACACAGGAGGAGATACAAAAATTCTTCGTTCCGATATTCCA CATCAGCCAATGGGAATCATAGCTGTTGCCAATGACACCAATAGCTGTGAACTTTCTCCATGTGCATTATTGAATGGAGGCTGCCATGAC TTGTGCCTTTTAACTCCCAATGGGAGAGTGAATTGTTCCTGCAGAGGGGACCGAATATTGCTAGAGGACAACAGATGTGTGACTAAAAAT TCCTCCTGCAACGCTTATTCGGAGTTTGAATGTGGAAATGGTGAGTGCATTGACTACCAGCTCACCTGTGATGGCATTCCTCACTGTAAA GATAAATCAGATGAAAAACTGCTCTACTGTGAAAACAGAAGCTGTCGAAGAGGCTTCAAGCCATGCTATAATCGCCGCTGCATTCCTCAT GGCAAGTTATGTGATGGAGAAAATGACTGCGGAGACAACTCTGATGAATTAGATTGTAAAGTTTCAACCTGTGCCACGGTTGAGTTCCGC TGTGCAGATGGGACTTGTATTCCAAGATCAGCACGATGCAACCAGAACATAGATTGTGCAGATGCTTCAGATGAAAAGAACTGCAATAAC ACAGACTGCACACATTTCTATAAGCTTGGAGTGAAAACCACAGGGTTCATAAGATGTAATTCTACCTCACTGTGTGTTCTGCCAACCTGG ATATGCGACGGGTCTAATGACTGTGGAGACTATTCAGATGAATTAAAGTGCCCAGTTCAAAACAAACACAAATGTGAAGAAAATTATTTT AGTTGTCCTAGTGGAAGATGCATTTTGAATACCTGGATATGCGATGGTCAGAAAGATTGTGAGGATGGACGTGATGAATTCCACTGTGAT TCTTCTTGCTCTTGGAACCAATTTGCTTGTTCCGCACAAAAATGTATTTCTAAGCATTGGATTTGTGATGGAGAAGATGACTGTGGGGAT GGGTTAGATGAAAGTGACAGCATTTGTGGTGCCATAACCTGTGCTGCTGACATGTTCAGCTGCCAGGGCTCTCGTGCCTGCGTGCCCCGA CATTGGCTTTGTGATGGTGAAAGGGACTGTCCAGATGGAAGCGATGAGCTTTCCACAGCAGGCTGCGCTCCCAATAATACATGTGATGAA AATGCTTTCATGTGCCATAATAAAGTATGCATTCCCAAGCAATTTGTTTGTGACCATGATGACGACTGTGGAGATGGCTCTGATGAGTCA CCGCAGTGTGGATATCGACAGTGTGGTACAGAAGAATTTAGTTGTGCTGATGGGCGGTGTCTTCTAAATACTCAATGGCAGTGTGATGGA GACTTTGACTGTCCTGACCATTCTGATGAAGCACCTTTAAACCCAAAGTGTAAAAGTGCAGAACAGTCATGCAACAGTTCATTTTTTATG TGCAAAAATGGCAGGTGCATTCCCAGTGGAGGTCTTTGCGACAATAAGGATGACTGTGGCGATGGTTCAGATGAGAGAAACTGCCATATA AATGAATGTTTGAGTAAGAAAGTCAGTGGATGTTCTCAAGACTGTCAAGACCTTCCGGTCAGTTATAAGTGCAAATGCTGGCCTGGATTC CAACTGAAGGATGACGGCAAAACATGTGTAGACATTGATGAATGCTCTTCAGGCTTTCCCTGTAGCCAGCAATGCATCAATACATACGGG ACTTACAAGTGCCTCTGTACAGATGGGTATGAAATACAACCTGATAACCCAAATGGCTGCAAATCGCTCTCAGATGAAGAACCTTTTTTA ATTCTTGCTGATCATCATGAGATAAGGAAAATTAGCACTGATGGCTCCAACTACACACTTTTAAAACAGGGATTAAACAATGTTATTGCT ATAGACTTTGATTACAGAGAAGAATTCATCTATTGGATCGATTCTAGCCGACCCAATGGCAGTCGCATAAATAGAATGTGTTTAAATGGA AGTGACATTAAGGTAGTTCATAACACAGCGGTCCCCAATGCACTTGCTGTCGATTGGATTGGAAAAAACCTCTATTGGTCTGACACAGAA AAAAGAATCATTGAAGTATCCAAACTCAATGGCTTGTACCCTACTATACTCGTTAGCAAAAGGCTGAAGTTTCCCAGAGACTTGTCTTTA GATCCTCAAGCTGGATATTTGTATTGGATTGACTGCTGCGAGTATCCTCATATTGGCCGTGTTGGAATGGATGGAACCAATCAGAGTGTT GTCATAGAAACCAAGATTTCTAGACCTATGGCACTAACAATAGATTATGTTAATCGTAGACTCTACTGGGCCGATGAAAATCACATTGAA TTTAGCAACATGGATGGATCTCATAGACACAAAGTCCCTAATCAAGATATTCCAGGGGTGATTGCACTAACATTGTTTGAAGACTACATC TACTGGACTGATGGGAAAACCAAGTCACTCAGCCGTGCCCATAAAACATCGGGAGCAGACAGACTCTCACTGATTTACTCATGGCATGCC ATCACAGATATCCAGGTGTATCATTCTTATAGACAACCTGATGTCTCCAAACATCTCTGCATGATAAATAATGGTGGTTGCAGTCATTTG TGCCTTTTAGCCCCTGGAAAAACCCACACTTGTGCATGTCCCACTAACTTCTATCTGGCAGCTGATAATAGGACTTGCTTATCCAACTGC ACAGCCAGCCAGTTTCGTTGCAAAACTGACAAATGTATTCCATTCTGGTGGAAATGTGACACCGTGGATGACTGTGGTGATGGATCTGAT GAACCTGATGACTGTCCTGAATTTAGATGTCAGCCAGGCCGATTTCAGTGTGGGACTGGACTCTGTGCTCTACCAGCTTTCATCTGTGAT GGAGAGAATGATTGTGGAGACAATTCTGATGAACTCAACTGTGACACACATGTCTGCCTGTCAGGTCAATTCAAATGTACCAAGAACCAG AAATGTATCCCAGTAAACTTAAGATGTAATGGGCAAGATGACTGTGGTGATGAGGAAGATGAAAGAGACTGTCCTGAAAACAGCTGTTCT CCAGACTATTTCCAGTGTAAGACTACGAAGCATTGCATTTCCAAGCTGTGGGTTTGTGACGAGGATCCAGACTGTGCAGATGCATCAGAC GAGGCCAACTGCGATAAAAAGACTTGTGGACCTCATGAATTCCAGTGTAAAAACAACAACTGTATTCCCGATCACTGGCGGTGTGATAGC CAAAATGACTGCAGTGATAATTCAGATGAAGAAAACTGTAAGCCACAGACATGTACATTGAAAGATTTCCTCTGTGCCAATGGGGACTGT GTTTCTTCAAGGTTTTGGTGTGATGGAGATTTTGACTGTGCAGATGGCTCTGATGAGAGAAATTGTGAGACAAGTTGTTCCAAAGATCAG TTCCGGTGTTCCAATGGTCAGTGTATACCAGCAAAATGGAAATGTGATGGCCATGAAGACTGCAAATATGGGGAAGATGAGAAAAGCTGT GAGCCAGCTTCTCCTACTTGCTCATCACGTGAATATATATGTGCCAGTGATGGATGTATTTCAGCATCTTTGAAATGTAATGGAGAATAT GATTGTGCTGATGGTTCAGATGAGATGGACTGTGTGACTGAATGTAAGGAAGATCAGTTTCGGTGCAAAAATAAAGCCCACTGTATTCCA ATTAGATGGCTGTGTGATGGAATTCATGACTGTGTGGATGGCAGTGATGAAGAGAACTGTGAAAGAGGAGGAAATATATGTAGAGCTGAT GAGTTCCTTTGCAATAATTCTCTCTGCAAACTACATTTCTGGGTGTGTGATGGAGAGGACGACTGTGGAGACAACTCTGATGAAGCCCCT GATATGTGTGTCAAATTTCTTTGTCCATCCACGAGACCTCACAGATGCAGAAATAACAGAATATGCCTACAGTCGGAGCAAATGTGCAAT GGGATTGATGAATGCGGTGACAATTCAGATGAAGATCACTGTGGTGGTAAGCTGACATATAAAGCAAGGCCTTGTAAAAAGGATGAGTTT GCTTGTAGTAATAAAAAATGCATCCCTATGGATCTCCAGTGTGATCGACTTGATGACTGCGGAGATGGTTCAGATGAGCAAGGATGCAGA ATAGCTCCTACTGAATATACCTGTGAAGATAATGTGAATCCATGTGGAGATGATGCATATTGTAATCAAATAAAAACATCTGTTTTCTGT CGCTGTAAGCCTGGATTTCAGAGAAACATGAAAAACAGACAATGTGAAGACCTTAATGAATGTTTGGTGTTTGGCACATGTTCCCATCAA TGTATAAATGTGGAAGGATCATATAAATGTGTGTGTGACCAGAATTTTCAAGAAAGAAATAACACCTGCATAGCAGAAGGCTCTGAAGAT CAAGTTCTCTACATTGCTAATGACACTGATATCCTGGGTTTTATATATCCATTCAACTACAGTGGCGATCATCAACAAATTTCTCATATT GAACATAATTCAAGAATAACAGGGATGGATGTATATTATCAAAGAGATATGATTATTTGGAGTACTCAGTTTAATCCAGGCGGAATTTTC TACAAAAGGATCCATGGCAGAGAAAAAAGGCAAGCAAACAGTGGCTTGATTTGTCCTGAATTTAAAAGGCCCAGGGACATTGCAGTTGAC TGGGTGGCTGGAAACATTTACTGGACTGATCATTCTAGAATGCATTGGTTCAGTTACTACACTACTCACTGGACCAGTCTGAGGTACTCT ATCAACGTAGGGCAGCTGAATGGCCCCAACTGCACCAGACTCTTAACAAATATGGCTGGAGAACCCTATGCTATTGCAGTAAATCCTAAA AGAGGGATGATGTACTGGACTGTTGTTGGGGATCATTCCCATATAGAAGAAGCAGCCATGGATGGTACACTGAGAAGGATTTTAGTACAA AAGAACTTACAGAGACCCACAGGTTTGGCTGTGGATTATTTTAGTGAACGCATATATTGGGCTGACTTTGAGCTCTCCATCATTGGCAGT GTTCTGTATGATGGCTCTAATTCAGTAGTCTCTGTCAGCAGCAAACAAGGTTTATTACATCCACATAGGATCGATATCTTTGAAGATTAT ATATATGGAGCAGGACCTAAAAATGGTGTATTTCGAGTTCAAAAATTTGGCCATGGTTCAGTAGAGTACTTAGCTTTAAATATTGATAAA ACAAAAGGTGTTTTGATATCTCATCGTTATAAACAACTAGATTTACCCAATCCATGCTTGGATTTAGCATGCGAATTTCTTTGCTTGCTA AATCCTTCTGGGGCCACTTGTGTGTGTCCAGAAGGAAAATATTTGATTAATGGCACCTGCAATGATGACAGCCTGTTAGATGATTCATGT AAGTTAACTTGTGAAAATGGAGGAAGATGCATTTTAAATGAGAAAGGTGATTTGAGGTGTCACTGTTGGCCCAGTTATTCAGGAGAAAGA TGTGAAGTCAACCACTGTAGCAACTACTGCCAGAATGGAGGAACTTGCGTACCATCAGTTCTAGGGAGACCCACCTGCAGCTGTGCACTG GGTTTCACTGGGCCAAACTGTGGTAAGACAGTCTGTGAGGATTTTTGTCAAAATGGAGGAACCTGCATTGTGACTGCTGGAAACCAGCCT TACTGCCACTGCCAGCCGGAATACACCGGAGACAGATGTCAGTACTACGTGTGCCACCACTATTGTGTGAATTCTGAATCATGTACCATT GGGGATGATGGAAGTGTTGAATGTGTCTGTCCAACGCGCTATGAAGGACCAAAATGTGAGGTTGACAAGTGTGTAAGGTGCCATGGGGGG CACTGCATTATAAATAAAGACAGTGAAGATATATTTTGCAACTGCACTAATGGAAAGATTGCCTCTAGCTGTCAGTTATGTGATGGCTAC TGTTACAATGGTGGCACATGCCAGCTGGACCCCGAGACAAATGTACCTGTGTGTCTATGCTCCACCAACTGGTCAGGCACACAGTGTGAA AGGCCAGCCCCAAAGAGCAGCAAGTCTGATCATATCAGCACAAGAAGCATTGCCATCATTGTGCCTCTCGTCCTCTTGGTGACTTTGATA ACCACCTTAGTAATTGGTTTAGTGCTTTGTAAAAGAAAAAGAAGGACAAAAACAATTAGAAGACAACCTATTATCAATGGAGGAATAAAT GTAGAAATTGGCAATCCATCTTATAACATGTATGAGGTAGATCATGATCACAACGATGGAGGTCTTTTAGATCCTGGCTTTATGATAGAC CCAACAAAGGCCAGGTACATAGGGGGAGGACCCAGTGCTTTCAAGCTTCCACACACAGCGCCGCCCATCTACCTAAACTCTGATTTGAAA GGACCACTAACTGCTGGGCCAACAAATTACTCCAATCCGGTATATGCAAAATTATATATGGATGGGCAAAACTGTCGAAACTCCTTAGGA AGTGTTGATGAAAGGAAAGAACTGCTTCCAAAGAAAATAGAAATTGGTATAAGAGAGACAGTGGCATAATCAGTGATATCTTTTATATGC TGTATAAATGTATAAAATATAAGGATTACTTTTGTATGTTCCAACAGTATTATACTTGTTTTGGCATCAGCATTACCTCTTTCTTTATCT TTTTCCTGGTTAATTGTTTTCTGAGTTTTTTGGGTTTTATTTTTTGCTGATGACTATTGATTGACCATTTGTATGGTATTTTTATGAAAA AGAACTGCACTACAGTACAATTTACAACAATGCTGCTGATATGACACACCTTTGAATTTGTTAAAATTAAAAACAACGTATTCCTTTGTA GTGTGAATATGAGCAATCTATTTTATATGAACTTTTTTGGTTGTACTTAATCAACGAGGAGAATCTCTGCACTTTTCCATTATACGGTTT GAAGGCTGTAATACAGTGTCATATTATTTTTCTGTTTAAATTGATGGAAAAATGATTGAATGGTCAACTCTCTTCTTTGTGCCCATAAAG ATCGATTCAGACTCTGCTGAAAATATATAGCTCTCACAAGTTCAGCATCACCTGCTTTGAAATTAGCCTTAGATTGCCAACCAATAGATG AGAATTTTGAGGAAAAAAATTAAAAATATGTAAAATTAATAATTTGCATGAACACAGATGACTACATTTTCCAAAACTTAGTGGACTCTA TGTGATGTACTAAATGTATACACCTTGTAAGCAATAGTTATATTTAGGTGGTAGAACATAGCAAAAATATAACCGAAAGTTGGCCGACTG CACTTGCTATGGAATAAGACCTTTTATTCTCCCTCAGTCTCGAGATAAATAGCCAGCCTAGAGCACAACAGGGCATTGGGTACTTGCATC TTAGGTATTTCTTCCCAGTCACATCCATTTTGTGGAAGATTAACCCAACCCCTTACACTACACTGAACACTAAAGAATAATATATAAGCA CACAAATTGGTGACAGAATTTCAATTACGTGAACGCATCCTCTTTGCTAGGTCAAAAAACAAAGGGCAAAGCAGACATTTTAGTATACAG AGTGATTGGCAAATATTTTCAAGATTTAATATGAGCAACCCATTATTTGCCCTATCCAAAATATATTCAAGGGCCTTCCAAGTTGTAGAA GAACAATGATCTTCCCATAATCAAAAGTGGAGAGTCGAAATGCTGTGCCAGTTGCTCTGGTATTCAGGTTTCTCTGGGTTTTACAGAACG CATGGACCCCATTCACGTTTGGTTTGTTTATCTTCAAATTTGAGTTGAAACGAGTGCGATTTATTTAAGTTGTATATAAAAATAAAAGGA TAGCATTTTTATACAAATATCTTTAAAGGCACAAAAGATTTATTCACAAGTTTTGGAGGGCTTTTTGTTCCTCTGATAGACATGACTGAC TTTTAGCTGTCATAATGTATTAACCTAACAGATGAAATATGTTAAATATGTGGTTGCTCTTTATCCCTTTGTACAAGCATTAAAAAAACT GCTGTTTTATAAGAAGACTTTTTGTTGTACTATGTGCATGCATACTACCTATTTCTAAACTTTGCCATATTGAGGCCTTTATAAACTATT GATTTATGTAATACTAGTGCAATTTTGCTTGAACAATGTTATGCATATCATAAACTTTTTCAGGTTCTTGTTTAAGTACATTTTTTAAAT TGAACAGTATTTTTCATTTTGGTTATAATATAGTCATTTTGCCTATGTTTCTACAATGAAGTGTTAAATACTTTATAAAAAATTGTTGAC >95587_95587_1_TXNL1-LRP1B_TXNL1_chr18_54305574_ENST00000217515_LRP1B_chr2_141299540_ENST00000389484_length(amino acids)=2284AA_BP=464 MRVPGEDSAVATSSVQASAFLSISPSSPRSSSPSVSKPVASRPPCRALCEDGGGEARRERPGFPARAERRGLQTRRGQVHHERVIVKSGP GTFLSLAVYDNYIFWSDWGRRAILRSNKYTGGDTKILRSDIPHQPMGIIAVANDTNSCELSPCALLNGGCHDLCLLTPNGRVNCSCRGDR ILLEDNRCVTKNSSCNAYSEFECGNGECIDYQLTCDGIPHCKDKSDEKLLYCENRSCRRGFKPCYNRRCIPHGKLCDGENDCGDNSDELD CKVSTCATVEFRCADGTCIPRSARCNQNIDCADASDEKNCNNTDCTHFYKLGVKTTGFIRCNSTSLCVLPTWICDGSNDCGDYSDELKCP VQNKHKCEENYFSCPSGRCILNTWICDGQKDCEDGRDEFHCDSSCSWNQFACSAQKCISKHWICDGEDDCGDGLDESDSICGAITCAADM FSCQGSRACVPRHWLCDGERDCPDGSDELSTAGCAPNNTCDENAFMCHNKVCIPKQFVCDHDDDCGDGSDESPQCGYRQCGTEEFSCADG RCLLNTQWQCDGDFDCPDHSDEAPLNPKCKSAEQSCNSSFFMCKNGRCIPSGGLCDNKDDCGDGSDERNCHINECLSKKVSGCSQDCQDL PVSYKCKCWPGFQLKDDGKTCVDIDECSSGFPCSQQCINTYGTYKCLCTDGYEIQPDNPNGCKSLSDEEPFLILADHHEIRKISTDGSNY TLLKQGLNNVIAIDFDYREEFIYWIDSSRPNGSRINRMCLNGSDIKVVHNTAVPNALAVDWIGKNLYWSDTEKRIIEVSKLNGLYPTILV SKRLKFPRDLSLDPQAGYLYWIDCCEYPHIGRVGMDGTNQSVVIETKISRPMALTIDYVNRRLYWADENHIEFSNMDGSHRHKVPNQDIP GVIALTLFEDYIYWTDGKTKSLSRAHKTSGADRLSLIYSWHAITDIQVYHSYRQPDVSKHLCMINNGGCSHLCLLAPGKTHTCACPTNFY LAADNRTCLSNCTASQFRCKTDKCIPFWWKCDTVDDCGDGSDEPDDCPEFRCQPGRFQCGTGLCALPAFICDGENDCGDNSDELNCDTHV CLSGQFKCTKNQKCIPVNLRCNGQDDCGDEEDERDCPENSCSPDYFQCKTTKHCISKLWVCDEDPDCADASDEANCDKKTCGPHEFQCKN NNCIPDHWRCDSQNDCSDNSDEENCKPQTCTLKDFLCANGDCVSSRFWCDGDFDCADGSDERNCETSCSKDQFRCSNGQCIPAKWKCDGH EDCKYGEDEKSCEPASPTCSSREYICASDGCISASLKCNGEYDCADGSDEMDCVTECKEDQFRCKNKAHCIPIRWLCDGIHDCVDGSDEE NCERGGNICRADEFLCNNSLCKLHFWVCDGEDDCGDNSDEAPDMCVKFLCPSTRPHRCRNNRICLQSEQMCNGIDECGDNSDEDHCGGKL TYKARPCKKDEFACSNKKCIPMDLQCDRLDDCGDGSDEQGCRIAPTEYTCEDNVNPCGDDAYCNQIKTSVFCRCKPGFQRNMKNRQCEDL NECLVFGTCSHQCINVEGSYKCVCDQNFQERNNTCIAEGSEDQVLYIANDTDILGFIYPFNYSGDHQQISHIEHNSRITGMDVYYQRDMI IWSTQFNPGGIFYKRIHGREKRQANSGLICPEFKRPRDIAVDWVAGNIYWTDHSRMHWFSYYTTHWTSLRYSINVGQLNGPNCTRLLTNM AGEPYAIAVNPKRGMMYWTVVGDHSHIEEAAMDGTLRRILVQKNLQRPTGLAVDYFSERIYWADFELSIIGSVLYDGSNSVVSVSSKQGL LHPHRIDIFEDYIYGAGPKNGVFRVQKFGHGSVEYLALNIDKTKGVLISHRYKQLDLPNPCLDLACEFLCLLNPSGATCVCPEGKYLING TCNDDSLLDDSCKLTCENGGRCILNEKGDLRCHCWPSYSGERCEVNHCSNYCQNGGTCVPSVLGRPTCSCALGFTGPNCGKTVCEDFCQN GGTCIVTAGNQPYCHCQPEYTGDRCQYYVCHHYCVNSESCTIGDDGSVECVCPTRYEGPKCEVDKCVRCHGGHCIINKDSEDIFCNCTNG KIASSCQLCDGYCYNGGTCQLDPETNVPVCLCSTNWSGTQCERPAPKSSKSDHISTRSIAIIVPLVLLVTLITTLVIGLVLCKRKRRTKT IRRQPIINGGINVEIGNPSYNMYEVDHDHNDGGLLDPGFMIDPTKARYIGGGPSAFKLPHTAPPIYLNSDLKGPLTAGPTNYSNPVYAKL -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TXNL1-LRP1B |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TXNL1-LRP1B |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TXNL1-LRP1B |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
