
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:TYK2-ZNRF1 (FusionGDB2 ID:95660) |
Fusion Gene Summary for TYK2-ZNRF1 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: TYK2-ZNRF1 | Fusion gene ID: 95660 | Hgene | Tgene | Gene symbol | TYK2 | ZNRF1 | Gene ID | 7297 | 84937 |
| Gene name | tyrosine kinase 2 | zinc and ring finger 1 | |
| Synonyms | IMD35|JTK1 | NIN283 | |
| Cytomap | 19p13.2 | 16q23.1 | |
| Type of gene | protein-coding | protein-coding | |
| Description | non-receptor tyrosine-protein kinase TYK2 | E3 ubiquitin-protein ligase ZNRF1RING-type E3 ubiquitin transferase ZNRF1nerve injury gene 283nerve injury-induced gene 283 proteinzinc and ring finger 1, E3 ubiquitin protein ligasezinc and ring finger protein 1zinc/RING finger protein 1 | |
| Modification date | 20200329 | 20200313 | |
| UniProtAcc | . | . | |
| Ensembl transtripts involved in fusion gene | ENST00000264818, ENST00000525621, ENST00000529370, ENST00000524462, ENST00000529422, | ENST00000564320, ENST00000320619, ENST00000335325, ENST00000566250, ENST00000567962, | |
| Fusion gene scores | * DoF score | 13 X 11 X 8=1144 | 7 X 7 X 6=294 |
| # samples | 15 | 9 | |
| ** MAII score | log2(15/1144*10)=-2.93105264628251 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(9/294*10)=-1.70781924850669 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: TYK2 [Title/Abstract] AND ZNRF1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | TYK2(10488889)-ZNRF1(75127469), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | TYK2-ZNRF1 seems lost the major protein functional domain in Hgene partner, which is a IUPHAR drug target due to the frame-shifted ORF. TYK2-ZNRF1 seems lost the major protein functional domain in Hgene partner, which is a kinase due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
 Fusion gene breakpoints across TYK2 (5'-gene) Fusion gene breakpoints across TYK2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
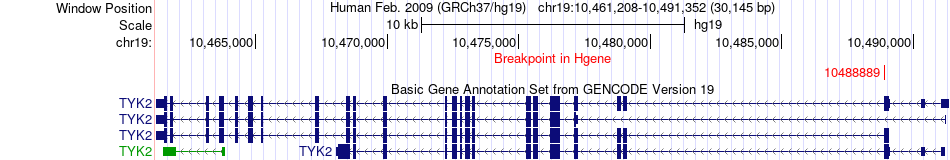 |
 Fusion gene breakpoints across ZNRF1 (3'-gene) Fusion gene breakpoints across ZNRF1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
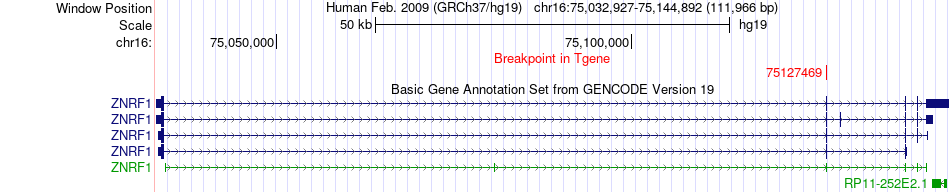 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-30-1866 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
Top |
Fusion Gene ORF analysis for TYK2-ZNRF1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-3UTR | ENST00000264818 | ENST00000564320 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| 5CDS-3UTR | ENST00000525621 | ENST00000564320 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| 5CDS-3UTR | ENST00000529370 | ENST00000564320 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| Frame-shift | ENST00000264818 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| Frame-shift | ENST00000264818 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| Frame-shift | ENST00000264818 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| Frame-shift | ENST00000264818 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000525621 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000525621 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000525621 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000525621 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000529370 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000529370 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000529370 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| In-frame | ENST00000529370 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000524462 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000524462 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000524462 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000524462 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000529422 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000529422 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000529422 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3CDS | ENST00000529422 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3UTR | ENST00000524462 | ENST00000564320 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
| intron-3UTR | ENST00000529422 | ENST00000564320 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000525621 | TYK2 | chr19 | 10488889 | - | ENST00000335325 | ZNRF1 | chr16 | 75127469 | + | 4229 | 675 | 2158 | 2625 | 155 |
| ENST00000525621 | TYK2 | chr19 | 10488889 | - | ENST00000320619 | ZNRF1 | chr16 | 75127469 | + | 2095 | 675 | 482 | 1087 | 201 |
| ENST00000525621 | TYK2 | chr19 | 10488889 | - | ENST00000566250 | ZNRF1 | chr16 | 75127469 | + | 993 | 675 | 482 | 880 | 132 |
| ENST00000525621 | TYK2 | chr19 | 10488889 | - | ENST00000567962 | ZNRF1 | chr16 | 75127469 | + | 1158 | 675 | 482 | 934 | 150 |
| ENST00000529370 | TYK2 | chr19 | 10488889 | - | ENST00000335325 | ZNRF1 | chr16 | 75127469 | + | 4078 | 524 | 2007 | 2474 | 155 |
| ENST00000529370 | TYK2 | chr19 | 10488889 | - | ENST00000320619 | ZNRF1 | chr16 | 75127469 | + | 1944 | 524 | 331 | 936 | 201 |
| ENST00000529370 | TYK2 | chr19 | 10488889 | - | ENST00000566250 | ZNRF1 | chr16 | 75127469 | + | 842 | 524 | 331 | 729 | 132 |
| ENST00000529370 | TYK2 | chr19 | 10488889 | - | ENST00000567962 | ZNRF1 | chr16 | 75127469 | + | 1007 | 524 | 331 | 783 | 150 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000525621 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.74950296 | 0.250497 |
| ENST00000525621 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.18169712 | 0.8183029 |
| ENST00000525621 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.0692551 | 0.9307449 |
| ENST00000525621 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.051803567 | 0.9481965 |
| ENST00000529370 | ENST00000335325 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.77538246 | 0.22461751 |
| ENST00000529370 | ENST00000320619 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.21977435 | 0.7802256 |
| ENST00000529370 | ENST00000566250 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.05344394 | 0.9465561 |
| ENST00000529370 | ENST00000567962 | TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.050379977 | 0.94962 |
Top |
Fusion Genomic Features for TYK2-ZNRF1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.010048061 | 0.9899519 |
| TYK2 | chr19 | 10488889 | - | ZNRF1 | chr16 | 75127469 | + | 0.010048061 | 0.9899519 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
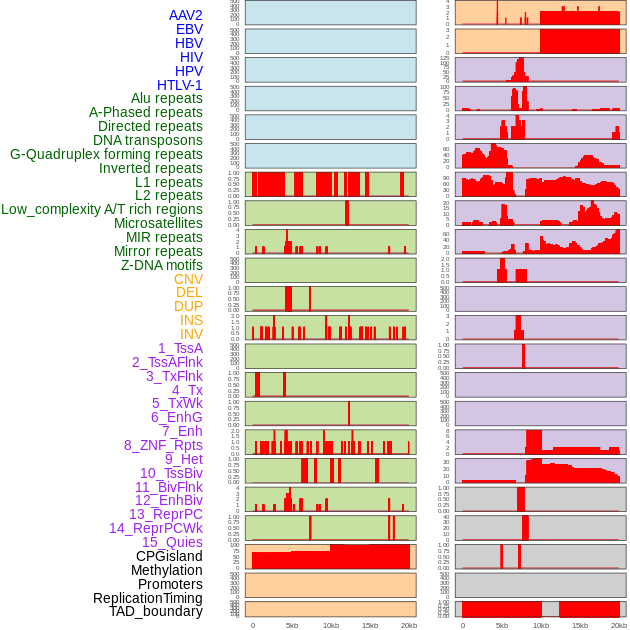 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
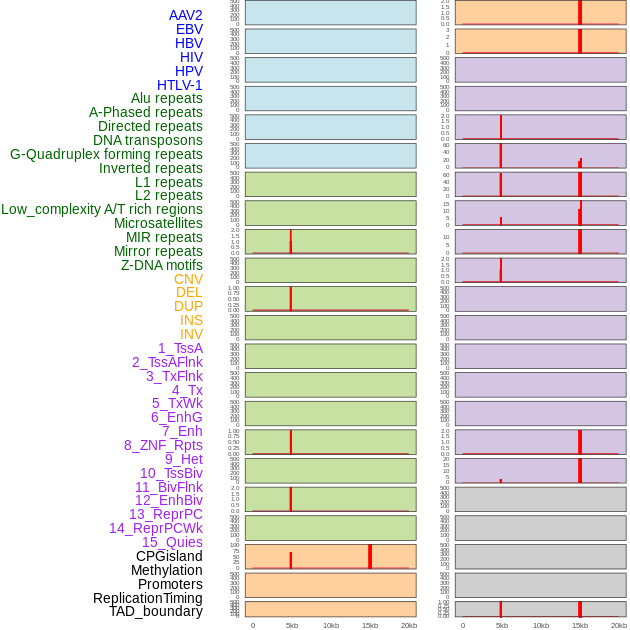 |
Top |
Fusion Protein Features for TYK2-ZNRF1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:10488889/chr16:75127469) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | . |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ZNRF1 | chr19:10488889 | chr16:75127469 | ENST00000320619 | 0 | 6 | 184_225 | 141 | 251.0 | Zinc finger | RING-type%3B atypical | |
| Tgene | ZNRF1 | chr19:10488889 | chr16:75127469 | ENST00000335325 | 0 | 5 | 184_225 | 141 | 960.0 | Zinc finger | RING-type%3B atypical | |
| Tgene | ZNRF1 | chr19:10488889 | chr16:75127469 | ENST00000567962 | 0 | 5 | 184_225 | 141 | 41.333333333333336 | Zinc finger | RING-type%3B atypical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000264818 | - | 1 | 23 | 26_431 | 64 | 1188.0 | Domain | FERM |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000264818 | - | 1 | 23 | 450_529 | 64 | 1188.0 | Domain | Note=SH2%3B atypical |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000264818 | - | 1 | 23 | 589_875 | 64 | 1188.0 | Domain | Protein kinase 1 |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000264818 | - | 1 | 23 | 897_1176 | 64 | 1188.0 | Domain | Protein kinase 2 |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000525621 | - | 3 | 25 | 26_431 | 64 | 1188.0 | Domain | FERM |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000525621 | - | 3 | 25 | 450_529 | 64 | 1188.0 | Domain | Note=SH2%3B atypical |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000525621 | - | 3 | 25 | 589_875 | 64 | 1188.0 | Domain | Protein kinase 1 |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000525621 | - | 3 | 25 | 897_1176 | 64 | 1188.0 | Domain | Protein kinase 2 |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000264818 | - | 1 | 23 | 903_911 | 64 | 1188.0 | Nucleotide binding | ATP |
| Hgene | TYK2 | chr19:10488889 | chr16:75127469 | ENST00000525621 | - | 3 | 25 | 903_911 | 64 | 1188.0 | Nucleotide binding | ATP |
| Tgene | ZNRF1 | chr19:10488889 | chr16:75127469 | ENST00000320619 | 0 | 6 | 2_10 | 141 | 251.0 | Region | Note=Required for endosomal and lysosomal localization and myristoylation | |
| Tgene | ZNRF1 | chr19:10488889 | chr16:75127469 | ENST00000335325 | 0 | 5 | 2_10 | 141 | 960.0 | Region | Note=Required for endosomal and lysosomal localization and myristoylation | |
| Tgene | ZNRF1 | chr19:10488889 | chr16:75127469 | ENST00000567962 | 0 | 5 | 2_10 | 141 | 41.333333333333336 | Region | Note=Required for endosomal and lysosomal localization and myristoylation |
Top |
Fusion Gene Sequence for TYK2-ZNRF1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >95660_95660_1_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000320619_length(transcript)=2095nt_BP=675nt AAAAAAAAAAAAATCCAATGGTGACGAGCAGGGAGAACAGAGCAGCTGCCAATGGGCGTGTGCGTTTCAGGCGGCCAATGGGAGGAGGCG TCTCGGCGGGGGACAAGCAGTAGCTACCCGCGGGAGCGGGGAGGGGTCCGGGTTCGAGCTTGTGTTCCCCCGGAAGGGTGAGTCTGGACG CGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCTTGACAGGCTGCACTTGGATGGGAGCACCT GGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATATTGAAACATTACAACCATCTTTTGATGGC AACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGCTTGCTTGAGTTGACACAGGGAGCTCCAGG ACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAGGGGCAGTAAGCCCGTTGGGGATGGAGCCC AGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGGGGAGCCCTGGGTCACTTTCAGTGAGTCAT CGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCATTTGCTCCAAGTCTGTGGCTTCTGACGAGA TGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGGAAGAATACAAAGAAGCCTCAGAGGGGCTCAGCCTGAGA AAACCTGGCTCTCAGGAAGAACTTTGCAAGAGCAGCCGTGGAGGACAGAGTGCTCTTGGGCCCCCATGGCCCAAATGCAGAGTTACCCCC ACTGCCCTGTGGAGAACCGGGAAGATGATGTGCTGACTAAAGACGCGGGTGAGTGTGTGATCTGCCTGGAGGAGCTGCTGCAGGGGGACA CGATAGCCAGGCTGCCCTGCCTGTGCATCTATCACAAAAGCTGCATAGACTCGTGGTTTGAAGTGAACAGATCTTGTCCGGAACACCCTG CGGACTGACCTGCGGGCTTGCTTGCTGACTCCTCTCAAAGGGACAGAGCGCCCCTGCTCCAGGGAGGAGGCTCACCGGACCCTGGGGCAG AGCTGAGCTTGGGACACCAGCGGGAACAGGGCACCCCTTCTGCACTGACTTCCAGATCATGGTTCTCCCTTCCTCCCTGAGGACACCAAA TTGGATGAGAGCAAGTTTGAGAGAAGAATGAATCAACTGCTATCCTTCCCCTCACCCCTCAGCCCAGGAGGGAAAGGGCATTTTCTTTTT CATCTTTGAAAGGCATTGTGGGTCTGTCTTTAAAGTGTTTACAAAAAAAAATTATATAAAAAAAAAGTCTAGTGTCGACTGGTGTTTTCC CTCGTGATGTTTACAGCTTGCTGTTTGCTGCCCAGCCATAACCCACTCAGTGACAGACGAACACAGCTAAGGCCTCGCTGCCAGCCTTCT GACGGCGGGCGAGCACACGGTCTCTTCCCCTGCCCCAACTGGCCCTCTCCACCACCAACTTCTCTACTTTGGGTTTGTTTGGTTTTTTCC CTCTATTTTTCTGGCTGGTTTTTTGCTCTTTTCTCCCCTTGAGACCCTAATATCTTGACTTCATTGCCAAAAAACAACCCATTGAGAACT TTCTTCCTACTGATCCCATCTCTTTTCCCTTCTTTCCTCCTGGTTCCGGTCAGTTCAGAGGATTTAACAATCACAAGTGTCCTGCAAAAA TGCCTGAACATTATTCTTAGGCCCTCGTGGATTTTTTTTTTCAGAAAACTTAAACAAAAAAAGACTTACTAAGAAATATGTACAGCTACC CCTGTTTTCAGGCACTATGTTTGAGAACATTTTAGCCATTGATGTTCACACGTGGCATCAGCCCATGCAAGATAGGTTTCTGTATTTATA TATTAAAATACAAAAAAAAACTTATAAAATGTTTAAAAAAATGTTCAAAGCTTGGGAGAAAAGCTTTCTTCATTAGTCAAGGTGTTTTGA >95660_95660_1_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000320619_length(amino acids)=201AA_BP=64 MPLRHWGMARGSKPVGDGAQPMAAMGGLKVLLHWAGPGGGEPWVTFSESSLTAEEVCIHIAHKVGFKCPICSKSVASDEMEMHFIMCLSK PRLSYNGRIQRSLRGAQPEKTWLSGRTLQEQPWRTECSWAPMAQMQSYPHCPVENREDDVLTKDAGECVICLEELLQGDTIARLPCLCIY -------------------------------------------------------------- >95660_95660_2_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000335325_length(transcript)=4229nt_BP=675nt AAAAAAAAAAAAATCCAATGGTGACGAGCAGGGAGAACAGAGCAGCTGCCAATGGGCGTGTGCGTTTCAGGCGGCCAATGGGAGGAGGCG TCTCGGCGGGGGACAAGCAGTAGCTACCCGCGGGAGCGGGGAGGGGTCCGGGTTCGAGCTTGTGTTCCCCCGGAAGGGTGAGTCTGGACG CGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCTTGACAGGCTGCACTTGGATGGGAGCACCT GGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATATTGAAACATTACAACCATCTTTTGATGGC AACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGCTTGCTTGAGTTGACACAGGGAGCTCCAGG ACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAGGGGCAGTAAGCCCGTTGGGGATGGAGCCC AGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGGGGAGCCCTGGGTCACTTTCAGTGAGTCAT CGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCATTTGCTCCAAGTCTGTGGCTTCTGACGAGA TGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGATGATGTGCTGACTAAAGACGCGGGTGAGTGTGTGATCT GCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCACAAAAGCTGCATAGACTCGTGGTTTGAAG TGAACAGATCTTGTCCGGAACACCCTGCGGACTGACCTGCGGGCTTGCTTGCTGACTCCTCTCAAAGGGACAGAGCGCCCCTGCTCCAGG GAGGAGGCTCACCGGACCCTGGGGCAGAGCTGAGCTTGGGACACCAGCGGGAACAGGGCACCCCTTCTGCACTGACTTCCAGATCATGGT TCTCCCTTCCTCCCTGAGGACACCAAATTGGATGAGAGCAAGTTTGAGAGAAGAATGAATCAACTGCTATCCTTCCCCTCACCCCTCAGC CCAGGAGGGAAAGGGCATTTTCTTTTTCATCTTTGAAAGGCATTGTGGGTCTGTCTTTAAAGTGTTTACAAAAAAAAATTATATAAAAAA AAAGTCTAGTGTCGACTGGTGTTTTCCCTCGTGATGTTTACAGCTTGCTGTTTGCTGCCCAGCCATAACCCACTCAGTGACAGACGAACA CAGCTAAGGCCTCGCTGCCAGCCTTCTGACGGCGGGCGAGCACACGGTCTCTTCCCCTGCCCCAACTGGCCCTCTCCACCACCAACTTCT CTACTTTGGGTTTGTTTGGTTTTTTCCCTCTATTTTTCTGGCTGGTTTTTTGCTCTTTTCTCCCCTTGAGACCCTAATATCTTGACTTCA TTGCCAAAAAACAACCCATTGAGAACTTTCTTCCTACTGATCCCATCTCTTTTCCCTTCTTTCCTCCTGGTTCCGGTCAGTTCAGAGGAT TTAACAATCACAAGTGTCCTGCAAAAATGCCTGAACATTATTCTTAGGCCCTCGTGGATTTTTTTTTTCAGAAAACTTAAACAAAAAAAG ACTTACTAAGAAATATGTACAGCTACCCCTGTTTTCAGGCACTATGTTTGAGAACATTTTAGCCATTGATGTTCACACGTGGCATCAGCC CATGCAAGATAGGTTTCTGTATTTATATATTAAAATACAAAAAAAAACTTATAAAATGTTTAAAAAAATGTTCAAAGCTTGGGAGAAAAG CTTTCTTCATTAGTCAAGGTGTTTTGATATGGTATTAAAATGTCTAATAAAAGATGGCACTGCGTGATTTTATTTTAATGAAGTGTTATA CAATCAAGAAATGGGGGCAAGGGCCTGCCCCCCACTCCCTCACCTACCTCCTTAGCATTCCTTCAGGCTGCTACTCGGGGCTCCAGGTGT GTGAATTGGTCCTCAGATAGTCAGGCCTGGGTGGAGGGAGTGGCAAGTCAGGCGTGCCTCCTACAAGCTTCCTAACCTCTTAAGCATCAT GGAACCAGTCAGCCCTCTGTGGAGTCATTGTGCTGGACCTCTGAAAAGATCTGCAGGGGCCAAGATGTTGCAGCCACCGGAGGCTGCAAG GATGTGGGTTCTTCCAGGTGTGGGCCCAGCCCCCCTCCTTCCAGCCTTTGCTCCCCATCCCACGTTCCACTCGCCCTGCCTGTTGTTCAG TTTGCGTCTCAGTGCTGACTCACGGGCATGCTTCATTGAGGCCCAGGAAGAGGCCCTGGTTTGGGGCTGTGCCAGCTCAGAGCCCTTGAA CCAGAACCAACTGCTCAGGCTCACAAAAGTTGGCAAAATGCGGGCTGGGCAGGCAGCTCGGGGCAGGGAGCAGCAGTGCAGGCCTGGCCT GTGTCCCCGTGCCCAGCGGGCTCCCGGAGCAGCATTCCAGAGCCTCCTGCAGAGCCAGCGAAGGAAAGCTCTAGAGGGAGACGACTCCAC CGCCTCTCTGCTCTGACCCTTCTCAGGCCCGCCTGATGTGCTGGACCATCCCCCTGCTGCCACGGCCCCTGGCTCCGCAGTCACCCTCCA TCAGATGCTTCCAGAAGCACCTACTCTGTGCAAGGGCATTCACAACAGGCCAGTGGGCAAACGTGAGCCAGGGCCAGCGGAGAAACACTA AAGAAGACTCGGTGGTTCAGAGCCTGGGTCTCAGCGCGGGACCCGTCTGGGGTGAAGACCTTGGGGCTGTTGTGAGTCGGTGGCAGGAAC GTGGGCTCTAGACTGTGCATTCAGGCTCTCCTACTTGGCAGAATGATCTTGGGGAAACGACTTCATCTGAACTTCAGATATTTCACATGT GAAGCGGGGACAAAACCATGCAGCTCAGAGGTCCCTGTGGGGGCTGGGGGAGCTGCCCTGCAGGTCTTGGCACATGCACAGCAGGCTCCC CATAGCTTTGTCACCACAAAGGGCACTGTTCTATTCACAGCACCTCCTGCTTCTGCCTGGCAACTGTGTCTCCCTGTGCTATATTTAATT CCACCAGCAAAGCTGGCGAGGCAGGGCCCAGCCCTGAAGGAGATCTCCTTGCCTGACCCCTGGACCTGGAAATGGAGGCTTCATGTGCCC GCCTTGGCGGCTTAAGCCTGCTGCTTTGGCAGTGCCATGGGTGAGCCGAGCAGCTGTGAGGTGGGTGGGGCAGGGCTGTAGCCCACGCCG GGTGCTATTCCAGGCTCTAGGGGCTGGTGCTCATCCCCACCCCCAGCGACTTCCGTCCTACCTGGCATGCTGCAGCCCTCTGCCGGCTGT GGTGTCCTCTGAATTTGGACCCAGGTTGCCTGCTCTTGTTGTGGGGGTGGGATTGGGGCTGTCTGTGTTTCCCAGGTGGAGCTCATGTTT GTGATCCTCTATCTGGACAGCTCTCCAGAATCCTGTGTTGCCCCTGTTCTGCCTTTCTGGGATGGGAGAGAAGGTGAGGGAGCCGGCGAG CAGGTTCTACCCTCTCTGCTAAGGGCCTAGCACACTTTGATGAGGTTCCTGGACGTGAGACCCTCACAAAGACCAACCATTCCTCTAATA GCCATTTAACGGTACTGCAGTTCATGGGAAAATGTCCTCATTCTTAGGAGATAGATGCTGGATTATATAGGTGAATGCCTTCAAGAAATG TGTAATTTAATTCCAAATAGTTCCCCCCAAAATACACAAAGCAAATAAGGCCAAAATGTTCATTGTTGAATCTGCAGTGGCTGGTATTTC AGTGAACAGTGTACTTGTTCTTTCAACTTTTCTGTATGTTTGGAATTTTTCTTAATAAAAGATTGTGGGAAATGGTCATTTGCTGAGCGC CTCCTAGTTGCCAGGCACTGGAGGGCGCATCAAAGAAGAGTCCTACCCAGGAGGGAGAGCGCAGCTGCTGCAGGATTCTCGCCTGGAAGA GTGGGTGGCTGGCTGGCTGGCTGGAAGGGGAGGGGCGAGAGGGCTGTCTCGGTGATGAGCACACGCGTGCTGGGGTGGTTTGGGGAGAGG >95660_95660_2_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000335325_length(amino acids)=155AA_BP= MEPVSPLWSHCAGPLKRSAGAKMLQPPEAARMWVLPGVGPAPLLPAFAPHPTFHSPCLLFSLRLSADSRACFIEAQEEALVWGCASSEPL -------------------------------------------------------------- >95660_95660_3_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000566250_length(transcript)=993nt_BP=675nt AAAAAAAAAAAAATCCAATGGTGACGAGCAGGGAGAACAGAGCAGCTGCCAATGGGCGTGTGCGTTTCAGGCGGCCAATGGGAGGAGGCG TCTCGGCGGGGGACAAGCAGTAGCTACCCGCGGGAGCGGGGAGGGGTCCGGGTTCGAGCTTGTGTTCCCCCGGAAGGGTGAGTCTGGACG CGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCTTGACAGGCTGCACTTGGATGGGAGCACCT GGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATATTGAAACATTACAACCATCTTTTGATGGC AACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGCTTGCTTGAGTTGACACAGGGAGCTCCAGG ACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAGGGGCAGTAAGCCCGTTGGGGATGGAGCCC AGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGGGGAGCCCTGGGTCACTTTCAGTGAGTCAT CGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCATTTGCTCCAAGTCTGTGGCTTCTGACGAGA TGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGATGATGTGCTGACTAAAGACGCGGGTGAGTGTGTGATCT GCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCACAAAAGGTAGGAGGGGTTGGCAAGGTAGC TTAGTGCACCCCACCTCCCAGAGGGGCGGACCCCCATCTCCAGCCATTCTGTCTCCTTTGGTTATGGGATGACCTCTCCCCCGACCTCTG >95660_95660_3_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000566250_length(amino acids)=132AA_BP=64 MPLRHWGMARGSKPVGDGAQPMAAMGGLKVLLHWAGPGGGEPWVTFSESSLTAEEVCIHIAHKVGFKCPICSKSVASDEMEMHFIMCLSK -------------------------------------------------------------- >95660_95660_4_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000567962_length(transcript)=1158nt_BP=675nt AAAAAAAAAAAAATCCAATGGTGACGAGCAGGGAGAACAGAGCAGCTGCCAATGGGCGTGTGCGTTTCAGGCGGCCAATGGGAGGAGGCG TCTCGGCGGGGGACAAGCAGTAGCTACCCGCGGGAGCGGGGAGGGGTCCGGGTTCGAGCTTGTGTTCCCCCGGAAGGGTGAGTCTGGACG CGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCTTGACAGGCTGCACTTGGATGGGAGCACCT GGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATATTGAAACATTACAACCATCTTTTGATGGC AACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGCTTGCTTGAGTTGACACAGGGAGCTCCAGG ACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAGGGGCAGTAAGCCCGTTGGGGATGGAGCCC AGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGGGGAGCCCTGGGTCACTTTCAGTGAGTCAT CGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCATTTGCTCCAAGTCTGTGGCTTCTGACGAGA TGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGATGATGTGCTGACTAAAGACGCGGGTGAGTGTGTGATCT GCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCACAAAAGCTGCATAGACTCGTGGTTTGAAG TGAACAGATCTTGTCCGGAACACCCTGCGGACTGACCTGCGGGCTTGCTTGCTGACTCCTCTCAAAGATCATGGTTCTCCCTTCCTCCCT GAGGACACCAAATTGGATGAGAGCAAGTTTGAGAGAAGAATGAATCAACTGCTATCCTTCCCCTCACCCCTCAGCCCAGGAGGGAAAGGG >95660_95660_4_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000525621_ZNRF1_chr16_75127469_ENST00000567962_length(amino acids)=150AA_BP=64 MPLRHWGMARGSKPVGDGAQPMAAMGGLKVLLHWAGPGGGEPWVTFSESSLTAEEVCIHIAHKVGFKCPICSKSVASDEMEMHFIMCLSK -------------------------------------------------------------- >95660_95660_5_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000320619_length(transcript)=1944nt_BP=524nt GTGTTCCCCCGGAAGGGTGAGTCTGGACGCGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCT TGACAGGCTGCACTTGGATGGGAGCACCTGGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATA TTGAAACATTACAACCATCTTTTGATGGCAACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGC TTGCTTGAGTTGACACAGGGAGCTCCAGGACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAG GGGCAGTAAGCCCGTTGGGGATGGAGCCCAGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGG GGAGCCCTGGGTCACTTTCAGTGAGTCATCGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCAT TTGCTCCAAGTCTGTGGCTTCTGACGAGATGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGGAAGAATACA AAGAAGCCTCAGAGGGGCTCAGCCTGAGAAAACCTGGCTCTCAGGAAGAACTTTGCAAGAGCAGCCGTGGAGGACAGAGTGCTCTTGGGC CCCCATGGCCCAAATGCAGAGTTACCCCCACTGCCCTGTGGAGAACCGGGAAGATGATGTGCTGACTAAAGACGCGGGTGAGTGTGTGAT CTGCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCACAAAAGCTGCATAGACTCGTGGTTTGA AGTGAACAGATCTTGTCCGGAACACCCTGCGGACTGACCTGCGGGCTTGCTTGCTGACTCCTCTCAAAGGGACAGAGCGCCCCTGCTCCA GGGAGGAGGCTCACCGGACCCTGGGGCAGAGCTGAGCTTGGGACACCAGCGGGAACAGGGCACCCCTTCTGCACTGACTTCCAGATCATG GTTCTCCCTTCCTCCCTGAGGACACCAAATTGGATGAGAGCAAGTTTGAGAGAAGAATGAATCAACTGCTATCCTTCCCCTCACCCCTCA GCCCAGGAGGGAAAGGGCATTTTCTTTTTCATCTTTGAAAGGCATTGTGGGTCTGTCTTTAAAGTGTTTACAAAAAAAAATTATATAAAA AAAAAGTCTAGTGTCGACTGGTGTTTTCCCTCGTGATGTTTACAGCTTGCTGTTTGCTGCCCAGCCATAACCCACTCAGTGACAGACGAA CACAGCTAAGGCCTCGCTGCCAGCCTTCTGACGGCGGGCGAGCACACGGTCTCTTCCCCTGCCCCAACTGGCCCTCTCCACCACCAACTT CTCTACTTTGGGTTTGTTTGGTTTTTTCCCTCTATTTTTCTGGCTGGTTTTTTGCTCTTTTCTCCCCTTGAGACCCTAATATCTTGACTT CATTGCCAAAAAACAACCCATTGAGAACTTTCTTCCTACTGATCCCATCTCTTTTCCCTTCTTTCCTCCTGGTTCCGGTCAGTTCAGAGG ATTTAACAATCACAAGTGTCCTGCAAAAATGCCTGAACATTATTCTTAGGCCCTCGTGGATTTTTTTTTTCAGAAAACTTAAACAAAAAA AGACTTACTAAGAAATATGTACAGCTACCCCTGTTTTCAGGCACTATGTTTGAGAACATTTTAGCCATTGATGTTCACACGTGGCATCAG CCCATGCAAGATAGGTTTCTGTATTTATATATTAAAATACAAAAAAAAACTTATAAAATGTTTAAAAAAATGTTCAAAGCTTGGGAGAAA >95660_95660_5_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000320619_length(amino acids)=201AA_BP=64 MPLRHWGMARGSKPVGDGAQPMAAMGGLKVLLHWAGPGGGEPWVTFSESSLTAEEVCIHIAHKVGFKCPICSKSVASDEMEMHFIMCLSK PRLSYNGRIQRSLRGAQPEKTWLSGRTLQEQPWRTECSWAPMAQMQSYPHCPVENREDDVLTKDAGECVICLEELLQGDTIARLPCLCIY -------------------------------------------------------------- >95660_95660_6_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000335325_length(transcript)=4078nt_BP=524nt GTGTTCCCCCGGAAGGGTGAGTCTGGACGCGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCT TGACAGGCTGCACTTGGATGGGAGCACCTGGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATA TTGAAACATTACAACCATCTTTTGATGGCAACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGC TTGCTTGAGTTGACACAGGGAGCTCCAGGACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAG GGGCAGTAAGCCCGTTGGGGATGGAGCCCAGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGG GGAGCCCTGGGTCACTTTCAGTGAGTCATCGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCAT TTGCTCCAAGTCTGTGGCTTCTGACGAGATGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGATGATGTGCT GACTAAAGACGCGGGTGAGTGTGTGATCTGCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCA CAAAAGCTGCATAGACTCGTGGTTTGAAGTGAACAGATCTTGTCCGGAACACCCTGCGGACTGACCTGCGGGCTTGCTTGCTGACTCCTC TCAAAGGGACAGAGCGCCCCTGCTCCAGGGAGGAGGCTCACCGGACCCTGGGGCAGAGCTGAGCTTGGGACACCAGCGGGAACAGGGCAC CCCTTCTGCACTGACTTCCAGATCATGGTTCTCCCTTCCTCCCTGAGGACACCAAATTGGATGAGAGCAAGTTTGAGAGAAGAATGAATC AACTGCTATCCTTCCCCTCACCCCTCAGCCCAGGAGGGAAAGGGCATTTTCTTTTTCATCTTTGAAAGGCATTGTGGGTCTGTCTTTAAA GTGTTTACAAAAAAAAATTATATAAAAAAAAAGTCTAGTGTCGACTGGTGTTTTCCCTCGTGATGTTTACAGCTTGCTGTTTGCTGCCCA GCCATAACCCACTCAGTGACAGACGAACACAGCTAAGGCCTCGCTGCCAGCCTTCTGACGGCGGGCGAGCACACGGTCTCTTCCCCTGCC CCAACTGGCCCTCTCCACCACCAACTTCTCTACTTTGGGTTTGTTTGGTTTTTTCCCTCTATTTTTCTGGCTGGTTTTTTGCTCTTTTCT CCCCTTGAGACCCTAATATCTTGACTTCATTGCCAAAAAACAACCCATTGAGAACTTTCTTCCTACTGATCCCATCTCTTTTCCCTTCTT TCCTCCTGGTTCCGGTCAGTTCAGAGGATTTAACAATCACAAGTGTCCTGCAAAAATGCCTGAACATTATTCTTAGGCCCTCGTGGATTT TTTTTTTCAGAAAACTTAAACAAAAAAAGACTTACTAAGAAATATGTACAGCTACCCCTGTTTTCAGGCACTATGTTTGAGAACATTTTA GCCATTGATGTTCACACGTGGCATCAGCCCATGCAAGATAGGTTTCTGTATTTATATATTAAAATACAAAAAAAAACTTATAAAATGTTT AAAAAAATGTTCAAAGCTTGGGAGAAAAGCTTTCTTCATTAGTCAAGGTGTTTTGATATGGTATTAAAATGTCTAATAAAAGATGGCACT GCGTGATTTTATTTTAATGAAGTGTTATACAATCAAGAAATGGGGGCAAGGGCCTGCCCCCCACTCCCTCACCTACCTCCTTAGCATTCC TTCAGGCTGCTACTCGGGGCTCCAGGTGTGTGAATTGGTCCTCAGATAGTCAGGCCTGGGTGGAGGGAGTGGCAAGTCAGGCGTGCCTCC TACAAGCTTCCTAACCTCTTAAGCATCATGGAACCAGTCAGCCCTCTGTGGAGTCATTGTGCTGGACCTCTGAAAAGATCTGCAGGGGCC AAGATGTTGCAGCCACCGGAGGCTGCAAGGATGTGGGTTCTTCCAGGTGTGGGCCCAGCCCCCCTCCTTCCAGCCTTTGCTCCCCATCCC ACGTTCCACTCGCCCTGCCTGTTGTTCAGTTTGCGTCTCAGTGCTGACTCACGGGCATGCTTCATTGAGGCCCAGGAAGAGGCCCTGGTT TGGGGCTGTGCCAGCTCAGAGCCCTTGAACCAGAACCAACTGCTCAGGCTCACAAAAGTTGGCAAAATGCGGGCTGGGCAGGCAGCTCGG GGCAGGGAGCAGCAGTGCAGGCCTGGCCTGTGTCCCCGTGCCCAGCGGGCTCCCGGAGCAGCATTCCAGAGCCTCCTGCAGAGCCAGCGA AGGAAAGCTCTAGAGGGAGACGACTCCACCGCCTCTCTGCTCTGACCCTTCTCAGGCCCGCCTGATGTGCTGGACCATCCCCCTGCTGCC ACGGCCCCTGGCTCCGCAGTCACCCTCCATCAGATGCTTCCAGAAGCACCTACTCTGTGCAAGGGCATTCACAACAGGCCAGTGGGCAAA CGTGAGCCAGGGCCAGCGGAGAAACACTAAAGAAGACTCGGTGGTTCAGAGCCTGGGTCTCAGCGCGGGACCCGTCTGGGGTGAAGACCT TGGGGCTGTTGTGAGTCGGTGGCAGGAACGTGGGCTCTAGACTGTGCATTCAGGCTCTCCTACTTGGCAGAATGATCTTGGGGAAACGAC TTCATCTGAACTTCAGATATTTCACATGTGAAGCGGGGACAAAACCATGCAGCTCAGAGGTCCCTGTGGGGGCTGGGGGAGCTGCCCTGC AGGTCTTGGCACATGCACAGCAGGCTCCCCATAGCTTTGTCACCACAAAGGGCACTGTTCTATTCACAGCACCTCCTGCTTCTGCCTGGC AACTGTGTCTCCCTGTGCTATATTTAATTCCACCAGCAAAGCTGGCGAGGCAGGGCCCAGCCCTGAAGGAGATCTCCTTGCCTGACCCCT GGACCTGGAAATGGAGGCTTCATGTGCCCGCCTTGGCGGCTTAAGCCTGCTGCTTTGGCAGTGCCATGGGTGAGCCGAGCAGCTGTGAGG TGGGTGGGGCAGGGCTGTAGCCCACGCCGGGTGCTATTCCAGGCTCTAGGGGCTGGTGCTCATCCCCACCCCCAGCGACTTCCGTCCTAC CTGGCATGCTGCAGCCCTCTGCCGGCTGTGGTGTCCTCTGAATTTGGACCCAGGTTGCCTGCTCTTGTTGTGGGGGTGGGATTGGGGCTG TCTGTGTTTCCCAGGTGGAGCTCATGTTTGTGATCCTCTATCTGGACAGCTCTCCAGAATCCTGTGTTGCCCCTGTTCTGCCTTTCTGGG ATGGGAGAGAAGGTGAGGGAGCCGGCGAGCAGGTTCTACCCTCTCTGCTAAGGGCCTAGCACACTTTGATGAGGTTCCTGGACGTGAGAC CCTCACAAAGACCAACCATTCCTCTAATAGCCATTTAACGGTACTGCAGTTCATGGGAAAATGTCCTCATTCTTAGGAGATAGATGCTGG ATTATATAGGTGAATGCCTTCAAGAAATGTGTAATTTAATTCCAAATAGTTCCCCCCAAAATACACAAAGCAAATAAGGCCAAAATGTTC ATTGTTGAATCTGCAGTGGCTGGTATTTCAGTGAACAGTGTACTTGTTCTTTCAACTTTTCTGTATGTTTGGAATTTTTCTTAATAAAAG ATTGTGGGAAATGGTCATTTGCTGAGCGCCTCCTAGTTGCCAGGCACTGGAGGGCGCATCAAAGAAGAGTCCTACCCAGGAGGGAGAGCG CAGCTGCTGCAGGATTCTCGCCTGGAAGAGTGGGTGGCTGGCTGGCTGGCTGGAAGGGGAGGGGCGAGAGGGCTGTCTCGGTGATGAGCA CACGCGTGCTGGGGTGGTTTGGGGAGAGGGCTGAGATTTGTCACCAGTGAGCCCTTGGCACGGTCTCTGCTCCCCACCAGGGCAGCACGG >95660_95660_6_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000335325_length(amino acids)=155AA_BP= MEPVSPLWSHCAGPLKRSAGAKMLQPPEAARMWVLPGVGPAPLLPAFAPHPTFHSPCLLFSLRLSADSRACFIEAQEEALVWGCASSEPL -------------------------------------------------------------- >95660_95660_7_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000566250_length(transcript)=842nt_BP=524nt GTGTTCCCCCGGAAGGGTGAGTCTGGACGCGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCT TGACAGGCTGCACTTGGATGGGAGCACCTGGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATA TTGAAACATTACAACCATCTTTTGATGGCAACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGC TTGCTTGAGTTGACACAGGGAGCTCCAGGACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAG GGGCAGTAAGCCCGTTGGGGATGGAGCCCAGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGG GGAGCCCTGGGTCACTTTCAGTGAGTCATCGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCAT TTGCTCCAAGTCTGTGGCTTCTGACGAGATGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGATGATGTGCT GACTAAAGACGCGGGTGAGTGTGTGATCTGCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCA CAAAAGGTAGGAGGGGTTGGCAAGGTAGCTTAGTGCACCCCACCTCCCAGAGGGGCGGACCCCCATCTCCAGCCATTCTGTCTCCTTTGG >95660_95660_7_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000566250_length(amino acids)=132AA_BP=64 MPLRHWGMARGSKPVGDGAQPMAAMGGLKVLLHWAGPGGGEPWVTFSESSLTAEEVCIHIAHKVGFKCPICSKSVASDEMEMHFIMCLSK -------------------------------------------------------------- >95660_95660_8_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000567962_length(transcript)=1007nt_BP=524nt GTGTTCCCCCGGAAGGGTGAGTCTGGACGCGGGCGCGGAAGGAGCGCGGCCGGAGGTCCTCAGGAAGAAGCCGCGGGGACTGGCTGCGCT TGACAGGCTGCACTTGGATGGGAGCACCTGGTGCCTCGGGACTGCTCCGATGCCCGGGTCTGTGCTGAATGTGTAATATGCGGAACTATA TTGAAACATTACAACCATCTTTTGATGGCAACACCCTGAGGACCTCCCTTTTCCAGATGGGGAAACTGAGGCCCAGAATTGCTAAGTGGC TTGCTTGAGTTGACACAGGGAGCTCCAGGACTCACCCTCAGCTGAGCCACCTGCCGGGAGCATGCCTCTGCGCCACTGGGGGATGGCCAG GGGCAGTAAGCCCGTTGGGGATGGAGCCCAGCCCATGGCTGCCATGGGAGGCCTGAAGGTGCTTCTGCACTGGGCTGGTCCAGGCGGCGG GGAGCCCTGGGTCACTTTCAGTGAGTCATCGCTGACAGCTGAGGAAGTCTGCATCCACATTGCACATAAAGTTGGTTTCAAGTGCCCCAT TTGCTCCAAGTCTGTGGCTTCTGACGAGATGGAAATGCACTTTATAATGTGTTTGAGCAAACCTCGCCTCTCCTACAACGATGATGTGCT GACTAAAGACGCGGGTGAGTGTGTGATCTGCCTGGAGGAGCTGCTGCAGGGGGACACGATAGCCAGGCTGCCCTGCCTGTGCATCTATCA CAAAAGCTGCATAGACTCGTGGTTTGAAGTGAACAGATCTTGTCCGGAACACCCTGCGGACTGACCTGCGGGCTTGCTTGCTGACTCCTC TCAAAGATCATGGTTCTCCCTTCCTCCCTGAGGACACCAAATTGGATGAGAGCAAGTTTGAGAGAAGAATGAATCAACTGCTATCCTTCC CCTCACCCCTCAGCCCAGGAGGGAAAGGGCATTTTCTTTTTCATCTTTGAAAGGCATTGTGGGTCTGTCTTTAAAGTGTTTACAAAAAAA >95660_95660_8_TYK2-ZNRF1_TYK2_chr19_10488889_ENST00000529370_ZNRF1_chr16_75127469_ENST00000567962_length(amino acids)=150AA_BP=64 MPLRHWGMARGSKPVGDGAQPMAAMGGLKVLLHWAGPGGGEPWVTFSESSLTAEEVCIHIAHKVGFKCPICSKSVASDEMEMHFIMCLSK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for TYK2-ZNRF1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for TYK2-ZNRF1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for TYK2-ZNRF1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
