
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:UBA2-LRP3 (FusionGDB2 ID:95811) |
Fusion Gene Summary for UBA2-LRP3 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: UBA2-LRP3 | Fusion gene ID: 95811 | Hgene | Tgene | Gene symbol | UBA2 | LRP3 | Gene ID | 10054 | 4037 |
| Gene name | ubiquitin like modifier activating enzyme 2 | LDL receptor related protein 3 | |
| Synonyms | ARX|HRIHFB2115|SAE2 | - | |
| Cytomap | 19q13.11 | 19q13.11 | |
| Type of gene | protein-coding | protein-coding | |
| Description | SUMO-activating enzyme subunit 2SUMO-1 activating enzyme subunit 2SUMO1 activating enzyme subunit 2UBA2, ubiquitin-activating enzyme E1 homologanthracycline-associated resistance ARXubiquitin-like 1-activating enzyme E1B | low-density lipoprotein receptor-related protein 3105 kDa low-density lipoprotein receptor-related proteinCTD-2540B15.13LRP-3hLRp105 | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | . | O75074 | |
| Ensembl transtripts involved in fusion gene | ENST00000246548, ENST00000439527, ENST00000592791, ENST00000588585, | ENST00000253193, | |
| Fusion gene scores | * DoF score | 14 X 11 X 8=1232 | 5 X 4 X 2=40 |
| # samples | 18 | 5 | |
| ** MAII score | log2(18/1232*10)=-2.77493344436523 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(5/40*10)=0.321928094887362 effective Gene in Pan-Cancer Fusion Genes (eGinPCFGs). DoF>8 and MAII>0 | |
| Context | PubMed: UBA2 [Title/Abstract] AND LRP3 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | UBA2(34957919)-LRP3(33687636), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | UBA2 | GO:0016925 | protein sumoylation | 20164921 |
| Hgene | UBA2 | GO:0033235 | positive regulation of protein sumoylation | 10187858 |
 Fusion gene breakpoints across UBA2 (5'-gene) Fusion gene breakpoints across UBA2 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
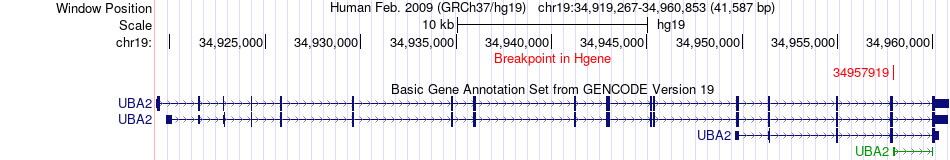 |
 Fusion gene breakpoints across LRP3 (3'-gene) Fusion gene breakpoints across LRP3 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
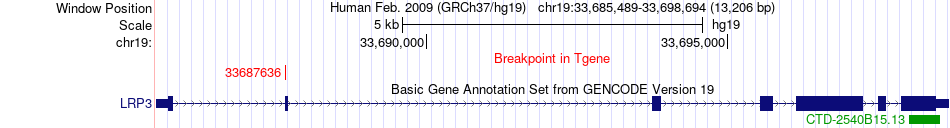 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | SARC | TCGA-DX-A8BN-01A | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + |
Top |
Fusion Gene ORF analysis for UBA2-LRP3 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| In-frame | ENST00000246548 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + |
| In-frame | ENST00000439527 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + |
| In-frame | ENST00000592791 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + |
| intron-3CDS | ENST00000588585 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000246548 | UBA2 | chr19 | 34957919 | + | ENST00000253193 | LRP3 | chr19 | 33687636 | + | 4264 | 1811 | 70 | 4050 | 1326 |
| ENST00000439527 | UBA2 | chr19 | 34957919 | + | ENST00000253193 | LRP3 | chr19 | 33687636 | + | 4404 | 1951 | 354 | 4190 | 1278 |
| ENST00000592791 | UBA2 | chr19 | 34957919 | + | ENST00000253193 | LRP3 | chr19 | 33687636 | + | 3004 | 551 | 31 | 2790 | 919 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000246548 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + | 0.005313941 | 0.994686 |
| ENST00000439527 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + | 0.006438548 | 0.9935615 |
| ENST00000592791 | ENST00000253193 | UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687636 | + | 0.019068986 | 0.980931 |
Top |
Fusion Genomic Features for UBA2-LRP3 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687635 | + | 2.06E-05 | 0.9999794 |
| UBA2 | chr19 | 34957919 | + | LRP3 | chr19 | 33687635 | + | 2.06E-05 | 0.9999794 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
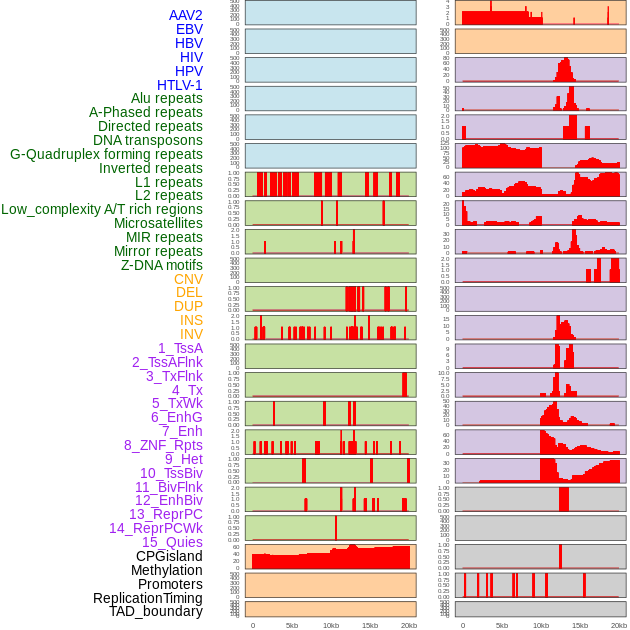 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
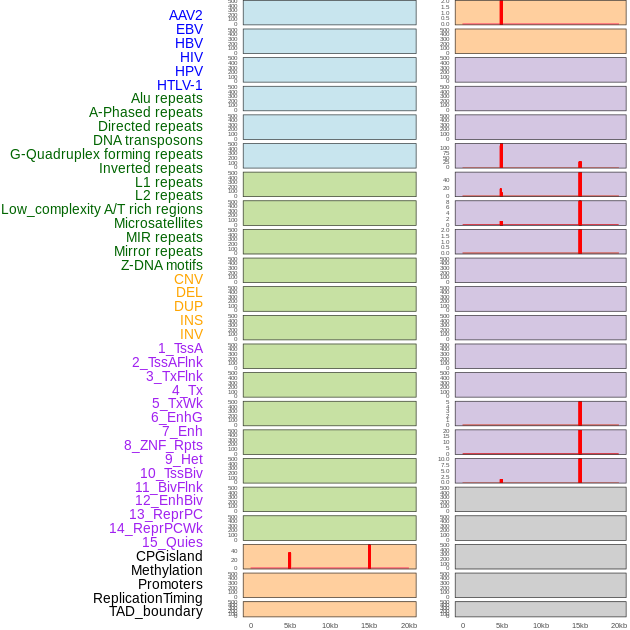 |
Top |
Fusion Protein Features for UBA2-LRP3 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:34957919/chr19:33687636) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| . | LRP3 |
| FUNCTION: Transcriptional activator which is required for calcium-dependent dendritic growth and branching in cortical neurons. Recruits CREB-binding protein (CREBBP) to nuclear bodies. Component of the CREST-BRG1 complex, a multiprotein complex that regulates promoter activation by orchestrating a calcium-dependent release of a repressor complex and a recruitment of an activator complex. In resting neurons, transcription of the c-FOS promoter is inhibited by BRG1-dependent recruitment of a phospho-RB1-HDAC1 repressor complex. Upon calcium influx, RB1 is dephosphorylated by calcineurin, which leads to release of the repressor complex. At the same time, there is increased recruitment of CREBBP to the promoter by a CREST-dependent mechanism, which leads to transcriptional activation. The CREST-BRG1 complex also binds to the NR2B promoter, and activity-dependent induction of NR2B expression involves a release of HDAC1 and recruitment of CREBBP (By similarity). {ECO:0000250}. | FUNCTION: Probable receptor, which may be involved in the internalization of lipophilic molecules and/or signal transduction. Its precise role is however unclear, since it does not bind to very low density lipoprotein (VLDL) or to LRPAP1 in vitro. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | UBA2 | chr19:34957919 | chr19:33687636 | ENST00000246548 | + | 16 | 17 | 117_122 | 580 | 641.0 | Nucleotide binding | ATP |
| Hgene | UBA2 | chr19:34957919 | chr19:33687636 | ENST00000246548 | + | 16 | 17 | 24_29 | 580 | 641.0 | Nucleotide binding | ATP |
| Hgene | UBA2 | chr19:34957919 | chr19:33687636 | ENST00000246548 | + | 16 | 17 | 56_59 | 580 | 641.0 | Nucleotide binding | ATP |
| Hgene | UBA2 | chr19:34957919 | chr19:33687636 | ENST00000246548 | + | 16 | 17 | 95_96 | 580 | 641.0 | Nucleotide binding | ATP |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 751_754 | 24 | 771.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 165_201 | 24 | 771.0 | Domain | LDL-receptor class A 1 | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 211_250 | 24 | 771.0 | Domain | LDL-receptor class A 2 | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 254_365 | 24 | 771.0 | Domain | CUB 2 | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 415_453 | 24 | 771.0 | Domain | LDL-receptor class A 3 | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 43_159 | 24 | 771.0 | Domain | CUB 1 | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 454_490 | 24 | 771.0 | Domain | LDL-receptor class A 4 | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 37_496 | 24 | 771.0 | Topological domain | Extracellular | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 518_770 | 24 | 771.0 | Topological domain | Cytoplasmic | |
| Tgene | LRP3 | chr19:34957919 | chr19:33687636 | ENST00000253193 | 0 | 7 | 497_517 | 24 | 771.0 | Transmembrane | Helical |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
Top |
Fusion Gene Sequence for UBA2-LRP3 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >95811_95811_1_UBA2-LRP3_UBA2_chr19_34957919_ENST00000246548_LRP3_chr19_33687636_ENST00000253193_length(transcript)=4264nt_BP=1811nt CCCCACCCGCTTCCGGCCGCGGCTCGGTTCTCCCGCCTCCGCCTCCGCCGCGGCTCGTGGTTGTCCCGCCATGGCACTGTCGCGGGGGCT GCCCCGGGAGCTGGCTGAGGCGGTGGCCGGGGGCCGGGTGCTGGTGGTGGGGGCGGGCGGCATCGGCTGCGAGCTCCTCAAGAATCTCGT GCTCACCGGTTTCTCCCACATCGACCTGATTGATCTGGATACTATTGATGTAAGCAACCTCAACAGACAGTTTTTGTTTCAAAAGAAACA TGTTGGAAGATCAAAGGCACAGGTTGCCAAGGAAAGTGTACTGCAGTTTTACCCGAAAGCTAATATCGTTGCCTACCATGACAGCATCAT GAACCCTGACTATAATGTGGAATTTTTCCGACAGTTTATACTGGTTATGAATGCTTTAGATAACAGAGCTGCCCGAAACCATGTTAATAG AATGTGCCTGGCAGCTGATGTTCCTCTTATTGAAAGTGGAACAGCTGGGTATCTTGGACAAGTAACTACTATCAAAAAGGGTGTGACCGA GTGTTATGAGTGTCATCCTAAGCCGACCCAGAGAACCTTTCCTGGCTGTACAATTCGTAACACACCTTCAGAACCTATACATTGCATCGT TTGGGCAAAGTACTTGTTCAACCAGTTGTTTGGGGAAGAAGATGCTGATCAAGAAGTATCTCCTGACAGAGCTGACCCTGAAGCTGCCTG GGAACCAACGGAAGCCGAAGCCAGAGCTAGAGCATCTAATGAAGATGGTGACATTAAACGTATTTCTACTAAGGAATGGGCTAAATCAAC TGGATATGATCCAGTTAAACTTTTTACCAAGCTTTTTAAAGATGACATCAGGTATCTGTTGACAATGGACAAACTATGGCGGAAAAGGAA ACCTCCAGTTCCGTTGGACTGGGCTGAAGTACAAAGTCAAGGAGAAGAAACGAATGCATCAGATCAACAGAATGAACCCCAGTTAGGCCT GAAAGACCAGCAGGTTCTAGATGTAAAGAGCTATGCACGTCTTTTTTCAAAGAGCATCGAGACTTTGAGAGTTCATTTAGCAGAAAAGGG GGATGGAGCTGAGCTCATATGGGATAAGGATGACCCATCTGCAATGGATTTTGTCACCTCTGCTGCAAACCTCAGGATGCATATTTTCAG TATGAATATGAAGAGTAGATTTGATATCAAATCAATGGCAGGGAACATTATTCCTGCTATTGCTACTACTAATGCAGTAATTGCTGGGTT GATAGTATTGGAAGGATTGAAGATTTTATCAGGAAAAATAGACCAGTGCAGAACAATTTTTTTGAATAAACAACCAAACCCAAGAAAGAA GCTTCTTGTGCCTTGTGCACTGGATCCTCCCAACCCCAATTGTTATGTATGTGCCAGCAAGCCAGAGGTGACTGTGCGGCTGAATGTCCA TAAAGTGACTGTTCTCACCTTACAAGACAAGATAGTGAAAGAAAAATTTGCTATGGTAGCACCAGATGTCCAAATTGAAGATGGGAAAGG AACAATCCTAATATCTTCCGAAGAGGGAGAGACGGAAGCTAATAATCACAAGAAGTTGTCAGAATTTGGAATTAGAAATGGCAGCCGGCT TCAAGCAGATGACTTCCTCCAGGACTATACTTTATTGATCAACATCCTTCATAGTGAAGACCTAGGAAAGGACGTTGAATTTGAAGTTGT TGGTGATGCCCCGGAAAAAGTGGGGCCCAAACAAGCTGAAGATGCTGCCAAAAGCATAACCAATGGCAGTGATGATGGAGCTCAGCCCTC CACCTCCACAGTGAACATCTTTCTCACCGGGAGACTCAGCAGTGCGGTTCCTGCCTTAGCGGCCTGCAGTGGGAAGCTGGAGCAGCACAC GGAGCGGCGTGGGGTCATCTACAGCCCGGCCTGGCCCCTCAACTACCCGCCAGGCACCAACTGCAGCTGGTACATCCAGGGCGACCGTGG TGACATGATTACCATCAGCTTCCGCAACTTTGACGTGGAGGAGTCCCACCAGTGCTCCCTGGACTGGCTCCTGCTGGGCCCAGCAGCCCC ACCCCGCCAGGAGGCCTTCCGCCTCTGTGGCTCCGCCATCCCACCTGCCTTCATCTCTGCCCGCGACCATGTCTGGATTTTCTTCCACTC AGACGCCTCCAGCTCCGGCCAGGCCCAGGGCTTCCGTCTGTCTTACATCCGAGGGAAGCTGGGCCAGGCATCCTGCCAGGCAGATGAGTT CCGCTGTGACAACGGCAAGTGCCTGCCCGGCCCGTGGCAGTGCAACACGGTGGACGAGTGTGGAGACGGCTCTGATGAGGGCAACTGCTC GGCGCCCGCCTCCGAGCCTCCAGGCAGCCTGTGCCCCGGGGGGACCTTCCCATGCAGCGGGGCGCGCTCCACGCGCTGCCTGCCTGTGGA GCGGCGCTGTGACGGCTTGCAGGACTGCGGCGACGGCTCGGATGAGGCGGGCTGCCCCGACCTGGCGTGCGGCCGGCGGCTGGGCAGCTT CTACGGCTCCTTTGCCTCCCCAGACCTGTTCGGCGCCGCTCGCGGGCCCTCAGACCTTCACTGCACGTGGCTGGTGGACACACAGGACTC CCGGCGGGTGCTGCTGCAGCTGGAACTGCGGCTGGGCTATGACGACTACGTGCAGGTATACGAGGGCCTGGGCGAGCGCGGGGACCGCCT GCTGCAGACGCTGTCCTACCGCAGCAACCACCGGCCCGTGAGCCTGGAGGCCGCCCAGGGCCGCCTCACTGTGGCCTACCACGCGCGCGC CCGCAGCGCCGGCCACGGCTTCAATGCCACCTACCAGGTGAAGGGCTATTGCCTCCCCTGGGAGCAGCCGTGCGGGAGCAGTAGTGACAG TGACGGGGGCAGCCTGGGCGACCAGGGCTGCTTCTCAGAGCCACAGCGCTGTGATGGCTGGTGGCATTGTGCCAGCGGCCGAGACGAGCA GGGCTGCCCTGCCTGCCCGCCCGACCAGTACCCCTGCGAGGGTGGCAGTGGTCTGTGCTACACGCCTGCCGACCGCTGCAACAACCAGAA AAGCTGTCCCGACGGCGCCGACGAGAAGAACTGCTTCTCCTGCCAGCCCGGCACCTTCCACTGCGGTACCAACCTGTGCATCTTCGAGAC GTGGCGCTGTGACGGCCAGGAAGACTGCCAGGACGGCAGCGATGAGCATGGGTGCCTGGCCGCCGTGCCCCGCAAGGTCATCACGGCGGC GCTCATTGGCAGCCTGGTGTGTGGCCTGCTGCTGGTCATCGCGCTGGGCTGCGCCTTCAAGCTCTACTCACTGCGCACGCAGGAATACAG GGCCTTCGAGACCCAGATGACGCGCCTGGAGGCTGAGTTCGTGCGGCGGGAGGCACCCCCATCCTATGGTCAGCTCATCGCCCAGGGCCT CATTCCACCCGTGGAGGACTTTCCTGTCTACAGTGCGTCCCAGGCCTCTGTGCTGCAGAATCTTCGCACAGCCATGCGGAGACAGATGCG TCGGCACGCCTCCCGCCGGGGGCCCTCCCGCCGCCGCCTCGGCCGCCTCTGGAACCGGCTCTTTCACCGGCCGCGGGCGCCCCGAGGCCA GATCCCACTGCTGACCGCAGCACGCCCCTCACAGACCGTGCTGGGCGATGGCTTCCTCCAGCCTGCTCCAGGGGCTGCCCCCGACCCCCC AGCACCGCTCATGGACACAGGCAGCACCAGGGCGGCCGGAGACAGGCCCCCCAGTGCCCCCGGCCGTGCACCGGAGGTGGGACCTTCAGG GCCACCCTTGCCCTCGGGCCTGCGAGACCCAGAGTGCAGGCCCGTGGACAAGGACAGAAAGGTCTGCAGGGAGCCACTGGTAGACGGCCC AGCTCCTGCAGATGCACCTCGGGAGCCCTGCTCAGCCCAGGACCCGCACCCCCAGGTCTCCACTGCCAGCAGCACCCTGGGCCCCCACTC GCCAGAGCCACTGGGGGTCTGCAGGAACCCCCCGCCCCCCTGCTCCCCAATGCTGGAGGCCAGCGATGATGAGGCCCTGTTGGTCTGTTG ACCGCTGGGCTCGCTGGTGACCGCCACAGCCCCGCTTTGTAACCAGGGAATACACAGTCATTTCTACCCTGCCTCTGCGTCCTTTCTTAT GGAGAGGCCCTCCGGGGACCCCAGCGGAGGGGCTGGCCCCTAAGCCAGCTGGCTGCACTGGTGGGCGGGAGCTGTGGGACTGAACGGCGG >95811_95811_1_UBA2-LRP3_UBA2_chr19_34957919_ENST00000246548_LRP3_chr19_33687636_ENST00000253193_length(amino acids)=1326AA_BP=579 MALSRGLPRELAEAVAGGRVLVVGAGGIGCELLKNLVLTGFSHIDLIDLDTIDVSNLNRQFLFQKKHVGRSKAQVAKESVLQFYPKANIV AYHDSIMNPDYNVEFFRQFILVMNALDNRAARNHVNRMCLAADVPLIESGTAGYLGQVTTIKKGVTECYECHPKPTQRTFPGCTIRNTPS EPIHCIVWAKYLFNQLFGEEDADQEVSPDRADPEAAWEPTEAEARARASNEDGDIKRISTKEWAKSTGYDPVKLFTKLFKDDIRYLLTMD KLWRKRKPPVPLDWAEVQSQGEETNASDQQNEPQLGLKDQQVLDVKSYARLFSKSIETLRVHLAEKGDGAELIWDKDDPSAMDFVTSAAN LRMHIFSMNMKSRFDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSGKIDQCRTIFLNKQPNPRKKLLVPCALDPPNPNCYVCASKPEV TVRLNVHKVTVLTLQDKIVKEKFAMVAPDVQIEDGKGTILISSEEGETEANNHKKLSEFGIRNGSRLQADDFLQDYTLLINILHSEDLGK DVEFEVVGDAPEKVGPKQAEDAAKSITNGSDDGAQPSTSTVNIFLTGRLSSAVPALAACSGKLEQHTERRGVIYSPAWPLNYPPGTNCSW YIQGDRGDMITISFRNFDVEESHQCSLDWLLLGPAAPPRQEAFRLCGSAIPPAFISARDHVWIFFHSDASSSGQAQGFRLSYIRGKLGQA SCQADEFRCDNGKCLPGPWQCNTVDECGDGSDEGNCSAPASEPPGSLCPGGTFPCSGARSTRCLPVERRCDGLQDCGDGSDEAGCPDLAC GRRLGSFYGSFASPDLFGAARGPSDLHCTWLVDTQDSRRVLLQLELRLGYDDYVQVYEGLGERGDRLLQTLSYRSNHRPVSLEAAQGRLT VAYHARARSAGHGFNATYQVKGYCLPWEQPCGSSSDSDGGSLGDQGCFSEPQRCDGWWHCASGRDEQGCPACPPDQYPCEGGSGLCYTPA DRCNNQKSCPDGADEKNCFSCQPGTFHCGTNLCIFETWRCDGQEDCQDGSDEHGCLAAVPRKVITAALIGSLVCGLLLVIALGCAFKLYS LRTQEYRAFETQMTRLEAEFVRREAPPSYGQLIAQGLIPPVEDFPVYSASQASVLQNLRTAMRRQMRRHASRRGPSRRRLGRLWNRLFHR PRAPRGQIPLLTAARPSQTVLGDGFLQPAPGAAPDPPAPLMDTGSTRAAGDRPPSAPGRAPEVGPSGPPLPSGLRDPECRPVDKDRKVCR -------------------------------------------------------------- >95811_95811_2_UBA2-LRP3_UBA2_chr19_34957919_ENST00000439527_LRP3_chr19_33687636_ENST00000253193_length(transcript)=4404nt_BP=1951nt AAAACAAATTCCCGGCTGTTAGGAGATGCGGGAGAGGATTGAGTCCCTCGCGGATGTTGTGACTTTAATTTTGAGGTCCCTGTCGTAACT CCGAAGTAACGACGCGGGGCGGAGGATGGCAATGATAGCGACCAACGCGTCCCGAGCGCTGGTCACGAGCGGGTTTACACACGGAACGCG CACCATCTTCATACCACCCGAGGAGTGCAAACGCTATTTCCACCCTGCTTTATTTTTAACAGATGAGGAAAGGGAGACGCAATGGGGTTG AGTAACTTACCCTGGGTCACGGAGCGAGTGGCAGGGCTCCCGCAGGCTGCGTGACTGTCATTGCTCTCACAGTAGACGATTGATCTGGAT ACTATTGATGTAAGCAACCTCAACAGACAGTTTTTGTTTCAAAAGAAACATGTTGGAAGATCAAAGGCACAGGTTGCCAAGGAAAGTGTA CTGCAGTTTTACCCGAAAGCTAATATCGTTGCCTACCATGACAGCATCATGAACCCTGACTATAATGTGGAATTTTTCCGACAGTTTATA CTGGTTATGAATGCTTTAGATAACAGAGCTGCCCGAAACCATGTTAATAGAATGTGCCTGGCAGCTGATGTTCCTCTTATTGAAAGTGGA ACAGCTGGGTATCTTGGACAAGTAACTACTATCAAAAAGGGTGTGACCGAGTGTTATGAGTGTCATCCTAAGCCGACCCAGAGAACCTTT CCTGGCTGTACAATTCGTAACACACCTTCAGAACCTATACATTGCATCGTTTGGGCAAAGTACTTGTTCAACCAGTTGTTTGGGGAAGAA GATGCTGATCAAGAAGTATCTCCTGACAGAGCTGACCCTGAAGCTGCCTGGGAACCAACGGAAGCCGAAGCCAGAGCTAGAGCATCTAAT GAAGATGGTGACATTAAACGTATTTCTACTAAGGAATGGGCTAAATCAACTGGATATGATCCAGTTAAACTTTTTACCAAGCTTTTTAAA GATGACATCAGGTATCTGTTGACAATGGACAAACTATGGCGGAAAAGGAAACCTCCAGTTCCGTTGGACTGGGCTGAAGTACAAAGTCAA GGAGAAGAAACGAATGCATCAGATCAACAGAATGAACCCCAGTTAGGCCTGAAAGACCAGCAGGTTCTAGATGTAAAGAGCTATGCACGT CTTTTTTCAAAGAGCATCGAGACTTTGAGAGTTCATTTAGCAGAAAAGGGGGATGGAGCTGAGCTCATATGGGATAAGGATGACCCATCT GCAATGGATTTTGTCACCTCTGCTGCAAACCTCAGGATGCATATTTTCAGTATGAATATGAAGAGTAGATTTGATATCAAATCAATGGCA GGGAACATTATTCCTGCTATTGCTACTACTAATGCAGTAATTGCTGGGTTGATAGTATTGGAAGGATTGAAGATTTTATCAGGAAAAATA GACCAGTGCAGAACAATTTTTTTGAATAAACAACCAAACCCAAGAAAGAAGCTTCTTGTGCCTTGTGCACTGGATCCTCCCAACCCCAAT TGTTATGTATGTGCCAGCAAGCCAGAGGTGACTGTGCGGCTGAATGTCCATAAAGTGACTGTTCTCACCTTACAAGACAAGATAGTGAAA GAAAAATTTGCTATGGTAGCACCAGATGTCCAAATTGAAGATGGGAAAGGAACAATCCTAATATCTTCCGAAGAGGGAGAGACGGAAGCT AATAATCACAAGAAGTTGTCAGAATTTGGAATTAGAAATGGCAGCCGGCTTCAAGCAGATGACTTCCTCCAGGACTATACTTTATTGATC AACATCCTTCATAGTGAAGACCTAGGAAAGGACGTTGAATTTGAAGTTGTTGGTGATGCCCCGGAAAAAGTGGGGCCCAAACAAGCTGAA GATGCTGCCAAAAGCATAACCAATGGCAGTGATGATGGAGCTCAGCCCTCCACCTCCACAGTGAACATCTTTCTCACCGGGAGACTCAGC AGTGCGGTTCCTGCCTTAGCGGCCTGCAGTGGGAAGCTGGAGCAGCACACGGAGCGGCGTGGGGTCATCTACAGCCCGGCCTGGCCCCTC AACTACCCGCCAGGCACCAACTGCAGCTGGTACATCCAGGGCGACCGTGGTGACATGATTACCATCAGCTTCCGCAACTTTGACGTGGAG GAGTCCCACCAGTGCTCCCTGGACTGGCTCCTGCTGGGCCCAGCAGCCCCACCCCGCCAGGAGGCCTTCCGCCTCTGTGGCTCCGCCATC CCACCTGCCTTCATCTCTGCCCGCGACCATGTCTGGATTTTCTTCCACTCAGACGCCTCCAGCTCCGGCCAGGCCCAGGGCTTCCGTCTG TCTTACATCCGAGGGAAGCTGGGCCAGGCATCCTGCCAGGCAGATGAGTTCCGCTGTGACAACGGCAAGTGCCTGCCCGGCCCGTGGCAG TGCAACACGGTGGACGAGTGTGGAGACGGCTCTGATGAGGGCAACTGCTCGGCGCCCGCCTCCGAGCCTCCAGGCAGCCTGTGCCCCGGG GGGACCTTCCCATGCAGCGGGGCGCGCTCCACGCGCTGCCTGCCTGTGGAGCGGCGCTGTGACGGCTTGCAGGACTGCGGCGACGGCTCG GATGAGGCGGGCTGCCCCGACCTGGCGTGCGGCCGGCGGCTGGGCAGCTTCTACGGCTCCTTTGCCTCCCCAGACCTGTTCGGCGCCGCT CGCGGGCCCTCAGACCTTCACTGCACGTGGCTGGTGGACACACAGGACTCCCGGCGGGTGCTGCTGCAGCTGGAACTGCGGCTGGGCTAT GACGACTACGTGCAGGTATACGAGGGCCTGGGCGAGCGCGGGGACCGCCTGCTGCAGACGCTGTCCTACCGCAGCAACCACCGGCCCGTG AGCCTGGAGGCCGCCCAGGGCCGCCTCACTGTGGCCTACCACGCGCGCGCCCGCAGCGCCGGCCACGGCTTCAATGCCACCTACCAGGTG AAGGGCTATTGCCTCCCCTGGGAGCAGCCGTGCGGGAGCAGTAGTGACAGTGACGGGGGCAGCCTGGGCGACCAGGGCTGCTTCTCAGAG CCACAGCGCTGTGATGGCTGGTGGCATTGTGCCAGCGGCCGAGACGAGCAGGGCTGCCCTGCCTGCCCGCCCGACCAGTACCCCTGCGAG GGTGGCAGTGGTCTGTGCTACACGCCTGCCGACCGCTGCAACAACCAGAAAAGCTGTCCCGACGGCGCCGACGAGAAGAACTGCTTCTCC TGCCAGCCCGGCACCTTCCACTGCGGTACCAACCTGTGCATCTTCGAGACGTGGCGCTGTGACGGCCAGGAAGACTGCCAGGACGGCAGC GATGAGCATGGGTGCCTGGCCGCCGTGCCCCGCAAGGTCATCACGGCGGCGCTCATTGGCAGCCTGGTGTGTGGCCTGCTGCTGGTCATC GCGCTGGGCTGCGCCTTCAAGCTCTACTCACTGCGCACGCAGGAATACAGGGCCTTCGAGACCCAGATGACGCGCCTGGAGGCTGAGTTC GTGCGGCGGGAGGCACCCCCATCCTATGGTCAGCTCATCGCCCAGGGCCTCATTCCACCCGTGGAGGACTTTCCTGTCTACAGTGCGTCC CAGGCCTCTGTGCTGCAGAATCTTCGCACAGCCATGCGGAGACAGATGCGTCGGCACGCCTCCCGCCGGGGGCCCTCCCGCCGCCGCCTC GGCCGCCTCTGGAACCGGCTCTTTCACCGGCCGCGGGCGCCCCGAGGCCAGATCCCACTGCTGACCGCAGCACGCCCCTCACAGACCGTG CTGGGCGATGGCTTCCTCCAGCCTGCTCCAGGGGCTGCCCCCGACCCCCCAGCACCGCTCATGGACACAGGCAGCACCAGGGCGGCCGGA GACAGGCCCCCCAGTGCCCCCGGCCGTGCACCGGAGGTGGGACCTTCAGGGCCACCCTTGCCCTCGGGCCTGCGAGACCCAGAGTGCAGG CCCGTGGACAAGGACAGAAAGGTCTGCAGGGAGCCACTGGTAGACGGCCCAGCTCCTGCAGATGCACCTCGGGAGCCCTGCTCAGCCCAG GACCCGCACCCCCAGGTCTCCACTGCCAGCAGCACCCTGGGCCCCCACTCGCCAGAGCCACTGGGGGTCTGCAGGAACCCCCCGCCCCCC TGCTCCCCAATGCTGGAGGCCAGCGATGATGAGGCCCTGTTGGTCTGTTGACCGCTGGGCTCGCTGGTGACCGCCACAGCCCCGCTTTGT AACCAGGGAATACACAGTCATTTCTACCCTGCCTCTGCGTCCTTTCTTATGGAGAGGCCCTCCGGGGACCCCAGCGGAGGGGCTGGCCCC >95811_95811_2_UBA2-LRP3_UBA2_chr19_34957919_ENST00000439527_LRP3_chr19_33687636_ENST00000253193_length(amino acids)=1278AA_BP=531 MDTIDVSNLNRQFLFQKKHVGRSKAQVAKESVLQFYPKANIVAYHDSIMNPDYNVEFFRQFILVMNALDNRAARNHVNRMCLAADVPLIE SGTAGYLGQVTTIKKGVTECYECHPKPTQRTFPGCTIRNTPSEPIHCIVWAKYLFNQLFGEEDADQEVSPDRADPEAAWEPTEAEARARA SNEDGDIKRISTKEWAKSTGYDPVKLFTKLFKDDIRYLLTMDKLWRKRKPPVPLDWAEVQSQGEETNASDQQNEPQLGLKDQQVLDVKSY ARLFSKSIETLRVHLAEKGDGAELIWDKDDPSAMDFVTSAANLRMHIFSMNMKSRFDIKSMAGNIIPAIATTNAVIAGLIVLEGLKILSG KIDQCRTIFLNKQPNPRKKLLVPCALDPPNPNCYVCASKPEVTVRLNVHKVTVLTLQDKIVKEKFAMVAPDVQIEDGKGTILISSEEGET EANNHKKLSEFGIRNGSRLQADDFLQDYTLLINILHSEDLGKDVEFEVVGDAPEKVGPKQAEDAAKSITNGSDDGAQPSTSTVNIFLTGR LSSAVPALAACSGKLEQHTERRGVIYSPAWPLNYPPGTNCSWYIQGDRGDMITISFRNFDVEESHQCSLDWLLLGPAAPPRQEAFRLCGS AIPPAFISARDHVWIFFHSDASSSGQAQGFRLSYIRGKLGQASCQADEFRCDNGKCLPGPWQCNTVDECGDGSDEGNCSAPASEPPGSLC PGGTFPCSGARSTRCLPVERRCDGLQDCGDGSDEAGCPDLACGRRLGSFYGSFASPDLFGAARGPSDLHCTWLVDTQDSRRVLLQLELRL GYDDYVQVYEGLGERGDRLLQTLSYRSNHRPVSLEAAQGRLTVAYHARARSAGHGFNATYQVKGYCLPWEQPCGSSSDSDGGSLGDQGCF SEPQRCDGWWHCASGRDEQGCPACPPDQYPCEGGSGLCYTPADRCNNQKSCPDGADEKNCFSCQPGTFHCGTNLCIFETWRCDGQEDCQD GSDEHGCLAAVPRKVITAALIGSLVCGLLLVIALGCAFKLYSLRTQEYRAFETQMTRLEAEFVRREAPPSYGQLIAQGLIPPVEDFPVYS ASQASVLQNLRTAMRRQMRRHASRRGPSRRRLGRLWNRLFHRPRAPRGQIPLLTAARPSQTVLGDGFLQPAPGAAPDPPAPLMDTGSTRA AGDRPPSAPGRAPEVGPSGPPLPSGLRDPECRPVDKDRKVCREPLVDGPAPADAPREPCSAQDPHPQVSTASSTLGPHSPEPLGVCRNPP -------------------------------------------------------------- >95811_95811_3_UBA2-LRP3_UBA2_chr19_34957919_ENST00000592791_LRP3_chr19_33687636_ENST00000253193_length(transcript)=3004nt_BP=551nt GGCGTATAAAATTACAGCACGTTTCCGATTTCTGCCTGTTATTTCTCCTCCAAAGATTTTTTTGAATAAACAACCAAACCCAAGAAAGAA GCTTCTTGTGCCTTGTGCACTGGATCCTCCCAACCCCAATTGTTATGTATGTGCCAGCAAGCCAGAGGTGACTGTGCGGCTGAATGTCCA TAAAGTGACTGTTCTCACCTTACAAGACAAGATAGTGAAAGAAAAATTTGCTATGGTAGCACCAGATGTCCAAATTGAAGATGGGAAAGG AACAATCCTAATATCTTCCGAAGAGGGAGAGACGGAAGCTAATAATCACAAGAAGTTGTCAGAATTTGGAATTAGAAATGGCAGCCGGCT TCAAGCAGATGACTTCCTCCAGGACTATACTTTATTGATCAACATCCTTCATAGTGAAGACCTAGGAAAGGACGTTGAATTTGAAGTTGT TGGTGATGCCCCGGAAAAAGTGGGGCCCAAACAAGCTGAAGATGCTGCCAAAAGCATAACCAATGGCAGTGATGATGGAGCTCAGCCCTC CACCTCCACAGTGAACATCTTTCTCACCGGGAGACTCAGCAGTGCGGTTCCTGCCTTAGCGGCCTGCAGTGGGAAGCTGGAGCAGCACAC GGAGCGGCGTGGGGTCATCTACAGCCCGGCCTGGCCCCTCAACTACCCGCCAGGCACCAACTGCAGCTGGTACATCCAGGGCGACCGTGG TGACATGATTACCATCAGCTTCCGCAACTTTGACGTGGAGGAGTCCCACCAGTGCTCCCTGGACTGGCTCCTGCTGGGCCCAGCAGCCCC ACCCCGCCAGGAGGCCTTCCGCCTCTGTGGCTCCGCCATCCCACCTGCCTTCATCTCTGCCCGCGACCATGTCTGGATTTTCTTCCACTC AGACGCCTCCAGCTCCGGCCAGGCCCAGGGCTTCCGTCTGTCTTACATCCGAGGGAAGCTGGGCCAGGCATCCTGCCAGGCAGATGAGTT CCGCTGTGACAACGGCAAGTGCCTGCCCGGCCCGTGGCAGTGCAACACGGTGGACGAGTGTGGAGACGGCTCTGATGAGGGCAACTGCTC GGCGCCCGCCTCCGAGCCTCCAGGCAGCCTGTGCCCCGGGGGGACCTTCCCATGCAGCGGGGCGCGCTCCACGCGCTGCCTGCCTGTGGA GCGGCGCTGTGACGGCTTGCAGGACTGCGGCGACGGCTCGGATGAGGCGGGCTGCCCCGACCTGGCGTGCGGCCGGCGGCTGGGCAGCTT CTACGGCTCCTTTGCCTCCCCAGACCTGTTCGGCGCCGCTCGCGGGCCCTCAGACCTTCACTGCACGTGGCTGGTGGACACACAGGACTC CCGGCGGGTGCTGCTGCAGCTGGAACTGCGGCTGGGCTATGACGACTACGTGCAGGTATACGAGGGCCTGGGCGAGCGCGGGGACCGCCT GCTGCAGACGCTGTCCTACCGCAGCAACCACCGGCCCGTGAGCCTGGAGGCCGCCCAGGGCCGCCTCACTGTGGCCTACCACGCGCGCGC CCGCAGCGCCGGCCACGGCTTCAATGCCACCTACCAGGTGAAGGGCTATTGCCTCCCCTGGGAGCAGCCGTGCGGGAGCAGTAGTGACAG TGACGGGGGCAGCCTGGGCGACCAGGGCTGCTTCTCAGAGCCACAGCGCTGTGATGGCTGGTGGCATTGTGCCAGCGGCCGAGACGAGCA GGGCTGCCCTGCCTGCCCGCCCGACCAGTACCCCTGCGAGGGTGGCAGTGGTCTGTGCTACACGCCTGCCGACCGCTGCAACAACCAGAA AAGCTGTCCCGACGGCGCCGACGAGAAGAACTGCTTCTCCTGCCAGCCCGGCACCTTCCACTGCGGTACCAACCTGTGCATCTTCGAGAC GTGGCGCTGTGACGGCCAGGAAGACTGCCAGGACGGCAGCGATGAGCATGGGTGCCTGGCCGCCGTGCCCCGCAAGGTCATCACGGCGGC GCTCATTGGCAGCCTGGTGTGTGGCCTGCTGCTGGTCATCGCGCTGGGCTGCGCCTTCAAGCTCTACTCACTGCGCACGCAGGAATACAG GGCCTTCGAGACCCAGATGACGCGCCTGGAGGCTGAGTTCGTGCGGCGGGAGGCACCCCCATCCTATGGTCAGCTCATCGCCCAGGGCCT CATTCCACCCGTGGAGGACTTTCCTGTCTACAGTGCGTCCCAGGCCTCTGTGCTGCAGAATCTTCGCACAGCCATGCGGAGACAGATGCG TCGGCACGCCTCCCGCCGGGGGCCCTCCCGCCGCCGCCTCGGCCGCCTCTGGAACCGGCTCTTTCACCGGCCGCGGGCGCCCCGAGGCCA GATCCCACTGCTGACCGCAGCACGCCCCTCACAGACCGTGCTGGGCGATGGCTTCCTCCAGCCTGCTCCAGGGGCTGCCCCCGACCCCCC AGCACCGCTCATGGACACAGGCAGCACCAGGGCGGCCGGAGACAGGCCCCCCAGTGCCCCCGGCCGTGCACCGGAGGTGGGACCTTCAGG GCCACCCTTGCCCTCGGGCCTGCGAGACCCAGAGTGCAGGCCCGTGGACAAGGACAGAAAGGTCTGCAGGGAGCCACTGGTAGACGGCCC AGCTCCTGCAGATGCACCTCGGGAGCCCTGCTCAGCCCAGGACCCGCACCCCCAGGTCTCCACTGCCAGCAGCACCCTGGGCCCCCACTC GCCAGAGCCACTGGGGGTCTGCAGGAACCCCCCGCCCCCCTGCTCCCCAATGCTGGAGGCCAGCGATGATGAGGCCCTGTTGGTCTGTTG ACCGCTGGGCTCGCTGGTGACCGCCACAGCCCCGCTTTGTAACCAGGGAATACACAGTCATTTCTACCCTGCCTCTGCGTCCTTTCTTAT GGAGAGGCCCTCCGGGGACCCCAGCGGAGGGGCTGGCCCCTAAGCCAGCTGGCTGCACTGGTGGGCGGGAGCTGTGGGACTGAACGGCGG >95811_95811_3_UBA2-LRP3_UBA2_chr19_34957919_ENST00000592791_LRP3_chr19_33687636_ENST00000253193_length(amino acids)=919AA_BP=172 MPVISPPKIFLNKQPNPRKKLLVPCALDPPNPNCYVCASKPEVTVRLNVHKVTVLTLQDKIVKEKFAMVAPDVQIEDGKGTILISSEEGE TEANNHKKLSEFGIRNGSRLQADDFLQDYTLLINILHSEDLGKDVEFEVVGDAPEKVGPKQAEDAAKSITNGSDDGAQPSTSTVNIFLTG RLSSAVPALAACSGKLEQHTERRGVIYSPAWPLNYPPGTNCSWYIQGDRGDMITISFRNFDVEESHQCSLDWLLLGPAAPPRQEAFRLCG SAIPPAFISARDHVWIFFHSDASSSGQAQGFRLSYIRGKLGQASCQADEFRCDNGKCLPGPWQCNTVDECGDGSDEGNCSAPASEPPGSL CPGGTFPCSGARSTRCLPVERRCDGLQDCGDGSDEAGCPDLACGRRLGSFYGSFASPDLFGAARGPSDLHCTWLVDTQDSRRVLLQLELR LGYDDYVQVYEGLGERGDRLLQTLSYRSNHRPVSLEAAQGRLTVAYHARARSAGHGFNATYQVKGYCLPWEQPCGSSSDSDGGSLGDQGC FSEPQRCDGWWHCASGRDEQGCPACPPDQYPCEGGSGLCYTPADRCNNQKSCPDGADEKNCFSCQPGTFHCGTNLCIFETWRCDGQEDCQ DGSDEHGCLAAVPRKVITAALIGSLVCGLLLVIALGCAFKLYSLRTQEYRAFETQMTRLEAEFVRREAPPSYGQLIAQGLIPPVEDFPVY SASQASVLQNLRTAMRRQMRRHASRRGPSRRRLGRLWNRLFHRPRAPRGQIPLLTAARPSQTVLGDGFLQPAPGAAPDPPAPLMDTGSTR AAGDRPPSAPGRAPEVGPSGPPLPSGLRDPECRPVDKDRKVCREPLVDGPAPADAPREPCSAQDPHPQVSTASSTLGPHSPEPLGVCRNP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for UBA2-LRP3 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for UBA2-LRP3 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for UBA2-LRP3 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
