
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:URI1-ARHGAP33 (FusionGDB2 ID:97064) |
Fusion Gene Summary for URI1-ARHGAP33 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: URI1-ARHGAP33 | Fusion gene ID: 97064 | Hgene | Tgene | Gene symbol | URI1 | ARHGAP33 | Gene ID | 8725 | 115703 |
| Gene name | URI1 prefoldin like chaperone | Rho GTPase activating protein 33 | |
| Synonyms | C19orf2|NNX3|PPP1R19|RMP|URI | NOMA-GAP|SNX26|TCGAP | |
| Cytomap | 19q12 | 19q13.12 | |
| Type of gene | protein-coding | protein-coding | |
| Description | unconventional prefoldin RPB5 interactor 1RNA polymerase II subunit 5-mediating proteinRPB5-mediating proteinprotein phosphatase 1, regulatory subunit 19 | rho GTPase-activating protein 33neurite outgrowth multiadaptor RhoGAP proteinrho-type GTPase-activating protein 33sorting nexin 26tc10/CDC42 GTPase-activating protein | |
| Modification date | 20200313 | 20200313 | |
| UniProtAcc | O94763 | O14559 | |
| Ensembl transtripts involved in fusion gene | ENST00000312051, ENST00000542441, ENST00000392271, ENST00000360605, ENST00000574176, | ENST00000221905, ENST00000007510, ENST00000314737, ENST00000378944, | |
| Fusion gene scores | * DoF score | 30 X 13 X 12=4680 | 1 X 1 X 1=1 |
| # samples | 40 | 1 | |
| ** MAII score | log2(40/4680*10)=-3.54843662469604 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(1/1*10)=3.32192809488736 | |
| Context | PubMed: URI1 [Title/Abstract] AND ARHGAP33 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | URI1(30433571)-ARHGAP33(36275074), # samples:3 | ||
| Anticipated loss of major functional domain due to fusion event. | URI1-ARHGAP33 seems lost the major protein functional domain in Hgene partner, which is a essential gene due to the frame-shifted ORF. | ||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Hgene | URI1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 12737519|15367675|21730289 |
| Hgene | URI1 | GO:0001558 | regulation of cell growth | 21730289 |
| Hgene | URI1 | GO:0071363 | cellular response to growth factor stimulus | 17936702 |
| Hgene | URI1 | GO:0071383 | cellular response to steroid hormone stimulus | 21730289 |
 Fusion gene breakpoints across URI1 (5'-gene) Fusion gene breakpoints across URI1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
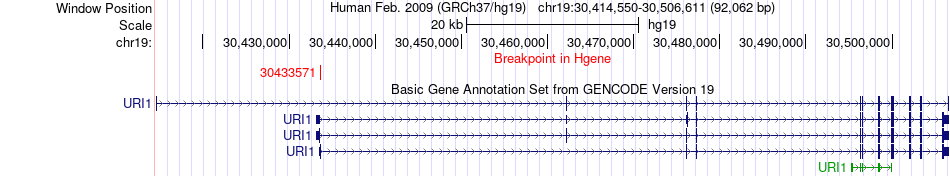 |
 Fusion gene breakpoints across ARHGAP33 (3'-gene) Fusion gene breakpoints across ARHGAP33 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
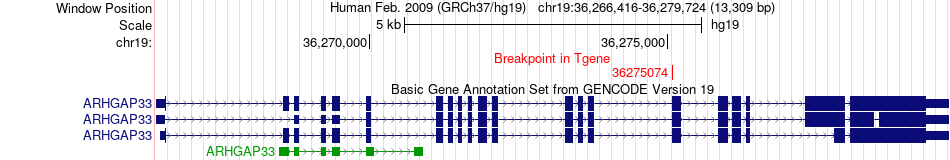 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | UCEC | TCGA-BK-A0CC-01A | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
Top |
Fusion Gene ORF analysis for URI1-ARHGAP33 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 5CDS-intron | ENST00000312051 | ENST00000221905 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| 5CDS-intron | ENST00000542441 | ENST00000221905 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| 5UTR-3CDS | ENST00000392271 | ENST00000007510 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| 5UTR-3CDS | ENST00000392271 | ENST00000314737 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| 5UTR-3CDS | ENST00000392271 | ENST00000378944 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| 5UTR-intron | ENST00000392271 | ENST00000221905 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| Frame-shift | ENST00000542441 | ENST00000007510 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| Frame-shift | ENST00000542441 | ENST00000314737 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| Frame-shift | ENST00000542441 | ENST00000378944 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| In-frame | ENST00000312051 | ENST00000007510 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| In-frame | ENST00000312051 | ENST00000314737 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| In-frame | ENST00000312051 | ENST00000378944 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-3CDS | ENST00000360605 | ENST00000007510 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-3CDS | ENST00000360605 | ENST00000314737 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-3CDS | ENST00000360605 | ENST00000378944 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-3CDS | ENST00000574176 | ENST00000007510 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-3CDS | ENST00000574176 | ENST00000314737 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-3CDS | ENST00000574176 | ENST00000378944 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-intron | ENST00000360605 | ENST00000221905 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
| intron-intron | ENST00000574176 | ENST00000221905 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000312051 | URI1 | chr19 | 30433571 | + | ENST00000378944 | ARHGAP33 | chr19 | 36275074 | + | 2899 | 147 | 10 | 2505 | 831 |
| ENST00000312051 | URI1 | chr19 | 30433571 | + | ENST00000007510 | ARHGAP33 | chr19 | 36275074 | + | 2983 | 147 | 10 | 2589 | 859 |
| ENST00000312051 | URI1 | chr19 | 30433571 | + | ENST00000314737 | ARHGAP33 | chr19 | 36275074 | + | 2500 | 147 | 10 | 2106 | 698 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000312051 | ENST00000378944 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + | 0.1538118 | 0.8461882 |
| ENST00000312051 | ENST00000007510 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + | 0.10302077 | 0.8969792 |
| ENST00000312051 | ENST00000314737 | URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275074 | + | 0.30401662 | 0.69598335 |
Top |
Fusion Genomic Features for URI1-ARHGAP33 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275073 | + | 5.24E-13 | 1 |
| URI1 | chr19 | 30433571 | + | ARHGAP33 | chr19 | 36275073 | + | 5.24E-13 | 1 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
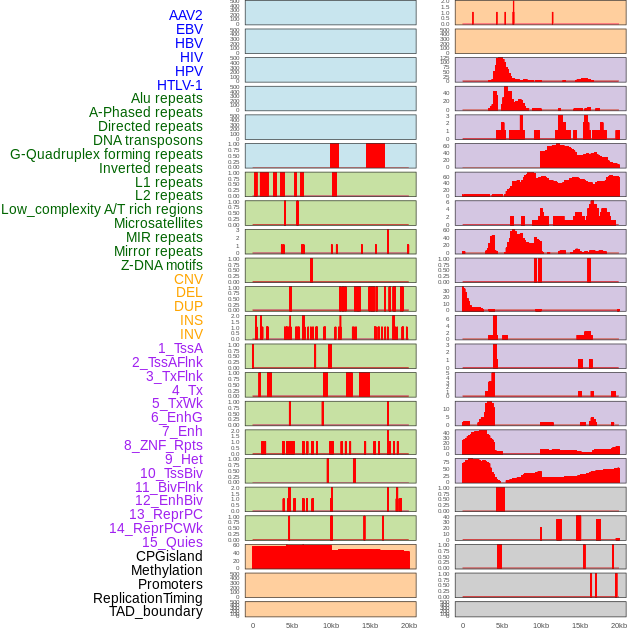 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
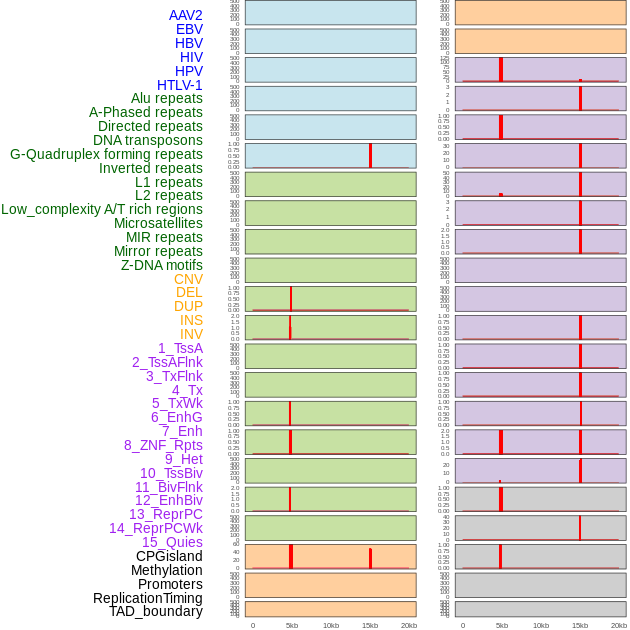 |
Top |
Fusion Protein Features for URI1-ARHGAP33 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr19:30433571/chr19:36275074) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| URI1 | ARHGAP33 |
| FUNCTION: Involved in gene transcription regulation. Acts as a transcriptional repressor in concert with the corepressor UXT to regulate androgen receptor (AR) transcription. May act as a tumor suppressor to repress AR-mediated gene transcription and to inhibit anchorage-independent growth in prostate cancer cells. Required for cell survival in ovarian cancer cells. Together with UXT, associates with chromatin to the NKX3-1 promoter region. Antagonizes transcriptional modulation via hepatitis B virus X protein.; FUNCTION: Plays a central role in maintaining S6K1 signaling and BAD phosphorylation under normal growth conditions thereby protecting cells from potential deleterious effects of sustained S6K1 signaling. The URI1-PPP1CC complex acts as a central component of a negative feedback mechanism that counteracts excessive S6K1 survival signaling to BAD in response to growth factors. Mediates inhibition of PPP1CC phosphatase activity in mitochondria. Coordinates the regulation of nutrient-sensitive gene expression availability in a mTOR-dependent manner. Seems to be a scaffolding protein able to assemble a prefoldin-like complex that contains PFDs and proteins with roles in transcription and ubiquitination. | FUNCTION: May be involved in several stages of intracellular trafficking. Could play an important role in the regulation of glucose transport by insulin. May act as a downstream effector of RHOQ/TC10 in the regulation of insulin-stimulated glucose transport (By similarity). {ECO:0000250}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 1006_1009 | 473 | 1288.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 1183_1188 | 473 | 1288.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 1266_1273 | 473 | 1288.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 677_699 | 473 | 1288.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 874_879 | 473 | 1288.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 889_893 | 473 | 1288.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 1006_1009 | 473 | 1127.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 1183_1188 | 473 | 1127.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 1266_1273 | 473 | 1127.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 677_699 | 473 | 1127.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 874_879 | 473 | 1127.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 889_893 | 473 | 1127.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 1006_1009 | 337 | 1124.0 | Compositional bias | Note=Poly-Gly | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 1183_1188 | 337 | 1124.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 1266_1273 | 337 | 1124.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 677_699 | 337 | 1124.0 | Compositional bias | Note=Poly-Ser | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 874_879 | 337 | 1124.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 889_893 | 337 | 1124.0 | Compositional bias | Note=Poly-Pro |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Hgene | URI1 | chr19:30433571 | chr19:36275074 | ENST00000360605 | + | 1 | 11 | 299_311 | 0 | 476.0 | Compositional bias | Note=Poly-Asp |
| Hgene | URI1 | chr19:30433571 | chr19:36275074 | ENST00000360605 | + | 1 | 11 | 314_321 | 0 | 476.0 | Compositional bias | Note=Poly-Asp |
| Hgene | URI1 | chr19:30433571 | chr19:36275074 | ENST00000392271 | + | 1 | 11 | 299_311 | 0 | 460.0 | Compositional bias | Note=Poly-Asp |
| Hgene | URI1 | chr19:30433571 | chr19:36275074 | ENST00000392271 | + | 1 | 11 | 314_321 | 0 | 460.0 | Compositional bias | Note=Poly-Asp |
| Hgene | URI1 | chr19:30433571 | chr19:36275074 | ENST00000542441 | + | 1 | 11 | 299_311 | 39 | 536.0 | Compositional bias | Note=Poly-Asp |
| Hgene | URI1 | chr19:30433571 | chr19:36275074 | ENST00000542441 | + | 1 | 11 | 314_321 | 39 | 536.0 | Compositional bias | Note=Poly-Asp |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 126_129 | 473 | 1288.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 126_129 | 473 | 1127.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 126_129 | 337 | 1124.0 | Compositional bias | Note=Poly-Pro | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 186_248 | 473 | 1288.0 | Domain | SH3 | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 315_510 | 473 | 1288.0 | Domain | Rho-GAP | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000007510 | 14 | 21 | 59_168 | 473 | 1288.0 | Domain | Note=PX%3B atypical | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 186_248 | 473 | 1127.0 | Domain | SH3 | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 315_510 | 473 | 1127.0 | Domain | Rho-GAP | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000314737 | 14 | 21 | 59_168 | 473 | 1127.0 | Domain | Note=PX%3B atypical | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 186_248 | 337 | 1124.0 | Domain | SH3 | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 315_510 | 337 | 1124.0 | Domain | Rho-GAP | |
| Tgene | ARHGAP33 | chr19:30433571 | chr19:36275074 | ENST00000378944 | 13 | 21 | 59_168 | 337 | 1124.0 | Domain | Note=PX%3B atypical |
Top |
Fusion Gene Sequence for URI1-ARHGAP33 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >97064_97064_1_URI1-ARHGAP33_URI1_chr19_30433571_ENST00000312051_ARHGAP33_chr19_36275074_ENST00000007510_length(transcript)=2983nt_BP=147nt ACACGCCGCGCTGAGGCCCGCGGGCCCGTCATGGAGGCGCCCACCGTGGAGACGCCCCCCGACCCCTCGCCCCCTTCGGCCCCGGCCCCT GCCCTGGTTCCGTTGCGCGCCCCGGATGTGGCGCGGCTGCGCGAGGAGCAGGAAAAGGTCCATGGAGCTGGAGTCAGTGGGAATGGGTGG CGCGGCGGCGTTCCGGGAAGTTCGGGTGCAGTCGGTGGTGGTGGAGTTTCTGCTCACCCATGTGGACGTCCTGTTCAGCGACACCTTCAC CTCCGCCGGCCTCGACCCTGCAGGCCGCTGCCTGCTCCCCAGGCCCAAGTCCCTTGCGGGCAGCTGCCCCTCCACCCGCCTGCTGACGCT GGAGGAAGCCCAGGCACGCACCCAGGGCCGGCTGGGGACGCCCACGGAGCCCACAACTCCCAAGGCCCCGGCCTCACCTGCGGAAAGGAG GAAAGGGGAGAGAGGGGAGAAGCAGCGGAAGCCAGGGGGCAGCAGCTGGAAGACGTTCTTTGCACTGGGCCGGGGCCCCAGTGTCCCTCG AAAGAAGCCCCTGCCCTGGCTGGGGGGCACCCGTGCCCCACCGCAGCCTTCAGGCAGCAGACCCGACACCGTCACACTGAGATCTGCCAA GAGCGAGGAGTCTCTGTCATCGCAGGCCAGCGGGGCTGGCCTCCAGAGGCTGCACAGGCTGCGGCGACCCCACTCCAGCAGCGACGCTTT CCCTGTGGGCCCAGCACCTGCTGGCTCCTGCGAGAGCCTGTCCTCGTCCTCCTCCTCCGAGTCCTCCTCCTCTGAGTCCTCCTCTTCCTC CTCTGAGTCCTCAGCAGCTGGGCTGGGGGCACTCTCTGGGTCTCCCTCACACCGTACCTCAGCCTGGCTAGATGATGGTGATGAGCTGGA CTTCAGCCCACCCCGCTGCCTGGAGGGACTCCGGGGGCTGGACTTTGATCCCTTAACCTTCCGCTGCAGCAGCCCCACCCCAGGGGATCC CGCACCTCCCGCCAGCCCAGCACCCCCCGCCCCTGCCTCTGCCTTCCCACCCAGGGTGACCCCCCAGGCCATCTCGCCCCGGGGGCCCAC CAGCCCCGCCTCGCCTGCTGCCCTAGACATCTCAGAGCCCCTGGCTGTATCAGTGCCACCCGCTGTCCTAGAACTGCTGGGGGCTGGGGG AGCACCTGCCTCAGCCACCCCAACACCAGCTCTCAGCCCCGGCCGGAGCCTGCGCCCCCATCTCATACCCCTGCTGCTGCGAGGAGCCGA GGCCCCGCTGACTGACGCCTGCCAGCAGGAGATGTGCAGCAAGCTCCGGGGAGCCCAGGGCCCACTCGGTCCTGATATGGAGTCACCACT GCCACCCCCTCCCCTGTCTCTCCTGCGCCCTGGGGGTGCCCCACCCCCGCCCCCTAAGAACCCAGCACGCCTCATGGCCCTGGCCCTGGC TGAGCGGGCTCAGCAGGTGGCCGAGCAACAGAGCCAGCAGGAGTGTGGGGGCACCCCACCTGCTTCCCAATCCCCCTTCCACCGCTCGCT GTCTCTGGAGGTGGGCGGGGAGCCCCTGGGGACCTCAGGGAGTGGGCCACCTCCCAACTCCCTAGCACACCCGGGTGCCTGGGTCCCGGG ACCCCCACCCTACTTACCAAGGCAACAAAGTGATGGGAGCCTGCTGAGGAGCCAGCGGCCCATGGGGACCTCAAGGAGGGGACTCCGAGG CCCTGCCCAGGTCAGTGCCCAGCTCAGGGCAGGTGGCGGGGGCAGGGATGCGCCAGAGGCAGCAGCCCAGTCCCCATGTTCTGTCCCCTC ACAGGTTCCTACCCCCGGCTTCTTCTCCCCAGCCCCCAGGGAGTGCCTGCCACCCTTCCTCGGGGTCCCCAAGCCAGGCTTGTACCCCCT GGGCCCCCCATCCTTCCAGCCCAGTTCCCCAGCCCCAGTCTGGAGGAGCTCTCTGGGCCCCCCTGCACCACTCGACAGGGGAGAGAACCT GTACTATGAGATCGGGGCAAGTGAGGGGTCCCCCTATTCTGGCCCCACCCGCTCCTGGAGTCCCTTTCGCTCCATGCCCCCCGACAGGCT CAATGCCTCCTACGGCATGCTTGGCCAATCACCCCCACTCCACAGGTCCCCCGACTTCCTGCTCAGCTACCCGCCAGCCCCCTCCTGCTT TCCCCCTGACCACCTTGGCTACTCAGCCCCCCAGCACCCTGCTCGGCGCCCTACACCGCCTGAGCCCCTCTACGTCAACCTAGCTCTAGG GCCCAGGGGTCCCTCACCTGCCTCTTCCTCCTCCTCTTCCCCTCCTGCCCACCCCCGAAGCCGTTCAGATCCCGGTCCCCCAGTCCCCCG CCTTCCCCAGAAACAACGGGCACCCTGGGGACCCCGTACCCCTCATAGGGTGCCGGGTCCCTGGGGCCCTCCTGAGCCTCTCCTGCTCTA CAGGGCAGCCCCGCCAGCCTACGGAAGGGGGGGCGAGCTCCACCGAGGGTCCTTGTACAGAAATGGAGGGCAAAGAGGGGAGGGGGCTGG TCCCCCACCCCCTTACCCCACTCCCAGCTGGTCCCTCCACTCTGAGGGCCAGACCCGAAGCTACTGCTGAGCACCAGCTGGGAGGGGCCG TCCTTCCTTCCCTTCACCCTCACTGGATCTTGGCCCAACCAAATCCCTTGTTTTGTATTTTCTTGAACCCCGACCACTACCCCAGGTTTC TAACTTTGTAACTTGCTTCTGATGTGGGTCCCTAACCTATAATCTCAGCTTCCCTACCCTGGACTGAAGGGTCTGCCCATCCCCCCACCA CCCTCCATCCTGGGGGCCCTCGCACAAATCTGGGGTGGGAGGGGCTAGGCTGACCCCATCCTCCTCTCCCTCCAGGAGCCCCCAGCATGT CCTGACCTGTGCACGGGGATGGGGGGACAACTCCTACCCTTCTTTCCCCACATGCCCCACTAAACCATCTGACAACATTAATGAATAAAA >97064_97064_1_URI1-ARHGAP33_URI1_chr19_30433571_ENST00000312051_ARHGAP33_chr19_36275074_ENST00000007510_length(amino acids)=859AA_BP=45 MRPAGPSWRRPPWRRPPTPRPLRPRPLPWFRCAPRMWRGCARSRKRSMELESVGMGGAAAFREVRVQSVVVEFLLTHVDVLFSDTFTSAG LDPAGRCLLPRPKSLAGSCPSTRLLTLEEAQARTQGRLGTPTEPTTPKAPASPAERRKGERGEKQRKPGGSSWKTFFALGRGPSVPRKKP LPWLGGTRAPPQPSGSRPDTVTLRSAKSEESLSSQASGAGLQRLHRLRRPHSSSDAFPVGPAPAGSCESLSSSSSSESSSSESSSSSSES SAAGLGALSGSPSHRTSAWLDDGDELDFSPPRCLEGLRGLDFDPLTFRCSSPTPGDPAPPASPAPPAPASAFPPRVTPQAISPRGPTSPA SPAALDISEPLAVSVPPAVLELLGAGGAPASATPTPALSPGRSLRPHLIPLLLRGAEAPLTDACQQEMCSKLRGAQGPLGPDMESPLPPP PLSLLRPGGAPPPPPKNPARLMALALAERAQQVAEQQSQQECGGTPPASQSPFHRSLSLEVGGEPLGTSGSGPPPNSLAHPGAWVPGPPP YLPRQQSDGSLLRSQRPMGTSRRGLRGPAQVSAQLRAGGGGRDAPEAAAQSPCSVPSQVPTPGFFSPAPRECLPPFLGVPKPGLYPLGPP SFQPSSPAPVWRSSLGPPAPLDRGENLYYEIGASEGSPYSGPTRSWSPFRSMPPDRLNASYGMLGQSPPLHRSPDFLLSYPPAPSCFPPD HLGYSAPQHPARRPTPPEPLYVNLALGPRGPSPASSSSSSPPAHPRSRSDPGPPVPRLPQKQRAPWGPRTPHRVPGPWGPPEPLLLYRAA -------------------------------------------------------------- >97064_97064_2_URI1-ARHGAP33_URI1_chr19_30433571_ENST00000312051_ARHGAP33_chr19_36275074_ENST00000314737_length(transcript)=2500nt_BP=147nt ACACGCCGCGCTGAGGCCCGCGGGCCCGTCATGGAGGCGCCCACCGTGGAGACGCCCCCCGACCCCTCGCCCCCTTCGGCCCCGGCCCCT GCCCTGGTTCCGTTGCGCGCCCCGGATGTGGCGCGGCTGCGCGAGGAGCAGGAAAAGGTCCATGGAGCTGGAGTCAGTGGGAATGGGTGG CGCGGCGGCGTTCCGGGAAGTTCGGGTGCAGTCGGTGGTGGTGGAGTTTCTGCTCACCCATGTGGACGTCCTGTTCAGCGACACCTTCAC CTCCGCCGGCCTCGACCCTGCAGGCCGCTGCCTGCTCCCCAGGCCCAAGTCCCTTGCGGGCAGCTGCCCCTCCACCCGCCTGCTGACGCT GGAGGAAGCCCAGGCACGCACCCAGGGCCGGCTGGGGACGCCCACGGAGCCCACAACTCCCAAGGCCCCGGCCTCACCTGCGGAAAGGAG GAAAGGGGAGAGAGGGGAGAAGCAGCGGAAGCCAGGGGGCAGCAGCTGGAAGACGTTCTTTGCACTGGGCCGGGGCCCCAGTGTCCCTCG AAAGAAGCCCCTGCCCTGGCTGGGGGGCACCCGTGCCCCACCGCAGCCTTCAGGCAGCAGACCCGACACCGTCACACTGAGATCTGCCAA GAGCGAGGAGTCTCTGTCATCGCAGGCCAGCGGGGCTGAACTGCTGGGGGCTGGGGGAGCACCTGCCTCAGCCACCCCAACACCAGCTCT CAGCCCCGGCCGGAGCCTGCGCCCCCATCTCATACCCCTGCTGCTGCGAGGAGCCGAGGCCCCGCTGACTGACGCCTGCCAGCAGGAGAT GTGCAGCAAGCTCCGGGGAGCCCAGGGCCCACTCGGTCCTGATATGGAGTCACCACTGCCACCCCCTCCCCTGTCTCTCCTGCGCCCTGG GGGTGCCCCACCCCCGCCCCCTAAGAACCCAGCACGCCTCATGGCCCTGGCCCTGGCTGAGCGGGCTCAGCAGGTGGCCGAGCAACAGAG CCAGCAGGAGTGTGGGGGCACCCCACCTGCTTCCCAATCCCCCTTCCACCGCTCGCTGTCTCTGGAGGTGGGCGGGGAGCCCCTGGGGAC CTCAGGGAGTGGGCCACCTCCCAACTCCCTAGCACACCCGGGTGCCTGGGTCCCGGGACCCCCACCCTACTTACCAAGGCAACAAAGTGA TGGGAGCCTGCTGAGGAGCCAGCGGCCCATGGGGACCTCAAGGAGGGGACTCCGAGGCCCTGCCCAGGTCAGTGCCCAGCTCAGGGCAGG TGGCGGGGGCAGGGATGCGCCAGAGGCAGCAGCCCAGTCCCCATGTTCTGTCCCCTCACAGGTTCCTACCCCCGGCTTCTTCTCCCCAGC CCCCAGGGAGTGCCTGCCACCCTTCCTCGGGGTCCCCAAGCCAGGCTTGTACCCCCTGGGCCCCCCATCCTTCCAGCCCAGTTCCCCAGC CCCAGTCTGGAGGAGCTCTCTGGGCCCCCCTGCACCACTCGACAGGGGAGAGAACCTGTACTATGAGATCGGGGCAAGTGAGGGGTCCCC CTATTCTGGCCCCACCCGCTCCTGGAGTCCCTTTCGCTCCATGCCCCCCGACAGGCTCAATGCCTCCTACGGCATGCTTGGCCAATCACC CCCACTCCACAGGTCCCCCGACTTCCTGCTCAGCTACCCGCCAGCCCCCTCCTGCTTTCCCCCTGACCACCTTGGCTACTCAGCCCCCCA GCACCCTGCTCGGCGCCCTACACCGCCTGAGCCCCTCTACGTCAACCTAGCTCTAGGGCCCAGGGGTCCCTCACCTGCCTCTTCCTCCTC CTCTTCCCCTCCTGCCCACCCCCGAAGCCGTTCAGATCCCGGTCCCCCAGTCCCCCGCCTTCCCCAGAAACAACGGGCACCCTGGGGACC CCGTACCCCTCATAGGGTGCCGGGTCCCTGGGGCCCTCCTGAGCCTCTCCTGCTCTACAGGGCAGCCCCGCCAGCCTACGGAAGGGGGGG CGAGCTCCACCGAGGGTCCTTGTACAGAAATGGAGGGCAAAGAGGGGAGGGGGCTGGTCCCCCACCCCCTTACCCCACTCCCAGCTGGTC CCTCCACTCTGAGGGCCAGACCCGAAGCTACTGCTGAGCACCAGCTGGGAGGGGCCGTCCTTCCTTCCCTTCACCCTCACTGGATCTTGG CCCAACCAAATCCCTTGTTTTGTATTTTCTTGAACCCCGACCACTACCCCAGGTTTCTAACTTTGTAACTTGCTTCTGATGTGGGTCCCT AACCTATAATCTCAGCTTCCCTACCCTGGACTGAAGGGTCTGCCCATCCCCCCACCACCCTCCATCCTGGGGGCCCTCGCACAAATCTGG GGTGGGAGGGGCTAGGCTGACCCCATCCTCCTCTCCCTCCAGGAGCCCCCAGCATGTCCTGACCTGTGCACGGGGATGGGGGGACAACTC >97064_97064_2_URI1-ARHGAP33_URI1_chr19_30433571_ENST00000312051_ARHGAP33_chr19_36275074_ENST00000314737_length(amino acids)=698AA_BP=45 MRPAGPSWRRPPWRRPPTPRPLRPRPLPWFRCAPRMWRGCARSRKRSMELESVGMGGAAAFREVRVQSVVVEFLLTHVDVLFSDTFTSAG LDPAGRCLLPRPKSLAGSCPSTRLLTLEEAQARTQGRLGTPTEPTTPKAPASPAERRKGERGEKQRKPGGSSWKTFFALGRGPSVPRKKP LPWLGGTRAPPQPSGSRPDTVTLRSAKSEESLSSQASGAELLGAGGAPASATPTPALSPGRSLRPHLIPLLLRGAEAPLTDACQQEMCSK LRGAQGPLGPDMESPLPPPPLSLLRPGGAPPPPPKNPARLMALALAERAQQVAEQQSQQECGGTPPASQSPFHRSLSLEVGGEPLGTSGS GPPPNSLAHPGAWVPGPPPYLPRQQSDGSLLRSQRPMGTSRRGLRGPAQVSAQLRAGGGGRDAPEAAAQSPCSVPSQVPTPGFFSPAPRE CLPPFLGVPKPGLYPLGPPSFQPSSPAPVWRSSLGPPAPLDRGENLYYEIGASEGSPYSGPTRSWSPFRSMPPDRLNASYGMLGQSPPLH RSPDFLLSYPPAPSCFPPDHLGYSAPQHPARRPTPPEPLYVNLALGPRGPSPASSSSSSPPAHPRSRSDPGPPVPRLPQKQRAPWGPRTP -------------------------------------------------------------- >97064_97064_3_URI1-ARHGAP33_URI1_chr19_30433571_ENST00000312051_ARHGAP33_chr19_36275074_ENST00000378944_length(transcript)=2899nt_BP=147nt ACACGCCGCGCTGAGGCCCGCGGGCCCGTCATGGAGGCGCCCACCGTGGAGACGCCCCCCGACCCCTCGCCCCCTTCGGCCCCGGCCCCT GCCCTGGTTCCGTTGCGCGCCCCGGATGTGGCGCGGCTGCGCGAGGAGCAGGAAAAGGTCCATGGAGCTGGAGTCAGTGGGAATGGGTGG CGCGGCGGCGTTCCGGGAAGTTCGGGTGCAGTCGGTGGTGGTGGAGTTTCTGCTCACCCATGTGGACGTCCTGTTCAGCGACACCTTCAC CTCCGCCGGCCTCGACCCTGCAGGCCGCTGCCTGCTCCCCAGGCCCAAGTCCCTTGCGGGCAGCTGCCCCTCCACCCGCCTGCTGACGCT GGAGGAAGCCCAGGCACGCACCCAGGGCCGGCTGGGGACGCCCACGGAGCCCACAACTCCCAAGGCCCCGGCCTCACCTGCGGAAAGGAG GAAAGGGGAGAGAGGGGAGAAGCAGCGGAAGCCAGGGGGCAGCAGCTGGAAGACGTTCTTTGCACTGGGCCGGGGCCCCAGTGTCCCTCG AAAGAAGCCCCTGCCCTGGCTGGGGGGCACCCGTGCCCCACCGCAGCCTTCAGGCAGCAGACCCGACACCGTCACACTGAGATCTGCCAA GAGCGAGGAGTCTCTGTCATCGCAGGCCAGCGGGGCTGGCCTCCAGAGGCTGCACAGGCTGCGGCGACCCCACTCCAGCAGCGACGCTTT CCCTGTGGGCCCAGCACCTGCTGGCTCCTGCGAGAGCCTGTCCTCGTCCTCCTCCTCCGAGTCCTCCTCCTCTGAGTCCTCCTCTTCCTC CTCTGAGTCCTCAGCAGCTGGGCTGGGGGCACTCTCTGGGTCTCCCTCACACCGTACCTCAGCCTGGCTAGATGATGGTGATGAGCTGGA CTTCAGCCCACCCCGCTGCCTGGAGGGACTCCGGGGGCTGGACTTTGATCCCTTAACCTTCCGCTGCAGCAGCCCCACCCCAGGGGATCC CGCACCTCCCGCCAGCCCAGCACCCCCCGCCCCTGCCTCTGCCTTCCCACCCAGGGTGACCCCCCAGGCCATCTCGCCCCGGGGGCCCAC CAGCCCCGCCTCGCCTGCTGCCCTAGACATCTCAGAGCCCCTGGCTGTATCAGTGCCACCCGCTGTCCTAGAACTGCTGGGGGCTGGGGG AGCACCTGCCTCAGCCACCCCAACACCAGCTCTCAGCCCCGGCCGGAGCCTGCGCCCCCATCTCATACCCCTGCTGCTGCGAGGAGCCGA GGCCCCGCTGACTGACGCCTGCCAGCAGGAGATGTGCAGCAAGCTCCGGGGAGCCCAGGGCCCACTCGGTCCTGATATGGAGTCACCACT GCCACCCCCTCCCCTGTCTCTCCTGCGCCCTGGGGGTGCCCCACCCCCGCCCCCTAAGAACCCAGCACGCCTCATGGCCCTGGCCCTGGC TGAGCGGGCTCAGCAGGTGGCCGAGCAACAGAGCCAGCAGGAGTGTGGGGGCACCCCACCTGCTTCCCAATCCCCCTTCCACCGCTCGCT GTCTCTGGAGGTGGGCGGGGAGCCCCTGGGGACCTCAGGGAGTGGGCCACCTCCCAACTCCCTAGCACACCCGGGTGCCTGGGTCCCGGG ACCCCCACCCTACTTACCAAGGCAACAAAGTGATGGGAGCCTGCTGAGGAGCCAGCGGCCCATGGGGACCTCAAGGAGGGGACTCCGAGG CCCTGCCCAGGTTCCTACCCCCGGCTTCTTCTCCCCAGCCCCCAGGGAGTGCCTGCCACCCTTCCTCGGGGTCCCCAAGCCAGGCTTGTA CCCCCTGGGCCCCCCATCCTTCCAGCCCAGTTCCCCAGCCCCAGTCTGGAGGAGCTCTCTGGGCCCCCCTGCACCACTCGACAGGGGAGA GAACCTGTACTATGAGATCGGGGCAAGTGAGGGGTCCCCCTATTCTGGCCCCACCCGCTCCTGGAGTCCCTTTCGCTCCATGCCCCCCGA CAGGCTCAATGCCTCCTACGGCATGCTTGGCCAATCACCCCCACTCCACAGGTCCCCCGACTTCCTGCTCAGCTACCCGCCAGCCCCCTC CTGCTTTCCCCCTGACCACCTTGGCTACTCAGCCCCCCAGCACCCTGCTCGGCGCCCTACACCGCCTGAGCCCCTCTACGTCAACCTAGC TCTAGGGCCCAGGGGTCCCTCACCTGCCTCTTCCTCCTCCTCTTCCCCTCCTGCCCACCCCCGAAGCCGTTCAGATCCCGGTCCCCCAGT CCCCCGCCTTCCCCAGAAACAACGGGCACCCTGGGGACCCCGTACCCCTCATAGGGTGCCGGGTCCCTGGGGCCCTCCTGAGCCTCTCCT GCTCTACAGGGCAGCCCCGCCAGCCTACGGAAGGGGGGGCGAGCTCCACCGAGGGTCCTTGTACAGAAATGGAGGGCAAAGAGGGGAGGG GGCTGGTCCCCCACCCCCTTACCCCACTCCCAGCTGGTCCCTCCACTCTGAGGGCCAGACCCGAAGCTACTGCTGAGCACCAGCTGGGAG GGGCCGTCCTTCCTTCCCTTCACCCTCACTGGATCTTGGCCCAACCAAATCCCTTGTTTTGTATTTTCTTGAACCCCGACCACTACCCCA GGTTTCTAACTTTGTAACTTGCTTCTGATGTGGGTCCCTAACCTATAATCTCAGCTTCCCTACCCTGGACTGAAGGGTCTGCCCATCCCC CCACCACCCTCCATCCTGGGGGCCCTCGCACAAATCTGGGGTGGGAGGGGCTAGGCTGACCCCATCCTCCTCTCCCTCCAGGAGCCCCCA GCATGTCCTGACCTGTGCACGGGGATGGGGGGACAACTCCTACCCTTCTTTCCCCACATGCCCCACTAAACCATCTGACAACATTAATGA >97064_97064_3_URI1-ARHGAP33_URI1_chr19_30433571_ENST00000312051_ARHGAP33_chr19_36275074_ENST00000378944_length(amino acids)=831AA_BP=45 MRPAGPSWRRPPWRRPPTPRPLRPRPLPWFRCAPRMWRGCARSRKRSMELESVGMGGAAAFREVRVQSVVVEFLLTHVDVLFSDTFTSAG LDPAGRCLLPRPKSLAGSCPSTRLLTLEEAQARTQGRLGTPTEPTTPKAPASPAERRKGERGEKQRKPGGSSWKTFFALGRGPSVPRKKP LPWLGGTRAPPQPSGSRPDTVTLRSAKSEESLSSQASGAGLQRLHRLRRPHSSSDAFPVGPAPAGSCESLSSSSSSESSSSESSSSSSES SAAGLGALSGSPSHRTSAWLDDGDELDFSPPRCLEGLRGLDFDPLTFRCSSPTPGDPAPPASPAPPAPASAFPPRVTPQAISPRGPTSPA SPAALDISEPLAVSVPPAVLELLGAGGAPASATPTPALSPGRSLRPHLIPLLLRGAEAPLTDACQQEMCSKLRGAQGPLGPDMESPLPPP PLSLLRPGGAPPPPPKNPARLMALALAERAQQVAEQQSQQECGGTPPASQSPFHRSLSLEVGGEPLGTSGSGPPPNSLAHPGAWVPGPPP YLPRQQSDGSLLRSQRPMGTSRRGLRGPAQVPTPGFFSPAPRECLPPFLGVPKPGLYPLGPPSFQPSSPAPVWRSSLGPPAPLDRGENLY YEIGASEGSPYSGPTRSWSPFRSMPPDRLNASYGMLGQSPPLHRSPDFLLSYPPAPSCFPPDHLGYSAPQHPARRPTPPEPLYVNLALGP RGPSPASSSSSSPPAHPRSRSDPGPPVPRLPQKQRAPWGPRTPHRVPGPWGPPEPLLLYRAAPPAYGRGGELHRGSLYRNGGQRGEGAGP -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for URI1-ARHGAP33 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for URI1-ARHGAP33 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for URI1-ARHGAP33 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
