
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:USB1-CNOT1 (FusionGDB2 ID:97107) |
Fusion Gene Summary for USB1-CNOT1 |
 Fusion gene summary Fusion gene summary |
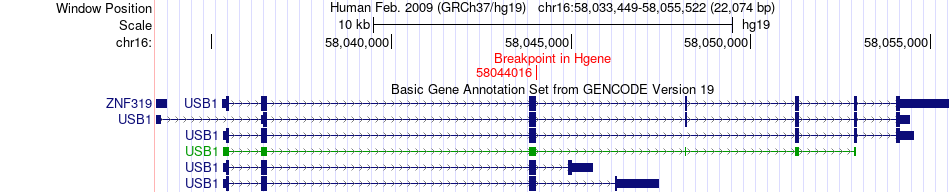
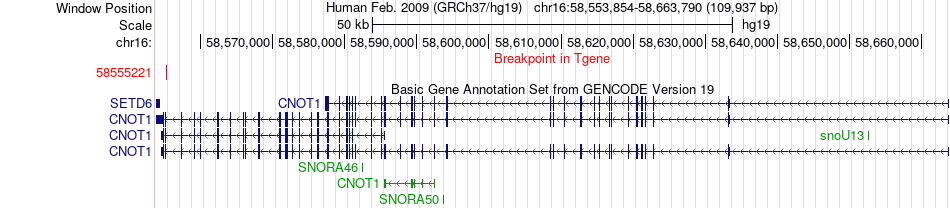
| Fusion gene information | Fusion gene name: USB1-CNOT1 | Fusion gene ID: 97107 | Hgene | Tgene | Gene symbol | USB1 | CNOT1 | Gene ID | 79650 | 23019 |
| Gene name | U6 snRNA biogenesis phosphodiesterase 1 | CCR4-NOT transcription complex subunit 1 | |
| Synonyms | C16orf57|HVSL1|Mpn1|PN|hUsb1 | AD-005|CDC39|HPE12|NOT1|NOT1H | |
| Cytomap | 16q21 | 16q21 | |
| Type of gene | protein-coding | protein-coding | |
| Description | U6 snRNA phosphodiesteraseHVSL motif containing 1U six biogenesis 1U6 snRNA biogenesis 1UPF0406 protein C16orf57mutated in poikiloderma with neutropenia protein 1putative U6 snRNA phosphodiesterase | CCR4-NOT transcription complex subunit 1CCR4-associated factor 1NOT1 (negative regulator of transcription 1, yeast) homologadrenal gland protein AD-005negative regulator of transcription subunit 1 homolog | |
| Modification date | 20200328 | 20200322 | |
| UniProtAcc | Q9BQ65 | Q9UKZ1 | |
| Ensembl transtripts involved in fusion gene | ENST00000565662, ENST00000219281, ENST00000423271, ENST00000539737, ENST00000561743, ENST00000563149, | ENST00000317147, ENST00000569240, ENST00000245138, ENST00000441024, ENST00000569732, | |
| Fusion gene scores | * DoF score | 6 X 6 X 5=180 | 13 X 11 X 8=1144 |
| # samples | 6 | 15 | |
| ** MAII score | log2(6/180*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(15/1144*10)=-2.93105264628251 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: USB1 [Title/Abstract] AND CNOT1 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | USB1(58044016)-CNOT1(58555221), # samples:1 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | CNOT1 | GO:0000122 | negative regulation of transcription by RNA polymerase II | 16778766 |
| Tgene | CNOT1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly | 21976065 |
| Tgene | CNOT1 | GO:0017148 | negative regulation of translation | 24736845 |
| Tgene | CNOT1 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway | 16778766 |
| Tgene | CNOT1 | GO:0035195 | gene silencing by miRNA | 23172285|24768540 |
| Tgene | CNOT1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway | 16778766 |
| Tgene | CNOT1 | GO:0060213 | positive regulation of nuclear-transcribed mRNA poly(A) tail shortening | 23644599 |
 Fusion gene breakpoints across USB1 (5'-gene) Fusion gene breakpoints across USB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene breakpoints across CNOT1 (3'-gene) Fusion gene breakpoints across CNOT1 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | GBM | TCGA-27-1831-01A | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
Top |
Fusion Gene ORF analysis for USB1-CNOT1 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000565662 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 3UTR-3CDS | ENST00000565662 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 3UTR-intron | ENST00000565662 | ENST00000245138 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 3UTR-intron | ENST00000565662 | ENST00000441024 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 3UTR-intron | ENST00000565662 | ENST00000569732 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000219281 | ENST00000245138 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000219281 | ENST00000441024 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000219281 | ENST00000569732 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000423271 | ENST00000245138 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000423271 | ENST00000441024 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000423271 | ENST00000569732 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000539737 | ENST00000245138 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000539737 | ENST00000441024 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000539737 | ENST00000569732 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000561743 | ENST00000245138 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000561743 | ENST00000441024 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000561743 | ENST00000569732 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000563149 | ENST00000245138 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000563149 | ENST00000441024 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| 5CDS-intron | ENST00000563149 | ENST00000569732 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000219281 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000219281 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000423271 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000423271 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000539737 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000539737 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000561743 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000561743 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000563149 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
| In-frame | ENST00000563149 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000561743 | USB1 | chr16 | 58044016 | + | ENST00000317147 | CNOT1 | chr16 | 58555221 | - | 1710 | 489 | 67 | 702 | 211 |
| ENST00000561743 | USB1 | chr16 | 58044016 | + | ENST00000569240 | CNOT1 | chr16 | 58555221 | - | 966 | 489 | 67 | 702 | 211 |
| ENST00000219281 | USB1 | chr16 | 58044016 | + | ENST00000317147 | CNOT1 | chr16 | 58555221 | - | 1781 | 560 | 111 | 773 | 220 |
| ENST00000219281 | USB1 | chr16 | 58044016 | + | ENST00000569240 | CNOT1 | chr16 | 58555221 | - | 1037 | 560 | 111 | 773 | 220 |
| ENST00000539737 | USB1 | chr16 | 58044016 | + | ENST00000317147 | CNOT1 | chr16 | 58555221 | - | 1749 | 528 | 79 | 741 | 220 |
| ENST00000539737 | USB1 | chr16 | 58044016 | + | ENST00000569240 | CNOT1 | chr16 | 58555221 | - | 1005 | 528 | 79 | 741 | 220 |
| ENST00000423271 | USB1 | chr16 | 58044016 | + | ENST00000317147 | CNOT1 | chr16 | 58555221 | - | 1747 | 526 | 77 | 739 | 220 |
| ENST00000423271 | USB1 | chr16 | 58044016 | + | ENST00000569240 | CNOT1 | chr16 | 58555221 | - | 1003 | 526 | 77 | 739 | 220 |
| ENST00000563149 | USB1 | chr16 | 58044016 | + | ENST00000317147 | CNOT1 | chr16 | 58555221 | - | 1734 | 513 | 64 | 726 | 220 |
| ENST00000563149 | USB1 | chr16 | 58044016 | + | ENST00000569240 | CNOT1 | chr16 | 58555221 | - | 990 | 513 | 64 | 726 | 220 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000561743 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.22119185 | 0.7788082 |
| ENST00000561743 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.08584296 | 0.9141571 |
| ENST00000219281 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.03701618 | 0.96298385 |
| ENST00000219281 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.030822037 | 0.9691779 |
| ENST00000539737 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.028979717 | 0.9710202 |
| ENST00000539737 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.029359365 | 0.9706406 |
| ENST00000423271 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.02630588 | 0.97369415 |
| ENST00000423271 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.027469862 | 0.9725301 |
| ENST00000563149 | ENST00000317147 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.026707225 | 0.97329277 |
| ENST00000563149 | ENST00000569240 | USB1 | chr16 | 58044016 | + | CNOT1 | chr16 | 58555221 | - | 0.027760945 | 0.9722391 |
Top |
Fusion Genomic Features for USB1-CNOT1 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
Top |
Fusion Protein Features for USB1-CNOT1 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:58044016/chr16:58555221) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| USB1 | CNOT1 |
| FUNCTION: Phosphodiesterase responsible for the U6 snRNA 3' end processing. Acts as an exoribonuclease (RNase) responsible for trimming the poly(U) tract of the last nucleotides in the pre-U6 snRNA molecule, leading to the formation of mature U6 snRNA 3' end-terminated with a 2',3'-cyclic phosphate. {ECO:0000255|HAMAP-Rule:MF_03040, ECO:0000269|PubMed:22899009, ECO:0000269|PubMed:23190533}. | FUNCTION: Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. Is required for the association of CNOT10 with the CCR4-NOT complex. Seems not to be required for complex deadenylase function. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 1333_1350 | 0 | 1552.0 | Compositional bias | Note=Thr-rich | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 153_157 | 0 | 1552.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 1639_1643 | 0 | 1552.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 181_185 | 0 | 1552.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 1942_1946 | 0 | 1552.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 2096_2100 | 0 | 1552.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 223_227 | 0 | 1552.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 570_574 | 0 | 1552.0 | Motif | Note=LXXLL |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 1333_1350 | 2305 | 2377.0 | Compositional bias | Note=Thr-rich | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 1333_1350 | 2300 | 2372.0 | Compositional bias | Note=Thr-rich | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 153_157 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 1639_1643 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 181_185 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 1942_1946 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 2096_2100 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 223_227 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 570_574 | 2305 | 2377.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 153_157 | 2300 | 2372.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 1639_1643 | 2300 | 2372.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 181_185 | 2300 | 2372.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 1942_1946 | 2300 | 2372.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 2096_2100 | 2300 | 2372.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 223_227 | 2300 | 2372.0 | Motif | Note=LXXLL | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 570_574 | 2300 | 2372.0 | Motif | Note=LXXLL |
Top |
Fusion Gene Sequence for USB1-CNOT1 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >97107_97107_1_USB1-CNOT1_USB1_chr16_58044016_ENST00000219281_CNOT1_chr16_58555221_ENST00000317147_length(transcript)=1781nt_BP=560nt GCGGTGCCAGCCCAGGCCCCGCCCCTGGGAGGGCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCT GGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGG ACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTC CCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACC CACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTG GTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCC CGTATGACCTCCTTCCACAGAGTTCTCTTGGAACGGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTG ATTAAAAACCCAGCGTTTAAGTTCTGGAACCATGAATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGC TGCATGGGACAGAAGCAGGCCCAGCAAGTAATGGAAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGA GGTCTCACTGCACCGAGTTCATAAACTGACTGAAGAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCC CAACTACTGTTGGGATCAGCCTCCTGTCTTATGTGGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGA TGCTGTGAATATCATTTTGAACTTGTGTAAATACATGAAAGAGAAAACCTTTGTCTGGAACTTCTTGGCTTTGTGCAAGCTGTGTCCAAG GCAAGTACATAAACTGGTACCTTGTAATGAAGAGGCAGCTGATGCCATGCACTTGTCTGAGGGCATAGCTCCATGTCTTCTGACATTCCT GGTGTCCCAAAGAATAGCAAAAAGCCAGTTTGAATATTATGTAACTTATTTTTTTAATGTGGACAGGGGACCTTGAAAATCACTAAGTTA TTAAAAATGTGGATGTGCTAGAATTGGATATGTCCAGGAACATGGGAAGGGCTCACTATTGGAATCCCATGAGTTTCCATTTTGTCTCTA CCCAAACGTATTCCAAAGCTGACTGCATTTGTACCATCTTATTTCTTTTGGGGATTATACACCTCAGCCGCCTGAGATGGGGGTCAGCTC TTTATATAAAGGGAAACCAGACCAGGCCTAAAGCCCACCCCTACCCTCACCCCCCCAATCCTCTCCTGAAACTTAAAAACAGTGGGAATA TAGGAAAGGGAACCAAATCTCATTAATTAATTGTTCTCCCCCATTACCCCACTGAATGAATGGCCATACAGGCTAAGCTGAATAATGACA AAGTTGAAAGGACCAATACAGCCCCTTTTATAAGGATTTTGAATGTTTTGCAAATGTATTGGTCCCTGTGTTGTATTTTGTAGCCTTTTC >97107_97107_1_USB1-CNOT1_USB1_chr16_58044016_ENST00000219281_CNOT1_chr16_58555221_ENST00000317147_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_2_USB1-CNOT1_USB1_chr16_58044016_ENST00000219281_CNOT1_chr16_58555221_ENST00000569240_length(transcript)=1037nt_BP=560nt GCGGTGCCAGCCCAGGCCCCGCCCCTGGGAGGGCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCT GGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGG ACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTC CCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACC CACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTG GTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCC CGTATGACCTCCTTCCACAGAGTTCTCTTGGAACGGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTG ATTAAAAACCCAGCGTTTAAGTTCTGGAACCATGAATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGC TGCATGGGACAGAAGCAGGCCCAGCAAGTAATGGAAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGA GGTCTCACTGCACCGAGTTCATAAACTGACTGAAGAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCC CAACTACTGTTGGGATCAGCCTCCTGTCTTATGTGGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGA >97107_97107_2_USB1-CNOT1_USB1_chr16_58044016_ENST00000219281_CNOT1_chr16_58555221_ENST00000569240_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_3_USB1-CNOT1_USB1_chr16_58044016_ENST00000423271_CNOT1_chr16_58555221_ENST00000317147_length(transcript)=1747nt_BP=526nt GCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGC CCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCC AGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCA CAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGT TCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCC AGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGAAC GGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCATG AATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAATGG AAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTGAA GAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTATGT GGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAATAC ATGAAAGAGAAAACCTTTGTCTGGAACTTCTTGGCTTTGTGCAAGCTGTGTCCAAGGCAAGTACATAAACTGGTACCTTGTAATGAAGAG GCAGCTGATGCCATGCACTTGTCTGAGGGCATAGCTCCATGTCTTCTGACATTCCTGGTGTCCCAAAGAATAGCAAAAAGCCAGTTTGAA TATTATGTAACTTATTTTTTTAATGTGGACAGGGGACCTTGAAAATCACTAAGTTATTAAAAATGTGGATGTGCTAGAATTGGATATGTC CAGGAACATGGGAAGGGCTCACTATTGGAATCCCATGAGTTTCCATTTTGTCTCTACCCAAACGTATTCCAAAGCTGACTGCATTTGTAC CATCTTATTTCTTTTGGGGATTATACACCTCAGCCGCCTGAGATGGGGGTCAGCTCTTTATATAAAGGGAAACCAGACCAGGCCTAAAGC CCACCCCTACCCTCACCCCCCCAATCCTCTCCTGAAACTTAAAAACAGTGGGAATATAGGAAAGGGAACCAAATCTCATTAATTAATTGT TCTCCCCCATTACCCCACTGAATGAATGGCCATACAGGCTAAGCTGAATAATGACAAAGTTGAAAGGACCAATACAGCCCCTTTTATAAG GATTTTGAATGTTTTGCAAATGTATTGGTCCCTGTGTTGTATTTTGTAGCCTTTTCCTGGGCTTCAGCTCCCCTACTTCTTGTATGTGTA >97107_97107_3_USB1-CNOT1_USB1_chr16_58044016_ENST00000423271_CNOT1_chr16_58555221_ENST00000317147_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_4_USB1-CNOT1_USB1_chr16_58044016_ENST00000423271_CNOT1_chr16_58555221_ENST00000569240_length(transcript)=1003nt_BP=526nt GCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGC CCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCC AGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCA CAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGT TCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCC AGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGAAC GGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCATG AATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAATGG AAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTGAA GAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTATGT GGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAATAC >97107_97107_4_USB1-CNOT1_USB1_chr16_58044016_ENST00000423271_CNOT1_chr16_58555221_ENST00000569240_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_5_USB1-CNOT1_USB1_chr16_58044016_ENST00000539737_CNOT1_chr16_58555221_ENST00000317147_length(transcript)=1749nt_BP=528nt GCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGC GCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGG CCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAG CACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGA GTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTC CCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGA ACGGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCA TGAATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAAT GGAAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTG AAGAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTAT GTGGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAAT ACATGAAAGAGAAAACCTTTGTCTGGAACTTCTTGGCTTTGTGCAAGCTGTGTCCAAGGCAAGTACATAAACTGGTACCTTGTAATGAAG AGGCAGCTGATGCCATGCACTTGTCTGAGGGCATAGCTCCATGTCTTCTGACATTCCTGGTGTCCCAAAGAATAGCAAAAAGCCAGTTTG AATATTATGTAACTTATTTTTTTAATGTGGACAGGGGACCTTGAAAATCACTAAGTTATTAAAAATGTGGATGTGCTAGAATTGGATATG TCCAGGAACATGGGAAGGGCTCACTATTGGAATCCCATGAGTTTCCATTTTGTCTCTACCCAAACGTATTCCAAAGCTGACTGCATTTGT ACCATCTTATTTCTTTTGGGGATTATACACCTCAGCCGCCTGAGATGGGGGTCAGCTCTTTATATAAAGGGAAACCAGACCAGGCCTAAA GCCCACCCCTACCCTCACCCCCCCAATCCTCTCCTGAAACTTAAAAACAGTGGGAATATAGGAAAGGGAACCAAATCTCATTAATTAATT GTTCTCCCCCATTACCCCACTGAATGAATGGCCATACAGGCTAAGCTGAATAATGACAAAGTTGAAAGGACCAATACAGCCCCTTTTATA AGGATTTTGAATGTTTTGCAAATGTATTGGTCCCTGTGTTGTATTTTGTAGCCTTTTCCTGGGCTTCAGCTCCCCTACTTCTTGTATGTG >97107_97107_5_USB1-CNOT1_USB1_chr16_58044016_ENST00000539737_CNOT1_chr16_58555221_ENST00000317147_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_6_USB1-CNOT1_USB1_chr16_58044016_ENST00000539737_CNOT1_chr16_58555221_ENST00000569240_length(transcript)=1005nt_BP=528nt GCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGC GCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGG CCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAG CACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGA GTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTC CCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGA ACGGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCA TGAATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAAT GGAAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTG AAGAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTAT GTGGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAAT >97107_97107_6_USB1-CNOT1_USB1_chr16_58044016_ENST00000539737_CNOT1_chr16_58555221_ENST00000569240_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_7_USB1-CNOT1_USB1_chr16_58044016_ENST00000561743_CNOT1_chr16_58555221_ENST00000317147_length(transcript)=1710nt_BP=489nt AGATGCAACCCACTTCCAGGTTCCCAGCTCATCTGCCTCTTACATTTAAAAGTTTGTAAGTTTTCTCCTGAGCAGAGGAAGAAGGAAGGA GAAGCCTTCACAGGACAGTCTGGCCTCTAACTGCTCTGCCAAAGAAATTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGA CAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGA GCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCA GACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTT CGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGAACGGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCT TATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCATGAATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATT CCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAATGGAAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTG TTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTGAAGAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTT GGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTATGTGGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTC TTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAATACATGAAAGAGAAAACCTTTGTCTGGAACTTCTTGGCTT TGTGCAAGCTGTGTCCAAGGCAAGTACATAAACTGGTACCTTGTAATGAAGAGGCAGCTGATGCCATGCACTTGTCTGAGGGCATAGCTC CATGTCTTCTGACATTCCTGGTGTCCCAAAGAATAGCAAAAAGCCAGTTTGAATATTATGTAACTTATTTTTTTAATGTGGACAGGGGAC CTTGAAAATCACTAAGTTATTAAAAATGTGGATGTGCTAGAATTGGATATGTCCAGGAACATGGGAAGGGCTCACTATTGGAATCCCATG AGTTTCCATTTTGTCTCTACCCAAACGTATTCCAAAGCTGACTGCATTTGTACCATCTTATTTCTTTTGGGGATTATACACCTCAGCCGC CTGAGATGGGGGTCAGCTCTTTATATAAAGGGAAACCAGACCAGGCCTAAAGCCCACCCCTACCCTCACCCCCCCAATCCTCTCCTGAAA CTTAAAAACAGTGGGAATATAGGAAAGGGAACCAAATCTCATTAATTAATTGTTCTCCCCCATTACCCCACTGAATGAATGGCCATACAG GCTAAGCTGAATAATGACAAAGTTGAAAGGACCAATACAGCCCCTTTTATAAGGATTTTGAATGTTTTGCAAATGTATTGGTCCCTGTGT TGTATTTTGTAGCCTTTTCCTGGGCTTCAGCTCCCCTACTTCTTGTATGTGTATGCATACTGTAGCTAACCATTAAAGTCATGACACACA >97107_97107_7_USB1-CNOT1_USB1_chr16_58044016_ENST00000561743_CNOT1_chr16_58555221_ENST00000317147_length(amino acids)=211AA_BP=138 MSRGRRKEKPSQDSLASNCSAKEIGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYEAKEEFLDLL DVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFKFWNHEFVHC -------------------------------------------------------------- >97107_97107_8_USB1-CNOT1_USB1_chr16_58044016_ENST00000561743_CNOT1_chr16_58555221_ENST00000569240_length(transcript)=966nt_BP=489nt AGATGCAACCCACTTCCAGGTTCCCAGCTCATCTGCCTCTTACATTTAAAAGTTTGTAAGTTTTCTCCTGAGCAGAGGAAGAAGGAAGGA GAAGCCTTCACAGGACAGTCTGGCCTCTAACTGCTCTGCCAAAGAAATTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGA CAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGA GCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCA GACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTT CGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGAACGGTTGATTGTAAATAGGCCACATCCTTGGGGTCTTCT TATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCATGAATTTGTACACTGTGCCCCAGAAATCGAAAAGTTATT CCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAATGGAAGGGACAGGTGCCAGTTAGACGAAACTGCATCTCTG TTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTGAAGAATCCTTTCAGCTCTTCCTGACTTTCCCAGCCCTTT GGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTATGTGGGCACGTTCCAAAGTTTAAATGCATTTTTTTGACTC >97107_97107_8_USB1-CNOT1_USB1_chr16_58044016_ENST00000561743_CNOT1_chr16_58555221_ENST00000569240_length(amino acids)=211AA_BP=138 MSRGRRKEKPSQDSLASNCSAKEIGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYEAKEEFLDLL DVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFKFWNHEFVHC -------------------------------------------------------------- >97107_97107_9_USB1-CNOT1_USB1_chr16_58044016_ENST00000563149_CNOT1_chr16_58555221_ENST00000317147_length(transcript)=1734nt_BP=513nt CGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGCCCCTGGTGGGCTA CAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCCAGAGCCCCCTTCC CAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGG ACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCT TGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCT GCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGAACGGTTGATTGTAAA TAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCATGAATTTGTACACTG TGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAATGGAAGGGACAGGTGC CAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTGAAGAATCCTTTCAGC TCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTATGTGGGCACGTTCCAA AGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAATACATGAAAGAGAAAA CCTTTGTCTGGAACTTCTTGGCTTTGTGCAAGCTGTGTCCAAGGCAAGTACATAAACTGGTACCTTGTAATGAAGAGGCAGCTGATGCCA TGCACTTGTCTGAGGGCATAGCTCCATGTCTTCTGACATTCCTGGTGTCCCAAAGAATAGCAAAAAGCCAGTTTGAATATTATGTAACTT ATTTTTTTAATGTGGACAGGGGACCTTGAAAATCACTAAGTTATTAAAAATGTGGATGTGCTAGAATTGGATATGTCCAGGAACATGGGA AGGGCTCACTATTGGAATCCCATGAGTTTCCATTTTGTCTCTACCCAAACGTATTCCAAAGCTGACTGCATTTGTACCATCTTATTTCTT TTGGGGATTATACACCTCAGCCGCCTGAGATGGGGGTCAGCTCTTTATATAAAGGGAAACCAGACCAGGCCTAAAGCCCACCCCTACCCT CACCCCCCCAATCCTCTCCTGAAACTTAAAAACAGTGGGAATATAGGAAAGGGAACCAAATCTCATTAATTAATTGTTCTCCCCCATTAC CCCACTGAATGAATGGCCATACAGGCTAAGCTGAATAATGACAAAGTTGAAAGGACCAATACAGCCCCTTTTATAAGGATTTTGAATGTT TTGCAAATGTATTGGTCCCTGTGTTGTATTTTGTAGCCTTTTCCTGGGCTTCAGCTCCCCTACTTCTTGTATGTGTATGCATACTGTAGC >97107_97107_9_USB1-CNOT1_USB1_chr16_58044016_ENST00000563149_CNOT1_chr16_58555221_ENST00000317147_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- >97107_97107_10_USB1-CNOT1_USB1_chr16_58044016_ENST00000563149_CNOT1_chr16_58555221_ENST00000569240_length(transcript)=990nt_BP=513nt CGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGCCCCTGGTGGGCTA CAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCCAGAGCCCCCTTCC CAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGG ACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCT TGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCT GCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGAGTTCTCTTGGAACGGTTGATTGTAAA TAGGCCACATCCTTGGGGTCTTCTTATTACCTTCATTGAGCTGATTAAAAACCCAGCGTTTAAGTTCTGGAACCATGAATTTGTACACTG TGCCCCAGAAATCGAAAAGTTATTCCAGTCGGTCGCACAGTGCTGCATGGGACAGAAGCAGGCCCAGCAAGTAATGGAAGGGACAGGTGC CAGTTAGACGAAACTGCATCTCTGTTGTACGTGTCAGTCTAGAGGTCTCACTGCACCGAGTTCATAAACTGACTGAAGAATCCTTTCAGC TCTTCCTGACTTTCCCAGCCCTTTGGTTTGTGGGTATCTGCCCCAACTACTGTTGGGATCAGCCTCCTGTCTTATGTGGGCACGTTCCAA AGTTTAAATGCATTTTTTTGACTCTTGGCCAAAATTTAGAAGATGCTGTGAATATCATTTTGAACTTGTGTAAATACATGAAAGAGAAAA >97107_97107_10_USB1-CNOT1_USB1_chr16_58044016_ENST00000563149_CNOT1_chr16_58555221_ENST00000569240_length(amino acids)=220AA_BP=147 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRVLLERLIVNRPHPWGLLITFIELIKNPAFK -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for USB1-CNOT1 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 1090_1605 | 0 | 1552.0 | CNOT6%2C CNOT6L%2C CNOT7 and CNOT8 | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000441024 | 0 | 31 | 800_1015 | 0 | 1552.0 | ZFP36 |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 1090_1605 | 2305.6666666666665 | 2377.0 | CNOT6%2C CNOT6L%2C CNOT7 and CNOT8 | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 1090_1605 | 2300.6666666666665 | 2372.0 | CNOT6%2C CNOT6L%2C CNOT7 and CNOT8 | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000317147 | 46 | 49 | 800_1015 | 2305.6666666666665 | 2377.0 | ZFP36 | |
| Tgene | CNOT1 | chr16:58044016 | chr16:58555221 | ENST00000569240 | 46 | 49 | 800_1015 | 2300.6666666666665 | 2372.0 | ZFP36 |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for USB1-CNOT1 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for USB1-CNOT1 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
