
|
||||||
|
 | Fusion Gene Summary |
 | Fusion Gene ORF analysis |
 | Fusion Genomic Features |
 | Fusion Protein Features |
 | Fusion Gene Sequence |
 | Fusion Gene PPI analysis |
 | Related Drugs |
 | Related Diseases |
Fusion gene:USB1-MAPKAPK5 (FusionGDB2 ID:97110) |
Fusion Gene Summary for USB1-MAPKAPK5 |
 Fusion gene summary Fusion gene summary |
| Fusion gene information | Fusion gene name: USB1-MAPKAPK5 | Fusion gene ID: 97110 | Hgene | Tgene | Gene symbol | USB1 | MAPKAPK5 | Gene ID | 79650 | 8550 |
| Gene name | U6 snRNA biogenesis phosphodiesterase 1 | MAPK activated protein kinase 5 | |
| Synonyms | C16orf57|HVSL1|Mpn1|PN|hUsb1 | MAPKAP-K5|MK-5|MK5|PRAK | |
| Cytomap | 16q21 | 12q24.12-q24.13 | |
| Type of gene | protein-coding | protein-coding | |
| Description | U6 snRNA phosphodiesteraseHVSL motif containing 1U six biogenesis 1U6 snRNA biogenesis 1UPF0406 protein C16orf57mutated in poikiloderma with neutropenia protein 1putative U6 snRNA phosphodiesterase | MAP kinase-activated protein kinase 5MAPKAP kinase 5MAPKAPK-5mitogen-activated protein kinase-activated protein kinase 5p38-regulated/activated protein kinase | |
| Modification date | 20200328 | 20200313 | |
| UniProtAcc | Q9BQ65 | Q8IW41 | |
| Ensembl transtripts involved in fusion gene | ENST00000565662, ENST00000219281, ENST00000539737, ENST00000561743, ENST00000423271, ENST00000563149, | ENST00000550735, ENST00000551404, ENST00000546394, ENST00000547305, | |
| Fusion gene scores | * DoF score | 6 X 6 X 5=180 | 8 X 10 X 3=240 |
| # samples | 6 | 11 | |
| ** MAII score | log2(6/180*10)=-1.58496250072116 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | log2(11/240*10)=-1.12553088208386 possibly effective Gene in Pan-Cancer Fusion Genes (peGinPCFGs). DoF>8 and MAII<0 | |
| Context | PubMed: USB1 [Title/Abstract] AND MAPKAPK5 [Title/Abstract] AND fusion [Title/Abstract] | ||
| Most frequent breakpoint | USB1(58051343)-MAPKAPK5(112308889), # samples:2 | ||
| Anticipated loss of major functional domain due to fusion event. | |||
| * DoF score (Degree of Frequency) = # partners X # break points X # cancer types ** MAII score (Major Active Isofusion Index) = log2(# samples/DoF score*10) |
 Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez Gene ontology of each fusion partner gene with evidence of Inferred from Direct Assay (IDA) from Entrez |
| Partner | Gene | GO ID | GO term | PubMed ID |
| Tgene | MAPKAPK5 | GO:0006417 | regulation of translation | 21329882 |
| Tgene | MAPKAPK5 | GO:0007265 | Ras protein signal transduction | 17254968 |
| Tgene | MAPKAPK5 | GO:0046777 | protein autophosphorylation | 21329882 |
| Tgene | MAPKAPK5 | GO:0090400 | stress-induced premature senescence | 17254968 |
 Fusion gene breakpoints across USB1 (5'-gene) Fusion gene breakpoints across USB1 (5'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
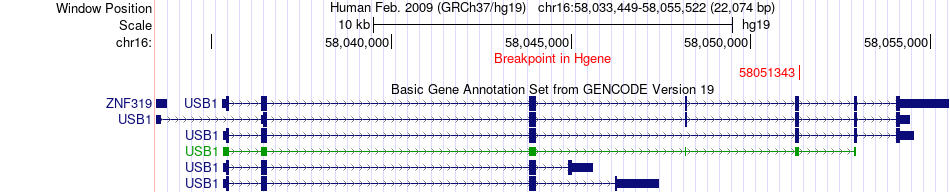 |
 Fusion gene breakpoints across MAPKAPK5 (3'-gene) Fusion gene breakpoints across MAPKAPK5 (3'-gene)* Click on the image to open the UCSC genome browser with custom track showing this image in a new window. |
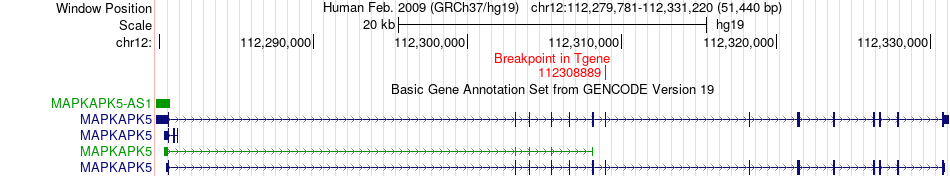 |
 Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0) Fusion gene information from two resources (ChiTars 5.0 and ChimerDB 4.0)* All genome coordinats were lifted-over on hg19. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| Source | Disease | Sample | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| ChimerDB4 | OV | TCGA-24-2262-01A | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
Top |
Fusion Gene ORF analysis for USB1-MAPKAPK5 |
 Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. Open reading frame (ORF) analsis of fusion genes based on Ensembl gene isoform structure. * Click on the break point to see the gene structure around the break point region using the UCSC Genome Browser. |
| ORF | Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand |
| 3UTR-3CDS | ENST00000565662 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 3UTR-3CDS | ENST00000565662 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 3UTR-intron | ENST00000565662 | ENST00000546394 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 3UTR-intron | ENST00000565662 | ENST00000547305 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 5CDS-intron | ENST00000219281 | ENST00000546394 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 5CDS-intron | ENST00000219281 | ENST00000547305 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 5CDS-intron | ENST00000539737 | ENST00000546394 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 5CDS-intron | ENST00000539737 | ENST00000547305 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 5CDS-intron | ENST00000561743 | ENST00000546394 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| 5CDS-intron | ENST00000561743 | ENST00000547305 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| In-frame | ENST00000219281 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| In-frame | ENST00000219281 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| In-frame | ENST00000539737 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| In-frame | ENST00000539737 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| In-frame | ENST00000561743 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| In-frame | ENST00000561743 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-3CDS | ENST00000423271 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-3CDS | ENST00000423271 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-3CDS | ENST00000563149 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-3CDS | ENST00000563149 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-intron | ENST00000423271 | ENST00000546394 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-intron | ENST00000423271 | ENST00000547305 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-intron | ENST00000563149 | ENST00000546394 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
| intron-intron | ENST00000563149 | ENST00000547305 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + |
 ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. ORFfinder result based on the fusion transcript sequence of in-frame fusion genes. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | Seq length (transcript) | BP loci (transcript) | Predicted start (transcript) | Predicted stop (transcript) | Seq length (amino acids) |
| ENST00000561743 | USB1 | chr16 | 58051343 | + | ENST00000550735 | MAPKAPK5 | chr12 | 112308889 | + | 1937 | 649 | 67 | 1581 | 504 |
| ENST00000561743 | USB1 | chr16 | 58051343 | + | ENST00000551404 | MAPKAPK5 | chr12 | 112308889 | + | 1688 | 649 | 67 | 1587 | 506 |
| ENST00000219281 | USB1 | chr16 | 58051343 | + | ENST00000550735 | MAPKAPK5 | chr12 | 112308889 | + | 2008 | 720 | 111 | 1652 | 513 |
| ENST00000219281 | USB1 | chr16 | 58051343 | + | ENST00000551404 | MAPKAPK5 | chr12 | 112308889 | + | 1759 | 720 | 111 | 1658 | 515 |
| ENST00000539737 | USB1 | chr16 | 58051343 | + | ENST00000550735 | MAPKAPK5 | chr12 | 112308889 | + | 1922 | 634 | 79 | 1566 | 495 |
| ENST00000539737 | USB1 | chr16 | 58051343 | + | ENST00000551404 | MAPKAPK5 | chr12 | 112308889 | + | 1673 | 634 | 79 | 1572 | 497 |
 DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. DeepORF prediction of the coding potential based on the fusion transcript sequence of in-frame fusion genes. DeepORF is a coding potential classifier based on convolutional neural network by comparing the real Ribo-seq data. If the no-coding score < 0.5 and coding score > 0.5, then the in-frame fusion transcript is predicted as being likely translated. |
| Henst | Tenst | Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | No-coding score | Coding score |
| ENST00000561743 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + | 0.000847419 | 0.99915254 |
| ENST00000561743 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + | 0.001160293 | 0.9988397 |
| ENST00000219281 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + | 0.000745137 | 0.9992549 |
| ENST00000219281 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + | 0.000989509 | 0.99901044 |
| ENST00000539737 | ENST00000550735 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + | 0.001092729 | 0.9989072 |
| ENST00000539737 | ENST00000551404 | USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308889 | + | 0.001460488 | 0.99853957 |
Top |
Fusion Genomic Features for USB1-MAPKAPK5 |
 FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. FusionAI prediction of the potential fusion gene breakpoint based on the pre-mature RNA sequence context (+/- 5kb of individual partner genes, total 20kb length sequence). FusionAI is a fusion gene breakpoint classifier based on convolutional neural network by comparing the fusion positive and negative sequence context of ~ 20K fusion gene data. From here, we can have the relative potentency of the 20K genomic sequence how individual sequnce will be likely used as the gene fusion breakpoints. |
| Hgene | Hchr | Hbp | Hstrand | Tgene | Tchr | Tbp | Tstrand | 1-p | p (fusion gene breakpoint) |
| USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308888 | + | 4.34E-05 | 0.9999566 |
| USB1 | chr16 | 58051343 | + | MAPKAPK5 | chr12 | 112308888 | + | 4.34E-05 | 0.9999566 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions. We integrated a total of 44 different types of human genomic feature loci information across five big categories including virus integration sites, repeats, structural variants, chromatin states, and gene expression regulation. More details are in help page. |
 |
 Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. Distribution of 44 human genomic features loci across 20kb length fusion breakpoint regions that are ovelapped with the top 1% feature importance score regions. More details are in help page. |
 |
Top |
Fusion Protein Features for USB1-MAPKAPK5 |
 Four levels of functional features of fusion genes Four levels of functional features of fusion genesGo to FGviewer search page for the most frequent breakpoint (https://ccsmweb.uth.edu/FGviewer/chr16:58051343/chr12:112308889) - FGviewer provides the online visualization of the retention search of the protein functional features across DNA, RNA, protein, and pathological levels. - How to search 1. Put your fusion gene symbol. 2. Press the tab key until there will be shown the breakpoint information filled. 4. Go down and press 'Search' tab twice. 4. Go down to have the hyperlink of the search result. 5. Click the hyperlink. 6. See the FGviewer result for your fusion gene. |
 |
 Main function of each fusion partner protein. (from UniProt) Main function of each fusion partner protein. (from UniProt) |
| Hgene | Tgene |
| USB1 | MAPKAPK5 |
| FUNCTION: Phosphodiesterase responsible for the U6 snRNA 3' end processing. Acts as an exoribonuclease (RNase) responsible for trimming the poly(U) tract of the last nucleotides in the pre-U6 snRNA molecule, leading to the formation of mature U6 snRNA 3' end-terminated with a 2',3'-cyclic phosphate. {ECO:0000255|HAMAP-Rule:MF_03040, ECO:0000269|PubMed:22899009, ECO:0000269|PubMed:23190533}. | FUNCTION: Tumor suppressor serine/threonine-protein kinase involved in mTORC1 signaling and post-transcriptional regulation. Phosphorylates FOXO3, ERK3/MAPK6, ERK4/MAPK4, HSP27/HSPB1, p53/TP53 and RHEB. Acts as a tumor suppressor by mediating Ras-induced senescence and phosphorylating p53/TP53. Involved in post-transcriptional regulation of MYC by mediating phosphorylation of FOXO3: phosphorylation of FOXO3 leads to promote nuclear localization of FOXO3, enabling expression of miR-34b and miR-34c, 2 post-transcriptional regulators of MYC that bind to the 3'UTR of MYC transcript and prevent MYC translation. Acts as a negative regulator of mTORC1 signaling by mediating phosphorylation and inhibition of RHEB. Part of the atypical MAPK signaling via its interaction with ERK3/MAPK6 or ERK4/MAPK4: the precise role of the complex formed with ERK3/MAPK6 or ERK4/MAPK4 is still unclear, but the complex follows a complex set of phosphorylation events: upon interaction with atypical MAPK (ERK3/MAPK6 or ERK4/MAPK4), ERK3/MAPK6 (or ERK4/MAPK4) is phosphorylated and then mediates phosphorylation and activation of MAPKAPK5, which in turn phosphorylates ERK3/MAPK6 (or ERK4/MAPK4). Mediates phosphorylation of HSP27/HSPB1 in response to PKA/PRKACA stimulation, inducing F-actin rearrangement. {ECO:0000269|PubMed:17254968, ECO:0000269|PubMed:17728103, ECO:0000269|PubMed:19166925, ECO:0000269|PubMed:21329882, ECO:0000269|PubMed:9628874}. |
 Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at Retention analysis result of each fusion partner protein across 39 protein features of UniProt such as six molecule processing features, 13 region features, four site features, six amino acid modification features, two natural variation features, five experimental info features, and 3 secondary structure features. Here, because of limited space for viewing, we only show the protein feature retention information belong to the 13 regional features. All retention annotation result can be downloaded at * Minus value of BPloci means that the break pointn is located before the CDS. |
| - In-frame and retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MAPKAPK5 | chr16:58051343 | chr12:112308889 | ENST00000550735 | 5 | 14 | 409_440 | 161 | 472.0 | Coiled coil | Ontology_term=ECO:0000255 | |
| Tgene | MAPKAPK5 | chr16:58051343 | chr12:112308889 | ENST00000551404 | 5 | 14 | 409_440 | 161 | 474.0 | Coiled coil | Ontology_term=ECO:0000255 |
| - In-frame and not-retained protein feature among the 13 regional features. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Protein feature | Protein feature note |
| Tgene | MAPKAPK5 | chr16:58051343 | chr12:112308889 | ENST00000550735 | 5 | 14 | 22_304 | 161 | 472.0 | Domain | Protein kinase | |
| Tgene | MAPKAPK5 | chr16:58051343 | chr12:112308889 | ENST00000551404 | 5 | 14 | 22_304 | 161 | 474.0 | Domain | Protein kinase | |
| Tgene | MAPKAPK5 | chr16:58051343 | chr12:112308889 | ENST00000550735 | 5 | 14 | 28_36 | 161 | 472.0 | Nucleotide binding | ATP | |
| Tgene | MAPKAPK5 | chr16:58051343 | chr12:112308889 | ENST00000551404 | 5 | 14 | 28_36 | 161 | 474.0 | Nucleotide binding | ATP |
Top |
Fusion Gene Sequence for USB1-MAPKAPK5 |
 For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. For in-frame fusion transcripts, we provide the fusion transcript sequences and fusion amino acid sequences. To have fusion amino acid sequence, we ran ORFfinder and chose the longest ORF among the all predicted ones. |
| >97110_97110_1_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000219281_MAPKAPK5_chr12_112308889_ENST00000550735_length(transcript)=2008nt_BP=720nt GCGGTGCCAGCCCAGGCCCCGCCCCTGGGAGGGCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCT GGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGG ACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTC CCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACC CACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTG GTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCC CGTATGACCTCCTTCCACAGATTCTTCTTTACTGCCAACCAGGTAAAGATTTACACCAATCAAGAGAAAACCAGGACCTTTATTGGGCTT GAGGTCACTTCAGGGCATGCCCAGTTCCTGGACCTGGTTTCAGAGGTGGACAGAGTCATGGAGGAATTCAACCTCACCACTTTCTACCAG GATGCCCCAGTGAAGTTGTGTGACTTTGGATTTGCCAAGATTGACCAAGGTGACTTGATGACACCCCAGTTCACCCCTTATTATGTAGCA CCCCAGGTACTGGAGGCGCAAAGAAGGCATCAGAAGGAGAAATCTGGCATCATACCTACCTCACCGACGCCCTACACTTACAACAAGAGC TGTGACTTGTGGTCCCTAGGGGTGATTATCTATGTGATGCTGTGCGGATACCCTCCTTTTTACTCCAAACACCACAGCCGGACTATCCCA AAGGATATGCGAAGAAAGATCATGACAGGCAGTTTTGAGTTCCCAGAGGAAGAGTGGAGTCAGATCTCAGAGATGGCCAAAGATGTTGTG AGGAAGCTCCTGAAGGTCAAACCGGAGGAGAGACTCACCATCGAGGGAGTGCTGGACCACCCCTGGCTCAATTCCACCGAGGCCCTGGAT AATGTGCTGCCTTCTGCTCAGCTGATGATGGACAAGGCAGTGGTTGCAGGAATCCAGCAGGCTCACGCGGAACAGTTGGCCAACATGAGA ATCCAGGATCTGAAAGTCAGCCTCAAACCCCTGCACTCAGTGAACAACCCCATTCTGCGGAAGAGGAAGTTACTTGGCACCAAGCCAAAG GACAGTGTCTATATCCACGACCATGAGAATGGAGCCGAGGATTCCAATGTTGCCTTGGAAAAACTCCGAGATGTGATTGCTCAGTGTATT CTCCCCCAGGCTGGAGAGAATGAAGATGAGAAACTGAATGAAGTAATGCAGGAGGCTTGGAAGTATAACCGGGAATGCAAACTCCTAAGA GATACTCTGCAGAGCTTCAGCTGGAATGGTCGTGGATTCACAGATAAAGTAGATCGACTAAAACTGGCAGAAATTGTGAAGCAGGTGATA GAAGAGCAAACCACGTCCCACGAATCCCAATAATGACAGCTTCAGACTTTGTTTTTTTAACAATTTGAAAAATTATTCTTTAATGTATAA AGTAATTTTATGTAAATTAATAAATCATAATTTCATTTCCACATTGATTAAAGCTGCTGTATAGATTTAGGGTGCAGGACTTAATAATAG TATAGTTATTGTTTGTTTTTAAGAAAAGCTCAGTTCTAGAGACATACTATTACTTTAGGACTGTGTAGTTGTATATTTGTAAGATGACAG ATGATGCTGTCAAGCAATATTGTTTTATTTGTAATAAAATATACAAAAATCACTTGCCAGCAGTAGAAAAAGGACCGACTATACCGACCT >97110_97110_1_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000219281_MAPKAPK5_chr12_112308889_ENST00000550735_length(amino acids)=513AA_BP=202 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRFFFTANQVKIYTNQEKTRTFIGLEVTSGHA QFLDLVSEVDRVMEEFNLTTFYQDAPVKLCDFGFAKIDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLG VIIYVMLCGYPPFYSKHHSRTIPKDMRRKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQ LMMDKAVVAGIQQAHAEQLANMRIQDLKVSLKPLHSVNNPILRKRKLLGTKPKDSVYIHDHENGAEDSNVALEKLRDVIAQCILPQAGEN -------------------------------------------------------------- >97110_97110_2_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000219281_MAPKAPK5_chr12_112308889_ENST00000551404_length(transcript)=1759nt_BP=720nt GCGGTGCCAGCCCAGGCCCCGCCCCTGGGAGGGCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCT GGTGGTCTTGGATGAGGCCCCATGAGCGCGGCGCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGG ACCAGGCCGGGGGATGGGAGCCACCGTCGTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTC CCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACC CACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTG GTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCC CGTATGACCTCCTTCCACAGATTCTTCTTTACTGCCAACCAGGTAAAGATTTACACCAATCAAGAGAAAACCAGGACCTTTATTGGGCTT GAGGTCACTTCAGGGCATGCCCAGTTCCTGGACCTGGTTTCAGAGGTGGACAGAGTCATGGAGGAATTCAACCTCACCACTTTCTACCAG GATGCCCCAGTGAAGTTGTGTGACTTTGGATTTGCCAAGATTGACCAAGGTGACTTGATGACACCCCAGTTCACCCCTTATTATGTAGCA CCCCAGGTACTGGAGGCGCAAAGAAGGCATCAGAAGGAGAAATCTGGCATCATACCTACCTCACCGACGCCCTACACTTACAACAAGAGC TGTGACTTGTGGTCCCTAGGGGTGATTATCTATGTGATGCTGTGCGGATACCCTCCTTTTTACTCCAAACACCACAGCCGGACTATCCCA AAGGATATGCGAAGAAAGATCATGACAGGCAGTTTTGAGTTCCCAGAGGAAGAGTGGAGTCAGATCTCAGAGATGGCCAAAGATGTTGTG AGGAAGCTCCTGAAGGTCAAACCGGAGGAGAGACTCACCATCGAGGGAGTGCTGGACCACCCCTGGCTCAATTCCACCGAGGCCCTGGAT AATGTGCTGCCTTCTGCTCAGCTGATGATGGACAAGGCAGTGGTTGCAGGAATCCAGCAGGCTCACGCGGAACAGTTGGCCAACATGAGA ATCCAGGATCTGAAAGTCAGCCTCAAACCCCTGCACTCAGTGAACAACCCCATTCTGCGGAAGAGGAAGTTACTTGGCACCAAGCCAAAG GACAGTGTCTATATCCACGACCATGAGAATGGAGCCGAGGATTCCAATGTTGCCTTGGAAAAACTCCGAGATGTGATTGCTCAGTGTATT CTCCCCCAGGCTGGTAAAGGAGAGAATGAAGATGAGAAACTGAATGAAGTAATGCAGGAGGCTTGGAAGTATAACCGGGAATGCAAACTC CTAAGAGATACTCTGCAGAGCTTCAGCTGGAATGGTCGTGGATTCACAGATAAAGTAGATCGACTAAAACTGGCAGAAATTGTGAAGCAG GTGATAGAAGAGCAAACCACGTCCCACGAATCCCAATAATGACAGCTTCAGACTTTGTTTTTTTAACAATTTGAAAAATTATTCTTTAAT >97110_97110_2_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000219281_MAPKAPK5_chr12_112308889_ENST00000551404_length(amino acids)=515AA_BP=202 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRFFFTANQVKIYTNQEKTRTFIGLEVTSGHA QFLDLVSEVDRVMEEFNLTTFYQDAPVKLCDFGFAKIDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLG VIIYVMLCGYPPFYSKHHSRTIPKDMRRKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQ LMMDKAVVAGIQQAHAEQLANMRIQDLKVSLKPLHSVNNPILRKRKLLGTKPKDSVYIHDHENGAEDSNVALEKLRDVIAQCILPQAGKG -------------------------------------------------------------- >97110_97110_3_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000539737_MAPKAPK5_chr12_112308889_ENST00000550735_length(transcript)=1922nt_BP=634nt GCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGC GCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGG CCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAG CACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGA GTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTC CCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGGACCTTTATTGG GCTTGAGGTCACTTCAGGGCATGCCCAGTTCCTGGACCTGGTTTCAGAGGTGGACAGAGTCATGGAGGAATTCAACCTCACCACTTTCTA CCAGGATGCCCCAGTGAAGTTGTGTGACTTTGGATTTGCCAAGATTGACCAAGGTGACTTGATGACACCCCAGTTCACCCCTTATTATGT AGCACCCCAGGTACTGGAGGCGCAAAGAAGGCATCAGAAGGAGAAATCTGGCATCATACCTACCTCACCGACGCCCTACACTTACAACAA GAGCTGTGACTTGTGGTCCCTAGGGGTGATTATCTATGTGATGCTGTGCGGATACCCTCCTTTTTACTCCAAACACCACAGCCGGACTAT CCCAAAGGATATGCGAAGAAAGATCATGACAGGCAGTTTTGAGTTCCCAGAGGAAGAGTGGAGTCAGATCTCAGAGATGGCCAAAGATGT TGTGAGGAAGCTCCTGAAGGTCAAACCGGAGGAGAGACTCACCATCGAGGGAGTGCTGGACCACCCCTGGCTCAATTCCACCGAGGCCCT GGATAATGTGCTGCCTTCTGCTCAGCTGATGATGGACAAGGCAGTGGTTGCAGGAATCCAGCAGGCTCACGCGGAACAGTTGGCCAACAT GAGAATCCAGGATCTGAAAGTCAGCCTCAAACCCCTGCACTCAGTGAACAACCCCATTCTGCGGAAGAGGAAGTTACTTGGCACCAAGCC AAAGGACAGTGTCTATATCCACGACCATGAGAATGGAGCCGAGGATTCCAATGTTGCCTTGGAAAAACTCCGAGATGTGATTGCTCAGTG TATTCTCCCCCAGGCTGGAGAGAATGAAGATGAGAAACTGAATGAAGTAATGCAGGAGGCTTGGAAGTATAACCGGGAATGCAAACTCCT AAGAGATACTCTGCAGAGCTTCAGCTGGAATGGTCGTGGATTCACAGATAAAGTAGATCGACTAAAACTGGCAGAAATTGTGAAGCAGGT GATAGAAGAGCAAACCACGTCCCACGAATCCCAATAATGACAGCTTCAGACTTTGTTTTTTTAACAATTTGAAAAATTATTCTTTAATGT ATAAAGTAATTTTATGTAAATTAATAAATCATAATTTCATTTCCACATTGATTAAAGCTGCTGTATAGATTTAGGGTGCAGGACTTAATA ATAGTATAGTTATTGTTTGTTTTTAAGAAAAGCTCAGTTCTAGAGACATACTATTACTTTAGGACTGTGTAGTTGTATATTTGTAAGATG ACAGATGATGCTGTCAAGCAATATTGTTTTATTTGTAATAAAATATACAAAAATCACTTGCCAGCAGTAGAAAAAGGACCGACTATACCG >97110_97110_3_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000539737_MAPKAPK5_chr12_112308889_ENST00000550735_length(amino acids)=495AA_BP=184 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRTFIGLEVTSGHAQFLDLVSEVDRVMEEFNL TTFYQDAPVKLCDFGFAKIDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLGVIIYVMLCGYPPFYSKHH SRTIPKDMRRKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQLMMDKAVVAGIQQAHAEQ LANMRIQDLKVSLKPLHSVNNPILRKRKLLGTKPKDSVYIHDHENGAEDSNVALEKLRDVIAQCILPQAGENEDEKLNEVMQEAWKYNRE -------------------------------------------------------------- >97110_97110_4_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000539737_MAPKAPK5_chr12_112308889_ENST00000551404_length(transcript)=1673nt_BP=634nt GCGCTTCCGGCACAGCGGAACTCCGGGTGCCGGTTGAGGTTGCTGGTGGACCTGCTCTGGTGGTCTTGGATGAGGCCCCATGAGCGCGGC GCCCCTGGTGGGCTACAGCAGCAGCGGCTCCGAGGATGAGTCCGAGGACGGGATGCGGACCAGGCCGGGGGATGGGAGCCACCGTCGTGG CCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGACAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAG CACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGAGCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGA GTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCAGACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTC CCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTTCGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGGACCTTTATTGG GCTTGAGGTCACTTCAGGGCATGCCCAGTTCCTGGACCTGGTTTCAGAGGTGGACAGAGTCATGGAGGAATTCAACCTCACCACTTTCTA CCAGGATGCCCCAGTGAAGTTGTGTGACTTTGGATTTGCCAAGATTGACCAAGGTGACTTGATGACACCCCAGTTCACCCCTTATTATGT AGCACCCCAGGTACTGGAGGCGCAAAGAAGGCATCAGAAGGAGAAATCTGGCATCATACCTACCTCACCGACGCCCTACACTTACAACAA GAGCTGTGACTTGTGGTCCCTAGGGGTGATTATCTATGTGATGCTGTGCGGATACCCTCCTTTTTACTCCAAACACCACAGCCGGACTAT CCCAAAGGATATGCGAAGAAAGATCATGACAGGCAGTTTTGAGTTCCCAGAGGAAGAGTGGAGTCAGATCTCAGAGATGGCCAAAGATGT TGTGAGGAAGCTCCTGAAGGTCAAACCGGAGGAGAGACTCACCATCGAGGGAGTGCTGGACCACCCCTGGCTCAATTCCACCGAGGCCCT GGATAATGTGCTGCCTTCTGCTCAGCTGATGATGGACAAGGCAGTGGTTGCAGGAATCCAGCAGGCTCACGCGGAACAGTTGGCCAACAT GAGAATCCAGGATCTGAAAGTCAGCCTCAAACCCCTGCACTCAGTGAACAACCCCATTCTGCGGAAGAGGAAGTTACTTGGCACCAAGCC AAAGGACAGTGTCTATATCCACGACCATGAGAATGGAGCCGAGGATTCCAATGTTGCCTTGGAAAAACTCCGAGATGTGATTGCTCAGTG TATTCTCCCCCAGGCTGGTAAAGGAGAGAATGAAGATGAGAAACTGAATGAAGTAATGCAGGAGGCTTGGAAGTATAACCGGGAATGCAA ACTCCTAAGAGATACTCTGCAGAGCTTCAGCTGGAATGGTCGTGGATTCACAGATAAAGTAGATCGACTAAAACTGGCAGAAATTGTGAA GCAGGTGATAGAAGAGCAAACCACGTCCCACGAATCCCAATAATGACAGCTTCAGACTTTGTTTTTTTAACAATTTGAAAAATTATTCTT >97110_97110_4_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000539737_MAPKAPK5_chr12_112308889_ENST00000551404_length(amino acids)=497AA_BP=184 MSAAPLVGYSSSGSEDESEDGMRTRPGDGSHRRGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYE AKEEFLDLLDVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRTFIGLEVTSGHAQFLDLVSEVDRVMEEFNL TTFYQDAPVKLCDFGFAKIDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLGVIIYVMLCGYPPFYSKHH SRTIPKDMRRKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQLMMDKAVVAGIQQAHAEQ LANMRIQDLKVSLKPLHSVNNPILRKRKLLGTKPKDSVYIHDHENGAEDSNVALEKLRDVIAQCILPQAGKGENEDEKLNEVMQEAWKYN -------------------------------------------------------------- >97110_97110_5_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000561743_MAPKAPK5_chr12_112308889_ENST00000550735_length(transcript)=1937nt_BP=649nt AGATGCAACCCACTTCCAGGTTCCCAGCTCATCTGCCTCTTACATTTAAAAGTTTGTAAGTTTTCTCCTGAGCAGAGGAAGAAGGAAGGA GAAGCCTTCACAGGACAGTCTGGCCTCTAACTGCTCTGCCAAAGAAATTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGA CAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGA GCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCA GACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTT CGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGATTCTTCTTTACTGCCAACCAGGTAAAGATTTACACCAATCAAGAGAAAAC CAGGACCTTTATTGGGCTTGAGGTCACTTCAGGGCATGCCCAGTTCCTGGACCTGGTTTCAGAGGTGGACAGAGTCATGGAGGAATTCAA CCTCACCACTTTCTACCAGGATGCCCCAGTGAAGTTGTGTGACTTTGGATTTGCCAAGATTGACCAAGGTGACTTGATGACACCCCAGTT CACCCCTTATTATGTAGCACCCCAGGTACTGGAGGCGCAAAGAAGGCATCAGAAGGAGAAATCTGGCATCATACCTACCTCACCGACGCC CTACACTTACAACAAGAGCTGTGACTTGTGGTCCCTAGGGGTGATTATCTATGTGATGCTGTGCGGATACCCTCCTTTTTACTCCAAACA CCACAGCCGGACTATCCCAAAGGATATGCGAAGAAAGATCATGACAGGCAGTTTTGAGTTCCCAGAGGAAGAGTGGAGTCAGATCTCAGA GATGGCCAAAGATGTTGTGAGGAAGCTCCTGAAGGTCAAACCGGAGGAGAGACTCACCATCGAGGGAGTGCTGGACCACCCCTGGCTCAA TTCCACCGAGGCCCTGGATAATGTGCTGCCTTCTGCTCAGCTGATGATGGACAAGGCAGTGGTTGCAGGAATCCAGCAGGCTCACGCGGA ACAGTTGGCCAACATGAGAATCCAGGATCTGAAAGTCAGCCTCAAACCCCTGCACTCAGTGAACAACCCCATTCTGCGGAAGAGGAAGTT ACTTGGCACCAAGCCAAAGGACAGTGTCTATATCCACGACCATGAGAATGGAGCCGAGGATTCCAATGTTGCCTTGGAAAAACTCCGAGA TGTGATTGCTCAGTGTATTCTCCCCCAGGCTGGAGAGAATGAAGATGAGAAACTGAATGAAGTAATGCAGGAGGCTTGGAAGTATAACCG GGAATGCAAACTCCTAAGAGATACTCTGCAGAGCTTCAGCTGGAATGGTCGTGGATTCACAGATAAAGTAGATCGACTAAAACTGGCAGA AATTGTGAAGCAGGTGATAGAAGAGCAAACCACGTCCCACGAATCCCAATAATGACAGCTTCAGACTTTGTTTTTTTAACAATTTGAAAA ATTATTCTTTAATGTATAAAGTAATTTTATGTAAATTAATAAATCATAATTTCATTTCCACATTGATTAAAGCTGCTGTATAGATTTAGG GTGCAGGACTTAATAATAGTATAGTTATTGTTTGTTTTTAAGAAAAGCTCAGTTCTAGAGACATACTATTACTTTAGGACTGTGTAGTTG TATATTTGTAAGATGACAGATGATGCTGTCAAGCAATATTGTTTTATTTGTAATAAAATATACAAAAATCACTTGCCAGCAGTAGAAAAA >97110_97110_5_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000561743_MAPKAPK5_chr12_112308889_ENST00000550735_length(amino acids)=504AA_BP=193 MSRGRRKEKPSQDSLASNCSAKEIGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYEAKEEFLDLL DVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRFFFTANQVKIYTNQEKTRTFIGLEVTSGHAQFLDLVSEV DRVMEEFNLTTFYQDAPVKLCDFGFAKIDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLGVIIYVMLCG YPPFYSKHHSRTIPKDMRRKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQLMMDKAVVA GIQQAHAEQLANMRIQDLKVSLKPLHSVNNPILRKRKLLGTKPKDSVYIHDHENGAEDSNVALEKLRDVIAQCILPQAGENEDEKLNEVM -------------------------------------------------------------- >97110_97110_6_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000561743_MAPKAPK5_chr12_112308889_ENST00000551404_length(transcript)=1688nt_BP=649nt AGATGCAACCCACTTCCAGGTTCCCAGCTCATCTGCCTCTTACATTTAAAAGTTTGTAAGTTTTCTCCTGAGCAGAGGAAGAAGGAAGGA GAAGCCTTCACAGGACAGTCTGGCCTCTAACTGCTCTGCCAAAGAAATTGGCCAGAGCCCCCTTCCCAGGCAGAGATTTCCAGTACCTGA CAGTGTGCTGAACATGTTCCCGGGCACCGAGGAGGGGCCTGAAGATGACAGCACAAAACACGGGGGACGGGTGCGCACCTTCCCCCACGA GCGAGGCAACTGGGCCACCCACGTCTATGTACCATATGAAGCCAAGGAGGAGTTCCTGGATCTGCTTGATGTGTTGCTGCCCCATGCCCA GACATATGTCCCCCGGCTGGTAAGGATGAAGGTGTTCCACCTCAGCCTGTCCCAGAGTGTGGTTCTGCGCCACCACTGGATCCTCCCCTT CGTGCAGGCTCTGAAAGCCCGTATGACCTCCTTCCACAGATTCTTCTTTACTGCCAACCAGGTAAAGATTTACACCAATCAAGAGAAAAC CAGGACCTTTATTGGGCTTGAGGTCACTTCAGGGCATGCCCAGTTCCTGGACCTGGTTTCAGAGGTGGACAGAGTCATGGAGGAATTCAA CCTCACCACTTTCTACCAGGATGCCCCAGTGAAGTTGTGTGACTTTGGATTTGCCAAGATTGACCAAGGTGACTTGATGACACCCCAGTT CACCCCTTATTATGTAGCACCCCAGGTACTGGAGGCGCAAAGAAGGCATCAGAAGGAGAAATCTGGCATCATACCTACCTCACCGACGCC CTACACTTACAACAAGAGCTGTGACTTGTGGTCCCTAGGGGTGATTATCTATGTGATGCTGTGCGGATACCCTCCTTTTTACTCCAAACA CCACAGCCGGACTATCCCAAAGGATATGCGAAGAAAGATCATGACAGGCAGTTTTGAGTTCCCAGAGGAAGAGTGGAGTCAGATCTCAGA GATGGCCAAAGATGTTGTGAGGAAGCTCCTGAAGGTCAAACCGGAGGAGAGACTCACCATCGAGGGAGTGCTGGACCACCCCTGGCTCAA TTCCACCGAGGCCCTGGATAATGTGCTGCCTTCTGCTCAGCTGATGATGGACAAGGCAGTGGTTGCAGGAATCCAGCAGGCTCACGCGGA ACAGTTGGCCAACATGAGAATCCAGGATCTGAAAGTCAGCCTCAAACCCCTGCACTCAGTGAACAACCCCATTCTGCGGAAGAGGAAGTT ACTTGGCACCAAGCCAAAGGACAGTGTCTATATCCACGACCATGAGAATGGAGCCGAGGATTCCAATGTTGCCTTGGAAAAACTCCGAGA TGTGATTGCTCAGTGTATTCTCCCCCAGGCTGGTAAAGGAGAGAATGAAGATGAGAAACTGAATGAAGTAATGCAGGAGGCTTGGAAGTA TAACCGGGAATGCAAACTCCTAAGAGATACTCTGCAGAGCTTCAGCTGGAATGGTCGTGGATTCACAGATAAAGTAGATCGACTAAAACT GGCAGAAATTGTGAAGCAGGTGATAGAAGAGCAAACCACGTCCCACGAATCCCAATAATGACAGCTTCAGACTTTGTTTTTTTAACAATT >97110_97110_6_USB1-MAPKAPK5_USB1_chr16_58051343_ENST00000561743_MAPKAPK5_chr12_112308889_ENST00000551404_length(amino acids)=506AA_BP=193 MSRGRRKEKPSQDSLASNCSAKEIGQSPLPRQRFPVPDSVLNMFPGTEEGPEDDSTKHGGRVRTFPHERGNWATHVYVPYEAKEEFLDLL DVLLPHAQTYVPRLVRMKVFHLSLSQSVVLRHHWILPFVQALKARMTSFHRFFFTANQVKIYTNQEKTRTFIGLEVTSGHAQFLDLVSEV DRVMEEFNLTTFYQDAPVKLCDFGFAKIDQGDLMTPQFTPYYVAPQVLEAQRRHQKEKSGIIPTSPTPYTYNKSCDLWSLGVIIYVMLCG YPPFYSKHHSRTIPKDMRRKIMTGSFEFPEEEWSQISEMAKDVVRKLLKVKPEERLTIEGVLDHPWLNSTEALDNVLPSAQLMMDKAVVA GIQQAHAEQLANMRIQDLKVSLKPLHSVNNPILRKRKLLGTKPKDSVYIHDHENGAEDSNVALEKLRDVIAQCILPQAGKGENEDEKLNE -------------------------------------------------------------- |
Top |
Fusion Gene PPI Analysis for USB1-MAPKAPK5 |
 Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in Go to ChiPPI (Chimeric Protein-Protein interactions) to see the chimeric PPI interaction in |
 Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) Protein-protein interactors with each fusion partner protein in wild-type (BIOGRID-3.4.160) |
| Hgene | Hgene's interactors | Tgene | Tgene's interactors |
 - Retained PPIs in in-frame fusion. - Retained PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Still interaction with |
 - Lost PPIs in in-frame fusion. - Lost PPIs in in-frame fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
 - Retained PPIs, but lost function due to frame-shift fusion. - Retained PPIs, but lost function due to frame-shift fusion. |
| Partner | Gene | Hbp | Tbp | ENST | Strand | BPexon | TotalExon | Protein feature loci | *BPloci | TotalLen | Interaction lost with |
Top |
Related Drugs for USB1-MAPKAPK5 |
 Drugs targeting genes involved in this fusion gene. Drugs targeting genes involved in this fusion gene. (DrugBank Version 5.1.8 2021-05-08) |
| Partner | Gene | UniProtAcc | DrugBank ID | Drug name | Drug activity | Drug type | Drug status |
Top |
Related Diseases for USB1-MAPKAPK5 |
 Diseases associated with fusion partners. Diseases associated with fusion partners. (DisGeNet 4.0) |
| Partner | Gene | Disease ID | Disease name | # pubmeds | Source |
